Abstract
Anaerobically grown cells of the metal-reducing bacterium Shewanella putrefaciens MR-1 contain multiple outer membrane (OM) cytochromes. A gene replacement mutant (strain OMCB1) lacking the OM cytochrome OmcB is markedly deficient in the reduction of MnO2 and exhibits reduced rates of Fe(III) reduction. The levels of other OM cytochromes are also decreased in OMCB1. Complementation of OMCB1 with wild-type omcB did not restore any of these defects. However, a 21-kb genomic fragment from MR-1, which included omcB and 19 kb of downstream DNA, fully restored MnO2 and Fe(III) reduction and the full complement of OM cytochromes to OMCB1. A 14.7-kb DNA fragment, including omcB and 12 kb of downstream DNA, provided only a modest increase in MnO2 reduction and OM cytochrome content, but it fully restored Fe(III) citrate reduction and partially restored FeOOH reduction. While omcB mRNA was readily detected in this complement, the OmcB protein was not detected in any cellular compartment. The restoration of Fe(III) reduction despite the absence of OmcB suggests that OmcB itself is not required for Fe(III) reduction. Another OM cytochrome, OmcA, was mislocalized to the cytoplasmic membrane of OMCB1. Only the 21-kb genomic fragment was able to restore proper localization of OmcA to the OM. This 21-kb fragment does not contain omcA, but it does contain several open reading frames (ORFs) downstream from omcB. The most downstream of these ORFs (altA) encodes a putative AraC-like transcriptional regulator. However, a gene replacement mutant of altA resembled the wild type with respect to MnO2 reduction, OM cytochrome content, and the localization of OmcA and OmcB to the OM. Since OMCB1 continues to express genes immediately downstream from omcB, the lack of expression of this downstream DNA does not explain its phenotype or the need for the large complementing fragment. The results suggest that the DNA downstream of omcB must be present in cis in order to restore Fe(III) reduction, MnO2 reduction, OM cytochrome content, and the localization of OmcA and OmcB to the OM.
Shewanella putrefaciens MR-1 can use a wide variety of terminal electron acceptors for anaerobic respiration, including insoluble manganese (Mn) and iron (Fe) oxides (22, 24, 28, 31, 33). When grown under anaerobic conditions, MR-1 localizes a majority of its membrane-bound cytochromes to the outer membrane (OM) (21). All of its OM cytochromes contain c-type hemes (27). Two of these OM cytochromes, OmcA and OmcB, have been purified, and sequence analysis predicts that both are lipoproteins, with each containing 10 heme c consensus-binding domains (CXXCH) (27, 32, 34). While omcB is immediately downstream of omcA, these genes are not cotranscribed as a polycistronic mRNA (32, 34). The OM cytochromes are localized where they could potentially make direct contact with extracellular metal oxides at the cell surface and could therefore represent a link between the electron transport systems and the reduction of extracellular insoluble metal oxides. A previous report described an omcB mutant (OMCB1) that was generated by gene replacement (34). OMCB1 is proficient at using many anaerobic electron acceptors, but it is markedly compromised with respect to Mn(IV) reduction, as well as its ability to properly localize other OM cytochromes to the OM (34).
To better understand the role of OM cytochromes in Mn(IV) reduction, additional studies were conducted on OMCB1. A variety of constructs were used to identify the minimum genetic material required to complement OMCB1 with respect to OmcB production, Mn(IV) reduction, OM cytochrome content, and proper subcellular localization of OmcA. Surprisingly, a 21-kb genomic fragment from MR-1, which included omcB and 19 kb of downstream DNA, was required to fully restore OmcB, Mn(IV) reduction, and the proper localization of OM cytochromes, including OmcA. The potential role of an open reading frame (ORF) that is localized within this downstream DNA and that encodes a putative transcriptional regulator was explored.
MATERIALS AND METHODS
All chemicals, enzymes, and other reagents used were obtained from sources described previously (25, 33).
Bacterial strains, plasmids, media, and growth conditions.
A list of the bacteria and plasmids used in this study is presented in Table 1. For molecular biology purposes, S. putrefaciens and Escherichia coli were grown aerobically on Luria-Bertani (LB) medium (39) supplemented, when required, with antibiotics at the following concentrations: ampicillin, 50 μg ml−1; kanamycin, 50 μg ml−1; and gentamicin, 10 μg ml−1. E. coli was grown at 37°C, whereas S. putrefaciens was grown either at 30°C or at room temperature (23 to 25°C).
TABLE 1.
Bacteria and plasmids used in this study
| Bacterial strain or plasmid | Description | Refer- ence | Source |
|---|---|---|---|
| S. putrefaciens strains | |||
| MR-1 | Manganese-reducing strain from Lake Oneida, N.Y., sediments | 28 | Previous study |
| OMCA1 | omcA mutant derived from MR-1; omcA::Kmr | 34 | Previous study |
| OMCB1 | omcB mutant derived from MR-1; omcB::Kmr | 34 | Previous study |
| ALTA1 | altA mutant derived from MR-1; altA::Kmr | This work | |
| E. coli strains | |||
| S17-1λpir | λ(pir) hsdR pro thi, chromosomal integrated RP4-2 Tc::Mu Km::Tn7; donor strain to mate pEP185.2-derived plasmids into MR-1 | 40 | V. L. Miller |
| TOP10 | F−mcrA Δ(mrr-hsdRMS-mcrBC) φ80lacZΔM15 ΔlacX74 deoR recA1 araD139 Δ(ara-leu)7697 galU galK rpsL (Smr) endA1 nupG; used as host for plasmids derived from pCR2.1-TOPO and pBAD/Thio-TOPO | Invitrogen | |
| Plasmids | |||
| pBAD/Thio-TOPO | 4.4-kb vector for generating OmcB antigen fusion; Apr | Invitrogen | |
| pCR2.1-TOPO | 3.9-kb vector for cloning PCR products; Apr | Invitrogen | |
| pUT/mini-Tn5Km | Apr; Tn5-based delivery plasmid; used as source of Kmr gene | 7 | D. Frank |
| pTOPO/altA | pCR2.1-TOPO with altA plus associated 5′ and 3′ DNA from MR-1; 5.2 kb | This study | |
| pTOPO/altA:Km | pTOPO/altA with the Kmr gene replacing 454 bp of altA; 6.8 kb | This study | |
| pDSEP/alt:Km | Kmr gene-interrupted altA gene from pTOPO/altA:Km cloned into the EcoRV site of pEP185.2; Cmr Kmr; 7.3 kb; used for gene replacement to generate ALTA1 mutant | This study | |
| pVK100 | 23-kb broad-host-range cosmid; Tcr Kmr Tra+ | 14 | ATCC 37156 |
| pJQ200KS | Mobilizable vector; P15A origin; Gmr | 37 | G. Reid |
| pEP185.2 | 4.28-kb mobilizable suicide vector derived from pEP184; oriR6K mobRP4 Cmr | 13 | V. L. Miller |
| pVKomcB | omcB of MR-1 cloned into the XhoI site of pVK100; used to complement OMCB1 | This study | |
| pVK21 | 21-kb MR-1 genomic DNA fragment including omcB and 19 kb of downstream DNA cloned into the HindIII site of pVK100; used to complement OMCB1 | This study | |
| pJQ10 | 12-kb MR-1 genomic DNA fragment including the most downstream 10-kb fragment of the pVK21 insert cloned into the PstI site of pJQ200KS; used to complement OMCB1 | This study | |
| pJQ14 | 14-kb MR-1 genomic DNA fragment including omcB and 12 kb of downstream DNA cloned into the SpeI site of pJQ200KS; used to complement OMCB1 | This study |
For preparation of subcellular fractions, S. putrefaciens was grown under anaerobic conditions as previously described (21) in M1 defined medium (31) supplemented with 15 mM lactate, 24 mM fumarate, and vitamin-free Casamino Acids (0.2 g liter−1). To test Mn(IV) or Fe(III) reduction, the defined medium was similarly supplemented with lactate and vitamin-free Casamino Acids, plus either 4.5 mM vernadite (δMnO2), 10 mM ferric citrate, or 2 mM FeOOH. Vernadite (28) and amorphous ferric oxyhydroxide (FeOOH) (16) were prepared as described previously.
To allow for growth of all strains under equivalent conditions (i.e., in the presence of kanamycin or gentamicin), the empty vectors pVK100 and pJQ200KS were introduced into the strains; the presence of these empty vectors did not alter electron acceptor phenotypes. To test Mn(IV) and Fe(III) reduction, inocula were prepared by using cells grown aerobically for 1 day on LB medium (except where noted otherwise) supplemented with the appropriate antibiotics; the cells were suspended in sterile distilled water, and the inoculum densities were adjusted to equalize turbidity (optical density at 500 nm as determined with a Beckman DU-64 spectrophotometer).
DNA manipulations.
A list of the synthetic oligonucleotides used is presented in Table 2. Restriction digestion, mapping, cloning, subcloning, and DNA electrophoresis were done by using standard techniques (39) and following manufacturers' recommendations as appropriate. The following procedures were done as previously reported (34): DNA ligation, isolation of plasmid and cosmid DNA, DNA sequencing, colony PCR, and determination of sizes of DNA fragments, RNA, and proteins. Computer-assisted sequence analysis and comparisons were done by using MacVector software (Accelrys, San Diego, Calif.).
TABLE 2.
Synthetic oligonucleotides used in this study
| Oligonucleotide | Sequencea |
|---|---|
| Oligonucleotides used for omcA experiments | |
| A5 | 5′-TTGATATCGGTGGCAGTGATGGRAAAGAT-3′ |
| A6 | 5′-TCAGTCGACTTAGTTACCGTGTGCTTCCAT-3′ |
| Oligonucleotides used for omcB experiments | |
| B1 | 5′-CATGTCGCCTTTAGTCACTTA-3′ |
| B2 | 5′-AACCAGTTTGCCAGCATCATT-3′ |
| B5 | 5′-GCAGTGGTAATAACGGCAATGAT-3′ |
| B6 | 5′-TCAACAACTGCACCGAAAGACTC-3′ |
| Oligonucleotides used for mtrA-mtrB experiments | |
| M1 | 5′-TACTGCCGGCACTTACCATCACA-3′ |
| M2 | 5′-GTGCGGTGTAGTCATGGCTGTTA-3′ |
| Oligonucleotides used for altA experiments | |
| C1 | 5′-TGGTCGACTTGGCTTTCTAGTGGGCTCAACC-3′ |
| C2 | 5′-TGGTCGACCCCGCTTGGCTGATGGAATACAC-3′ |
| C3 | 5′-CTGATCGATCGGCATTGACAAGAATAAT-3′ |
| C4 | 5′-CTGATCGATTCAGGCCAGAGTGAGACT-3′ |
| Oligonucleotides based on cat gene | |
| R1 | 5′-CAGACGGCATGATGAACCTGAATC-3′ |
| R2 | 5′-CCACCGTTGATATATCCCAATGGC-3′ |
| Oligonucleotide based on Kmr gene | |
| K1 | 5′-ATTGCGATCGATTTATGCTTGTAAACCGTT-3′ |
Underlined regions indicate the following restriction endonuclease sites engineered into the oligonulceotides: ClaI sites in C3, C4, and K1; SalI sites in C1, C2, and A6; and an EcoRV site in A5.
Electroporation and preparation of cells for electroporation were done as previously described (25, 34).
Construction of altA insertion mutation.
A gene replacement mutant of altA (AraC-like transcriptional regulator) was constructed from MR-1 by using a strategy analogous to that previously described for construction of mutants of omcA, omcB, and cymA (33, 34). A 1.3-kb fragment containing the entire altA gene plus 5′ and 3′ flanking sequences was generated by PCR of MR-1 genomic DNA by using primers C1 and C2 (Table 2). This PCR product was cloned into pCR2.1-TOPO, generating pTOPO/altA. Inverse PCR (35) of pTOPO/altA with primers C3 and C4 generated TOPO/altA(Δ454), a 4.8-kb fragment that is missing a 454-bp internal fragment of altA. The 2.1-kb Kmr gene from pUT/mini-Tn5Km was generated by PCR with primer K1. Following digestion of each PCR product with ClaI, the Kmr gene was ligated to the TOPO/altA(Δ454) fragment, generating pTOPO/altA:Km. A 3.0-kb DNA fragment containing the Kmr-interrupted altA gene was generated by PCR by using pTOPO/altA:Km as the template and primers C1 and C2. This fragment was blunt ended and ligated into the EcoRV site of suicide vector pEP185.2, generating pDSEP/alt:Km, which was electroporated into the donor strain E. coli S17-1λpir. Throughout this procedure, appropriate analyses (restriction digestion, PCR, DNA sequencing) were done to verify that the expected constructs were obtained.
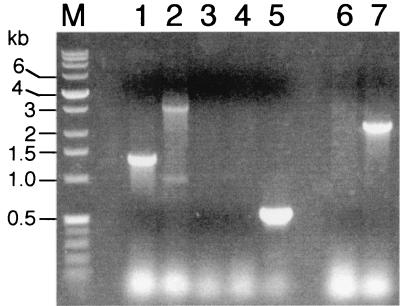
E. coli S17-1λpir(pDSEP/alt:Km) was mated with MR-1, and MR-1 exconjugants were selected by using kanamycin under aerobic conditions on defined medium with 15 mM lactate as the electron donor. Colonies were screened by colony PCR; one strain, designated ALTA1, lacked the expected 1.3-kb wild-type PCR product for altA, and it was positive for a 3.0-kb PCR product consistent with Kmr gene-interrupted altA (Fig. 1); these results were confirmed by PCR by using genomic DNA as a template. Strain ALTA1 was positive for the PCR product expected for the Kmr gene used to interrupt altA, and it was negative for a PCR product when primers specific to the cat gene of pEP185.2 were used (Fig. 1). Overall, these results are consistent with double-crossover gene replacement of altA, as a single-crossover insertion into the genome should have retained the cat gene of the suicide vector.
FIG. 1.
Colony PCRs comparing MR-1 and ALTA1. Lanes 1 and 2, reactions with primers C1 and C2 (Table 2) specific for the altA gene; lanes 3 to 5, reactions with primers R1 and R2 (Table 2) specific for the chloramphenicol acetyltransferase gene (cat) from pEP185.2; lanes 6 and 7, reactions with primer K1 (Table 2) specific for both ends of the kanamycin resistance (Kmr) gene. The templates for PCR were as follows: lanes 1, 3, and 6, MR-1; lanes 2, 4, and 7, ALTA1; and lane 5, pEP185.2. The sizes of the DNA markers (lane M) are indicated on the left.
RT-PCR and RNase protection.
Purification of total RNA and reverse transcription (RT)-PCR were done as previously described (25, 34). Total RNA (2 μg) from each strain served as the template, and sense and antisense primers were used as indicated below.
RNase protection assays were done by using the RPA III system (Ambion, Austin, Tex.). A 234-bp internal fragment of omcB was cloned into pCR2.1-TOPO; the desired orientation was verified by PCR. By using M13 primers, a DNA fragment containing the omcB fragment and flanking 5′ and 3′ vector DNAs was generated by PCR; by using this fragment as a template, the 435-base biotin-labeled antisense omcB RNA probe was generated with a MAXIscript kit (Ambion) and biotin-14-CTP. The probe was gel purified on a Tris-borate-EDTA-urea polyacrylamide gel. RNase protection assays were done by using total RNA (10 μg) and 400 pg of the probe.
Antibody specific for OmcB.
Recombinant technology was used to generate a protein fusion of thioredoxin (TR) to an internal 195-residue fragment of OmcB. Specifically, a 585-bp fragment of omcB was generated by PCR of MR-1 genomic DNA by using primers B1 and B2 and Platinum Taq DNA polymerase (Life Technologies, Rockville, Md.). This PCR product was cloned into pBAD/Thio-TOPO (Invitrogen, Carlsbad, Calif.) and transformed into E. coli TOP10. After identification of a clone in which the omcB insert was in the proper orientation, expression of the fusion protein was induced by incubation with 0.02% arabinose for 2 h at 37°C. The cells were harvested by centrifugation and lysed by using Bugbuster protein extraction reagent (Novagen, Madison, Wis.). The resulting fusion protein, which contained a 195-residue fragment of OmcB sandwiched between TR at the N terminus and a six-histidine tag at the C terminus, was localized primarily in inclusion bodies. After solubilization with 6 M urea, the TR-OmcB fusion was purified by using His·Bind Quick resin (Novagen) according to the manufacturer's instructions. The purified fusion was dialyzed at 4°C against 20 mM Tris-HCl (pH 7.5)-0.1 M glycine-5% (wt/vol) glycerol-1% (wt/vol) NaCl and then concentrated by ultrafiltration. The purified concentrated TR-OmcB fusion protein was used as an antigen to generate polyconal antisera in New Zealand White rabbits as previously described (27), except that Titermax (CytRx Corp., Norcross, Ga.) was used as an adjuvant.
Anti-peptide antibody specific for OmcA.
An internal peptide sequence from OmcA (32) was selected which was hydrophilic (according to the criteria of Hopp and Woods [12] as determined by MacVector software) and which was specific to OmcA when it was compared to likely proteins encoded by MR-1 based on the genomic sequence of MR-1 (The Institute for Genomic Research). An oligopeptide corresponding to amino acid residues 101 to 113 of immature OmcA was selected. The sequence of this peptide was CKEKVGETEADRGY; the amino-terminal cysteine was not part of the OmcA sequence but was added to facilitate coupling to the carrier protein. This oligopeptide was synthesized by ResGen (Huntsville, Ala.), which also verified the peptide composition by mass spectrometry. The oligopeptide was coupled to the carrier protein keyhole limpet hemocyanin (KLH) (Pierce, Inc., Rockford, Ill.) through the sulfhydryl group of the terminal cysteine residue of the peptide by using an Imject maleimide-activated immunogen conjugation kit (Pierce, Inc.). The coupling procedures were performed according to the instructions supplied by the manufacturer. The coupled peptide was divided into aliquots and stored at −80°C until it was used. The peptide-KLH conjugate was used as an immunogen to immunize rabbits as described above, which generated a polyclonal anti-OmcA-KLH antibody. The specificity of this antibody for OmcA was confirmed by using OMCA1, an omcA mutant, as a negative control (Myers and Myers, Abstr. 101st Gen. Meeting Am. Soc. Microbiol., abstr. K-99, 2001).
Miscellaneous procedures.
Purification of immunoglobulin G (IgG) from sera, removal of nonspecific antibodies by absorption with E. coli, and Western blotting were done as previously described (27).
Mn(II) in filtrates was determined by a formaldoxime method (1, 4). Fe(II) was determined by a ferrozine extraction procedure (16, 29).
Cytoplasmic membrane (CM), intermediate membrane (IM), and OM fractions, as well as soluble fractions (periplasm plus cytoplasm), were purified from cells by using an EDTA-lysozyme-Brij protocol as previously described (21). IM fractions have also been observed during subcellular fractionation of other gram-negative bacteria. Except for a buoyant density intermediate between that of the CM and that of the OM, the IM closely resembles the OM (27). The separation and purity of these subcellular fractions were assessed by spectral cytochrome content analysis (21), membrane buoyant density analysis (21), and sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) with gels (15, 19) stained for protein with Pro-Blue (Owl Separation Systems, Woburn, Mass.) or for heme as previously described (41). Protein was determined by a modified Lowry method, with bovine serum albumin as the standard (25).
Statistical analysis was performed by using analysis of variance and Tukey's multiple comparison post test (InStat software; GraphPad, San Diego, Calif.).
RESULTS
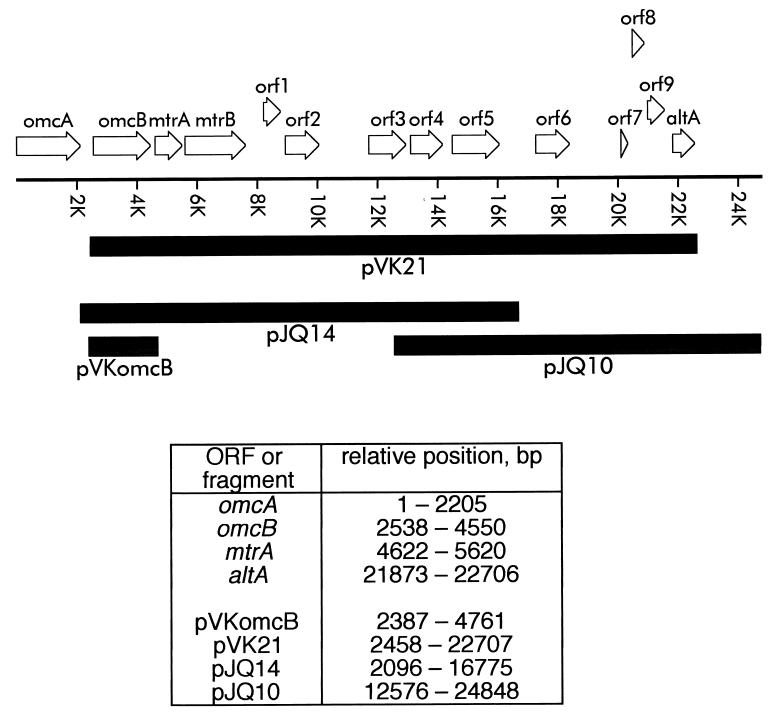
The linear arrangement of the genes and ORFs in the MR-1 genome from omcA downstream is shown in Fig. 2. omcA (32) and omcB (34) encode decaheme c-type cytochromes which are putative OM lipoproteins. altA encodes a putative transcriptional regulator whose role is discussed below. The best matches to these ORFs as identified by BLAST analyses are presented in Table 3.
FIG. 2.
Linear orientation of the gene cluster of MR-1 from omcA downstream, as derived from the preliminary genomic sequence data provided by The Institute for Genomic Research (http://www.tigr.org). The diagram is drawn to scale, and the transcription direction for all genes is from left to right (5′ → 3′). The relative positions of some of the ORFs and complementing fragments are indicated at the bottom; position 1 corresponded to the first base of the start codon of omcA. The solid boxes below the gene cluster indicate the DNA fragments used in complementation studies. omcA and omcB encode OM cytochromes involved in Mn(IV) reduction (32, 34), while mtrA and mtrB have putative roles in metal reduction (2, 3). Other possible ORFs (orf1 through orf9 and altA) with some sequence homology to known genes from other species are also shown. The best matches to these ORFs, as identified by BLAST analysis, are shown in Table 3.
TABLE 3.
Amino acid sequence homologies between sequences encoded by the downstream ORFs from MR-1 and the best matches from other organismsa
| ORF | Total no. of residuesb | Accession no.c | Organism | Proposed function | P(N)d | % Identity of aligned regionse |
|---|---|---|---|---|---|---|
| orf1 | 237 | A82343 (PIR) | Vibrio cholerae | Conserved hypothetical | 9.2 × 10−59 | 51 (88), 53 (58), 50 (42) |
| orf2 | 407 | B64740 (PIR) | E. coli | Sugar diacid regulator | 1.6 × 10−93 | 42 (182), 53 (166) |
| orf3 | 452 | F82402 (PIR) | V. cholerae | Probable permease | 0 | 78 (235), 82 (205) |
| orf4 | 384 | P44507 (SP) | Haemophilus influenzae | Glycerate kinase | 7.5 × 10−18 | 55 (288), 62 (81) |
| orf5 | 549 | H82234 (PIR) | V. cholerae | Probable glutamate decarboxylase | 0 | 65 (447), 54 (82) |
| orf6 | 400 | NP_046537.1 | Yersinia pestis | Probable transposase | 1.1 × 10−106 | 47 (309), 58 (43) |
| orf7 | 105 | AAF39790.1 (GB) | Caenorhabditis elegans | Ion channel | 0.41 | 42 (28), 40 (35) |
| orf8 | 171 | AAF60966.1 (GB) | E. coli | Unknown | 7.9 × 10−47 | 46 (166) |
| orf9 | 225 | AAF63432.1 (GB) | Yersinia enterocolitica | Streptogramin A acetyltransferase | 1.1 × 10−97 | 65 (212) |
| altA | 278 | F82388 (PIR) | V. cholerae | Trancriptional regulator, AraC/XylS family | 5.8 × 10−120 | 65 (266) |
The best matches were identified by searching the sequence databases on the National Center for Biotechnology Information server by using the predicted amino acid sequences encoded by the MR-1 ORFs as queries for BLASTP analysis (BLOSUM62 matrix).
Total number of amino acid residues encoded by each MR-1 ORF.
Accession number in the following databases: PIR-protein (PIR), Swiss-Protein (SP), and GenBank (GB).
P(N), smallest sum probability value of the match.
Percent identity in the amino acid sequence. Because the BLAST algorithm is optimized for ungapped sequences, each value is the percent identity for each aligned segment within the total protein. The values in parentheses are the total numbers of residues in the aligned segments.
Cytochrome content.
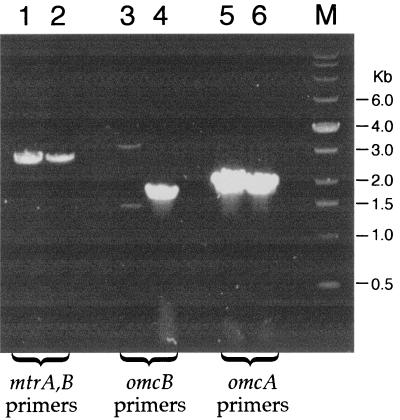
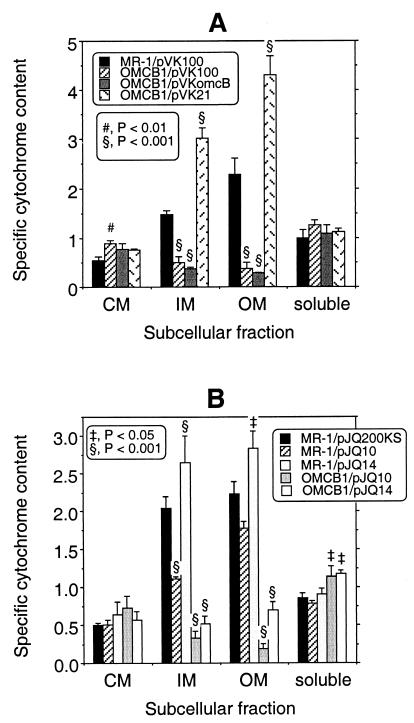
OMCB1, the omcB mutant, does not express omcB, but it expresses the upstream gene omcA and the downstream genes mtrA and mtrB (Fig. 3). Consistent with a previous report (34), OMCB1 lacked a heme-positive band corresponding to OmcB in the IM and OM, and the levels of all OM cytochromes were markedly depressed in OMCB1 (Fig. 4). When examined quantitatively, the specific cytochrome content of the OM of OMCB1/pVK100 was only about 17% of that of MR-1/pvK100 (Fig. 5A). Even though OMCB1 contains wild-type omcB, complementation of OMCB1 with pVKomcB did not restore OmcB or the other cytochromes to the IM or OM (Fig. 4 and 5A). However, complementation of OMCB1 with pVK21, which contains omcB plus approximately 19 kb of downstream DNA, did restore OmcB as well as the other OM cytochromes (Fig. 4); in fact, the specific OM cytochrome content of OMCB1/pVK21 was 188% that of wild-type MR-1/pVK100 (Fig. 5A). These results imply that downstream DNA must be present in cis with omcB to restore the normal OM cytochrome content in OMCB1. The results obtained for the IM paralleled those obtained for the OM; i.e., relative to MR-1/pVK100, the cytochrome content was markedly lower in OMCB1/pVK100 and OMCB1/pVKomcB, but it was significantly higher in OMCB1/pVK21 (Fig. 5A). While the exact nature of the IM is unknown, the IM closely resembles the OM except for its lower buoyant density and lower specific cytochrome content (27). Relative to MR-1/pVK100, there were only minor differences in the cytochrome contents of the CM and soluble fractions of MR-1 and OMCB1 (Fig. 4 and 5A). The large decreases in the cytochrome contents of the IM and OM of OMCB1 were not, therefore, the result of global declines in cytochrome synthesis.
FIG. 3.
RT-PCR products obtained by using total RNA from the following strains (grown anaerobically with fumarate) as templates: OMCB1 (lanes 1, 3, and 5) and MR-1 (lanes 2, 4, and 6). The following primer pairs (Table 2) were used: M1 and M2 specific for mtrA, B (lanes 1 and 2), B5 and B6 specific for omcB (lanes 3 and 4), and A5 and A6 specific for omcA (lanes 5 and 6). The sizes of the DNA markers (lane M) are indicated on the right. Controls without reverse transcriptase yielded no bands (data not shown), indicating that the products arose from mRNA.
FIG. 4.
Heme-stained SDS-PAGE profiles of subcellular fractions prepared from cells of the following strains grown anaerobically with fumarate as the electron acceptor: MR-1/pVK100 (lanes 1, 5, 9, and 13), OMCB1/pVK100 (lanes, 2, 6, 10, and 14), OMCB1/pVKomcB (lanes 3, 7, 11, and 15), and OMCB1/pVK21 (lanes 4, 8, 12, and 16). The lanes were loaded with 25-μg portions of protein from the following subcellular fractions: IM (lanes 1 to 4), OM (lanes 5 to 8), CM (lanes 9 to 12), and soluble fraction (lanes 13 to 16). The arrow on the left indicates the position of OmcB in the IM and OM. The positions and molecular masses of protein standards obtained from parallel gels containing the same samples but stained for protein are indicated on the left and right.
FIG. 5.
Specific cytochrome contents of the subcellular fractions prepared from strains MR-1 and OMCB1 that were grown anaerobically with fumarate as the electron acceptor. The strains contained various plasmids derived from either pVK100 (A) or pJQ200KS (B). The specific cytochrome content is the difference between the absorbance at the peak and the absorbance at the trough of the Soret region for reduced-minus-oxidized difference spectra per milligram of protein. The values are means ± standard deviations based on the results of independent experiments (n = 3 independent cultures of each strain). The section signs, number sign, and double daggers indicate that values are statistically significantly different from the values for MR-1/pVK100 (A) and MR-1/pJQ200KS (B) at P < 0.001, P < 0.01, and P < 0.05, respectively. Note the different y-axis scales for panels A and B.
Complementation of OMCB1 with pJQ14, which contains omcB plus approximately 12 kb of downstream DNA, was not sufficient to restore the OM cytochrome levels to the wild-type levels; the levels were 31% of those observed for MR-1/pJQ200KS (Fig. 5B). Interestingly, the presence of pJQ14 in MR-1 increased the specific OM cytochrome content relative to that of MR-1/pJQ200KS (Fig. 4B). However, while the OM cytochrome levels in OMCB1/pJQ14 were marginally higher than those in OMCB1/pVK100 (Fig. 5), the differences between these two strains were not significant (P > 0.05). The markedly different effects of pVK21 and pJQ14 on the OM cytochrome content suggest that at least a portion of the most downstream 5.9 kb of pVK21 is required to restore OM cytochromes to OMCB1. However, complementation of OMCB1 with just the most downstream DNA (pJQ10) did not increase the OM cytochrome contents of OMCB1 and MR-1 (Fig. 5B).
Expression and localization of OmcB and OmcA.
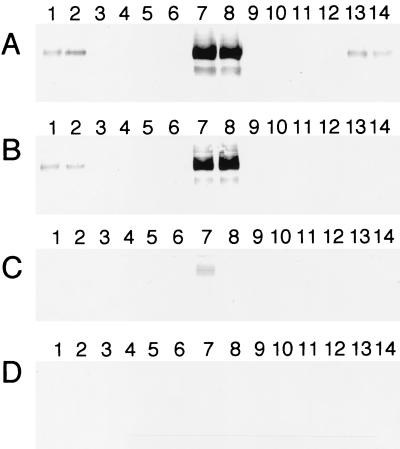
An antibody specific for OmcB was used to determine the presence of OmcB in the subcellular fractions of the various strains. OmcB was present in the OM and IM of MR-1/pVK100 and MR-1/pJQ200KS and at markedly increased levels in the OM and IM of OMCB1/pVK21 (Fig. 6). None of the other OMCB1 strains contained detectable OmcB in the OM and IM fractions (Fig. 6). OmcB was not detected in the CM or soluble fractions of any of these strains, with the exception of small amounts in the CM of OMCB1/pVK21 (Fig. 6). The OmcB band in Fig. 6 migrated with an apparent molecular mass of approximately 77 kDa, consistent with the predicted molecular mass of the mature protein (75.7 kDa). In previous studies, heme-positive bands representing OmcB were also observed at approximately 53 to 62 kDa (34), and these bands may have represented proteolytic products generated during the lengthy subcellular fractionation process or OmcB that was incompletely denatured by SDS; evidence for such lower-molecular-mass OmcB species was seen in the OM of OMCB1/pVK21 (Fig. 6). Overall, the OMCB1 strains containing pVK100, pVKomcB, pJQ10, and pJQ14 lack detectable OmcB protein; i.e., OmcB is not merely mislocalized to other subcellular fractions, and OmcB lacking heme c is not present.
FIG. 6.
Western blots obtained by using polyclonal IgG specific for OmcB. The lanes were loaded with 5-μg portions of protein from OM (A), IM (B), CM (C), and soluble (D) fractions that were isolated from cells of the following strains grown anaerobically with fumarate as the electron acceptor: MR-1/pVK100 (lane 1), MR-1/pJQ200KS (lane 2), OMCB1/pVK100 (lanes 3 and 4), OMCB1/pVKomcB (lanes 5 and 6), OMCB1/pVK21 (lanes 7 and 8), OMCB1/pJQ10 (lanes 9 and 10), OMCB1/pJQ14 (lanes 11 and 12), and ALTA1 (lanes 13 and 14). For the strains loaded in two lanes, each lane contained the fractions isolated from an independent culture. In total, fractions from three independent cultures of each strain were analyzed, and the results shown are representative of the results for the triplicate samples.
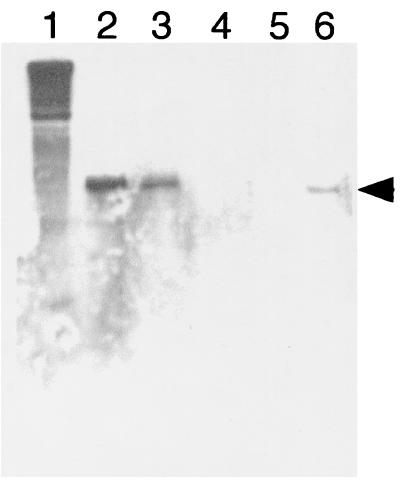
To determine if the lack of OmcB in most OMCB1 strains was due to a lack of transcription, an RNase protection assay analysis was done to examine omcB mRNA content. In agreement with the presence of OmcB protein, omcB transcript was detected in MR-1/pVK100 and OMCB1/pVK21 and was not detected in OMCB1/pVK100 and OMCB1/pVKomcB (Fig. 7). However, OMCB1/pJQ14 contained prominent levels of omcB transcript (Fig. 7) even though it lacked detectable levels of OmcB protein (Fig. 6).
FIG. 7.
RNase protection analysis of the omcB transcript in total RNA isolated from cells of the following strains grown anaerobically with fumarate as the electron acceptor: OMCB1/pJQ14 (lane 2), OMCB1/pVK21 (lane 3), OMCB1/pVKomcB (lane 4), OMCB1/pVK100 (lane 5), and MR-1/pVK100 (lane 6). Lane 1 contained the full-length untreated 435-base probe. The position of the 234-base protected probe specific for omcB is indicated by the arrowhead. The results shown are the results of a representative experiment.
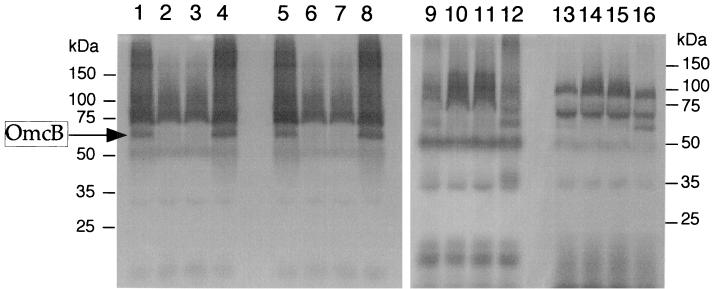
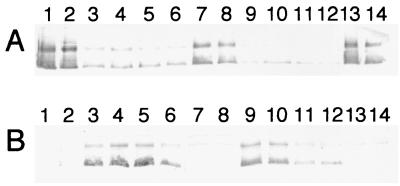
In a previous report, it was noted that OmcA was mislocalized to the CM of the omcB mutant OMCB1 (34). Therefore, Western blotting with anti-OmcA-KLH was used to examine how complementation of OMCB1 with the various constructs affected the localization of OmcA. The localization of OmcA in OMCB1/pVK21 resembled that in the MR-1 strains; i.e., OmcA was predominant in the OM, and there were minor amounts in the CM (Fig. 8). In contrast, OmcA was mislocalized to the CM of OMCB1 strains containing pVK100, pVKomcB, pJQ10, and pJQ14, and there were only minor amounts in the OM (Fig. 8). Therefore, only pVK21 restored normal localization of OmcA to the OM of OMCB1 (Fig. 8); similarly, only pVK21 restored OmcB to OMCB1 (Fig. 6). Previously, when we loaded 5-μg portions of subcellular fractions on Western blots probed with an antibody to purified SDS-denatured OmcA, OmcA similarly appeared as a broad band with greatest intensity in the upper and lower regions (34). This entire signal represents OmcA as it is absent in the omcA mutant OMCA1 (34). As shown in Fig. 8, the upper OmcA band migrated at about 83 kDa, which is consistent with the predicted mass of the mature protein, 82.7 kDa (32). The lower OmcA band in Fig. 8 migrated at about 66 kDa and could have resulted from partial proteolysis that may have occurred during the lengthy cellular subfractionation process. Alternatively, this lower OmcA band could have resulted from incomplete denaturation of OmcA by SDS.
FIG. 8.
Western blots obtained by using polyclonal IgG specific for OmcA. The lanes were loaded with 5-μg portions of protein from OM (A) and CM (B) fractions that were isolated from cells of the following strains grown anaerobically with fumarate as the electron acceptor: MR-1/pVK100 (lane 1), MR-1/pJQ200KS (lane 2), OMCB1/pVK100 (lanes 3 and 4), OMCB1/pVKomcB (lanes 5 and 6), OMCB1/pVK21 (lanes 7 and 8), OMCB1/pJQ10 (lanes 9 and 10), OMCB1/pJQ14 (lanes 11 and 12), and ALTA1 (lanes 13 and 14). For the strains loaded in two lanes, each lane contained the fractions isolated from an independent culture. In total, fractions from three independent cultures of each strain were analyzed, and the results shown are representative of the results for the triplicate samples.
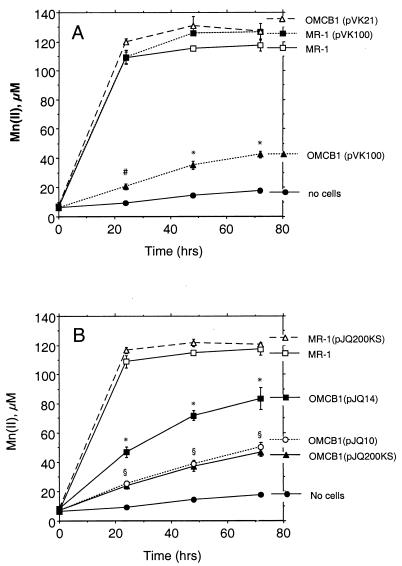
Mn(IV) reduction.
For wild-type strains (MR-1 with or without vectors), the vast majority of Mn(IV) reduction occurred during the first 24 h (Fig. 9). The Mn(IV) reduction by MR-1 was essentially the same as that by MR-1 containing the empty vector pVK100 or pJQ200KS (Fig. 9). In contrast, the rates of Mn(IV) reduction by OMCB1/pVK100 and OMCB1/pJQ200KS were 11 and 14% of the rate of Mn(IV) reduction by MR-1, when they were corrected for the no-cell background rate (Fig. 9). The rates of Mn(IV) reduction by OMCB1/pJQ10 (Fig. 9) and OMCB1/pVKomcB (data not shown) were the same as that catalyzed by OMCB1/pJQ200KS. Complementation of OMCB1 with pJQ14 only partially restored Mn(IV) reduction [36% of the rate of Mn(IV) reduction by MR-1], whereas complementation with pVK21 fully restored Mn(IV) reduction (Fig. 9).
FIG. 9.
Reduction of δMnO2 under anaerobic conditions as determined by the formation of Mn(II) over time. Strains were complemented with DNA fragments cloned in pVK100 (A) or pJQ200KS (B). All values represent the mean ± standard deviation (n = 3 independent cultures of each strain). For points lacking apparent error bars, the bars were smaller than the points. An asterisk indicates that a value is statistically significantly different from the values for all other strains in the panel at P < 0.001; a number sign indicates that a value is statistically significantly different from the value for no cells at P < 0.05 and from the values for all other strains in the panel at P < 0.001; and a section sign indicates that the values for strains OMCB1(pJQ10) and OMCB1(pJQ200KS) are statistically the same but are statistically significantly different from the values for all other strains in the panel at P < 0.001.
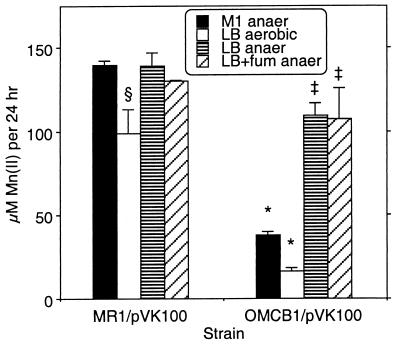
Effect of inoculum preparation on Mn(IV) reduction.
In the experiments whose results are shown in Fig. 9, as in previous studies in which electron acceptor use was tested, inocula were prepared by using cells grown aerobically on LB medium (23, 25, 26, 33, 34). However, some components for anaerobic electron transport (e.g., OM cytochromes, fumarate reductase) are expressed only under anaerobic conditions (20, 21, 23). Menaquinone, which is required for Mn(IV) reduction, is present at significantly higher levels in anaerobic cells (23). To determine if the deficiency in Mn(IV) reduction by OMCB1 might be due to slower adaptation to anaerobic conditions, the rates of anaerobic Mn(IV) reduction were compared by using inocula that were pregrown either aerobically (LB medium) or anaerobically (M1 defined medium plus fumarate). When preparations were compared to MR-1 inocula pregrown under identical conditions, the marked deficiency in Mn(IV) reduction by OMCB1 was observed with inocula grown in M1 defined medium anaerobically and in LB medium aerobically (Fig. 10). MR-1 cells grown anaerobically in M1 defined medium containing fumarate have been shown to be well adapted for Mn(IV) reduction; they have prominent levels of various anaerobic electron transport components and are capable of rapid reduction of MnO2 (21, 22, 31). Therefore, the deficiency in Mn(IV) reduction by OMCB1 cannot be explained by slower adaptation from aerobic to anaerobic conditions.
FIG. 10.
Reduction of δMnO2 under anaerobic conditions by inocula pregrown under various conditions as determined by the increase in Mn(II) concentration over the first 24 h. The inocula for the two strains were prepared by using cells grown under the following conditions: M1 defined medium containing 15 mM lactate plus 24 mM fumarate in the anaerobic chamber (M1 anaer); LB medium under room air (LB aerobic); LB medium in the anaerobic chamber (LB anaer); and LB medium containing 24 mM fumarate in the anaerobic chamber (LB+fum anaer). The values are means ± standard deviations based on the results of three parallel but independent experiments. Asterisks, the section sign, and double daggers indicate that values are statistically significantly different from the value for MR-1/pVK100 (M1 anaer) at P < 0.001, P < 0.01, and P < 0.05, respectively.
The inoculum studies were extended to also include cells that were pregrown anaerobically in LB medium with or without fumarate. For MR-1/pVK100, the rates of Mn(IV) reduction were similar for all LB medium inocula (Fig. 10). In contrast, inocula of OMCB1/pVK100 grown anaerobically in LB medium (with or without fumarate) exhibited significantly higher rates of Mn(IV) reduction than inocula grown in LB medium aerobically or in M1 defined medium anaerobically (Fig. 10), although these rates were still statistically lower than those observed for MR-1/pVK100 pregrown anaerobically in LB medium (P < 0.05) (Fig. 10). Since anaerobically grown MR-1 and OMCB1 cells have lower OM cytochrome contents when they are grown in LB medium than when they are grown in M1 defined medium containing fumarate (data not shown), the higher rates of Mn(IV) reduction by OMCB1 cells grown anaerobically in LB medium cannot be explained by OM cytochrome content.
However, after anaerobic growth in LB medium, a black deposit was noted on the glass in the necks of the culture bottles; this deposit was not observed with cultures grown anaerobically in M1 defined medium containing fumarate. This black deposit resembled a metal sulfide precipitate, suggesting possible H2S generation. To examine this possibility, MR-1/pVK100 and OMCB1/pVK100 cells were grown under anaerobic conditions in LB medium and M1 defined medium (both supplemented with fumarate); after 2 days of growth, 1 ml of each culture was added to 1 ml of a 1% ferrous ammonium sulfate solution. The LB medium cultures grown anaerobically yielded a prominent black precipitate (probably FeS) within 15 min, suggesting that H2S was present. No black precipitate was observed with anaerobically grown M1 defined medium cultures even after 5 h or with aerobically grown LB medium cultures. Since H2S is a potent reductant of Mn(IV) (5), its generation could explain the increased Mn(II) levels observed with OMCB1 cells pregrown anaerobically in LB medium (Fig. 10). Because the reduction of Mn(IV) by sulfide is relatively rapid (5), the studies were repeated in order to examine Mn(II) after both 5 and 24 h. For inocula of MR-1/pVK100 and OMCB1/pVK100 grown anaerobically in LB medium, the vast majority of Mn(IV) reduction occurred early; i.e., 69 to 70% of the 24-h Mn(II) was observed at the 5-h time point. In contrast, for inocula grown aerobically in LB medium, only 20 to 24% of the 24-h Mn(II) was observed at the 5-h time point.
Overall, the results obtained with inocula grown aerobically in LB medium and anaerobically in M1 defined medium demonstrate that OMCB1 is markedly deficient in Mn(IV) reduction. However, the generation of H2S in anaerobic LB medium cultures makes these inocula unsuitable for accurately assessing direct Mn(IV) reduction.
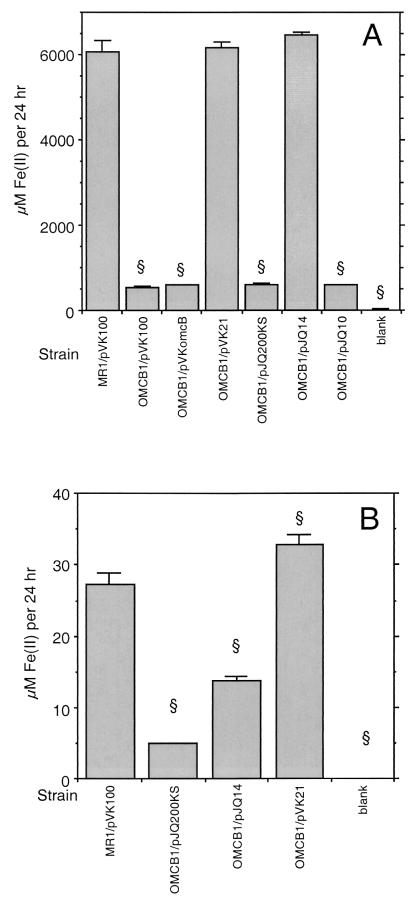
Fe(III) reduction.
We previously reported that OMCB1 was similar to MR-1 in its ability to reduce Fe(III) (34). This finding was based on examination of Fe(III) citrate reduction at 3 to 4 days when we used inoculum densities similar to those originally used to identify OMCB1 as deficient in Mn(IV) reduction. Beliaev et al. (3) have since reported that an omcB (mtrC) mutant (strain SR-8) isolated from MR-1R by transposon mutagenesis was deficient in Fe(III) reduction when it was examined over a few hours. This prompted a more thorough examination of Fe(III) reduction by OMCB1 in which lower inoculum densities were used. After 5 days, OMCB1/pVK100 and OMCB1/pJQ200KS cultures were strongly positive for Fe(III) citrate reduction, with Fe(II) levels of >4 mM (data not shown). However, when examined after only 24 h, OMCB1 strains containing pVK100, pVKomcB, pJQ200KS, and pJQ10 were all deficient in Fe(III) citrate reduction, with rates that were 9 to 10% of those observed for MR-1/pVK100 (Fig. 11A). Complementation of OMCB1 with either pVK21 or pJQ14 restored the Fe(III) citrate reduction level to that of the wild type (Fig. 11A).
FIG. 11.
Reduction of Fe(III) citrate (A) or αFeOOH (B) under anaerobic conditions as determined by the increase in Fe(II) concentration over the first 24 h. All values are means ± standard deviations based on the results for three independent cultures. Blank, cell-free control. A section sign indicates that a value is statistically significantly different from the value for MR-1/pVK100 (A) or MR-1/pJQ200KS (B) at P < 0.001. Note the different y-axis scales for panels A and B.
The rates of reduction of amorphous FeOOH were also examined by using a subset of these strains (Fig. 11B). While the rates of reduction of insoluble FeOOH (Fig. 11B) were considerably lower than those of soluble Fe(III) citrate, as expected, the overall trends were similar for the various strains. OMCB1/pJQ200KS was markedly deficient in FeOOH reduction, with rates that were 18% of those observed for MR-1/pJQ200KS (Fig. 11B). Complementation of OMCB1 with pVK21 restored FeOOH reduction to levels that were 20% greater than those of the wild type, consistent with the ability of this plasmid to fully restore Fe(III) citrate reduction (Fig. 11A). In contrast to its ability to fully restore Fe(III) citrate reduction, pJQ14 restored FeOOH reduction to levels that were only 50% of those of the wild type (Fig. 11B).
Analysis of the altA mutant.
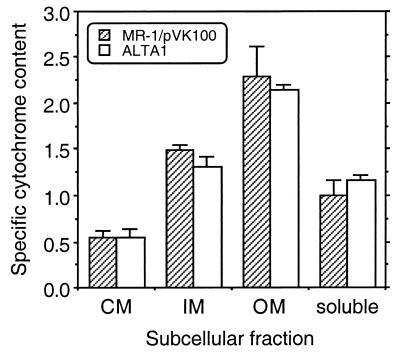
The marked phenotypic differences between the OM cytochrome content and Mn(IV) reduction associated with complementation of OMCB1 with pJQ14 and the OM cytochrome content and Mn(IV) reduction associated with complementation with pVK21 suggest that there is something essential in the most downstream region of the insert in pVK21 that is lacking in pJQ14. This downstream region of the pVK21 insert contains five possible ORFs, orf6 through orf9 and altA (Fig. 2); of these, the most downstream ORF, altA, is the only one which putatively encodes a positive transcriptional regulator. To determine if altA has a role in OM cytochrome expression, a gene replacement mutant (ALTA1) of altA was generated (Fig. 1) and characterized. When examined quantitatively, the specific cytochrome contents of the various subcellular fractions of ALTA1 were essentially the same as those of MR-1/pVK100 (Fig. 12). With the exception of a minor difference in the CM, the overall patterns of cytochromes as determined by heme-stained SDS-PAGE of the subcellular fractions were also similar for these two strains (data not shown). Western blot analysis demonstrated that OmcB and OmcA were present and localized to the OM of ALTA1 (Fig. 6 and 8). As determined with an antibody specific for the fumarate reductase of MR-1 (18), the levels of this soluble flavocytochrome were also similar in MR-1 and ALTA1 (data not shown). While the percentage of total cellular protein recovered in the OM was greater for ALTA1 than for MR-1 (data not shown), the difference was not statistically significant. The rate of Mn(IV) reduction by ALTA1 was statistically indistinguishable from that of MR-1 (data not shown). Therefore, altA is not required for expression or proper localization of the OM cytochromes or for wild-type levels of Mn(IV) reduction. It is, therefore, unlikely that the presence of altA in the downstream region of the pVK21 insert contributes to the ability of pVK21 to fully restore the phenotype of the omcB mutant OMCB1.
FIG. 12.
Specific cytochrome contents of the subcellular fractions prepared from strains MR-1/pVK100 and ALTA1 that were grown anaerobically with fumarate as the electron acceptor. The specific cytochrome content is the difference between the absorbance at the peak and the absorbance at the trough of the Soret region from reduced-minus-oxidized difference spectra per milligram of protein. The values are means ± standard deviations based on results of independent experiments (n = 3 independent cultures of each strain). The differences between the two strains were not statistically significant (P > 0.05).
DISCUSSION
The ability of MR-1 to obtain energy for growth via the reduction of insoluble Mn(IV) and Fe(III) oxides under anaerobic conditions implies the existence of a mechanism to link its electron transport machinery in the CM to the reduction of these extracellular metal oxides. It has been previously demonstrated that the tetraheme cytochrome CymA and menaquinone, which are associated with the CM, are required for reduction of Mn(IV) and Fe(III) (23, 25, 33). Since their localization to the CM would preclude direct contact with extracellular insoluble metal oxides, these CM components are likely part of electron transport chains that eventually transfer electrons to redox-active components in the OM. The localization of cytochromes (e.g., OmcA and OmcB) to the OM of anaerobically grown MR-1 (21, 27) suggests a possible mechanism by which electrons could be transferred to MnO2 at the cell surface. A preliminary characterization of OMCB1 demonstrated that there was a marked decline in OM cytochrome content and Mn(IV) reduction (34). The data reported here extend the examination of OMCB1. The low rate of Mn(IV) reduction (11 to 14% of the MR-1 rate) corresponded to the low OM cytochrome content (15 to 17% of the MR-1 content) in OMCB1. The low rate of Mn(IV) reduction might be mediated by the limited remaining cytochromes in the OM. Complementation with pVK21 simultaneously restored the rate of Mn(IV) reduction to that of the wild type and resulted in a OM cytochrome content greater than that of the wild type. These findings are consistent with a role for OM cytochromes in the reduction of Mn(IV).
OmcB and/or other OM cytochromes could serve as a terminal Mn(IV) reductase(s) or as intermediate electron transport components with some other as-yet-unidentified noncytochrome OM component(s) serving as the terminal Mn(IV) reductase(s). The pleiotropic effects on the proper localization of OmcA and other OM cytochromes, however, complicate interpretation of the exact role of OmcB in Mn(IV) reduction. The mislocalization of OmcA to the CM of OMCB1 does not fully account for the decline in Mn(IV) reduction because a mutant lacking only omcA reduces Mn(IV) at a rate that is 55% of the rate of MR-1 (34). The mechanism by which interruption of omcB results in decreased levels of all OM cytochromes is not known. omcB gene replacement does not prevent expression of the genes immediately downstream from omcB. RT-PCR indicates that mtrA and mtrB continue to be expressed in OMCB1, but a partial decline in expression of these genes cannot be ruled out at this point. However, it is clear that Mn(IV) reduction and proper localization of OM cytochromes are restored only when OmcB is itself restored. It is therefore possible that OmcB itself is somehow necessary for the proper localization of other cytochromes to the OM. Exactly how OmcB might do this is not known at this time, but one possibility is that it may act as some sort of chaperone which facilitates proper localization of other OM cytochromes. The possibility that OmcB may also be required for proper localization of other proteins to the OM must be considered. However, since OmcB is still localized to the OM in an omcA mutant (34), OmcA is not necessary for localization of OmcB to the OM.
The mislocalization of OmcA to the CM in the omcB mutant resembles the findings described in a recent report on a type II secretion mutant of S. putrefaciens strain 200 (9). A 91-kDa heme-containing protein which is apparently localized to the outer face of the OM in the wild type was not surface associated in this type II mutant and was at least partially mislocalized to the CM (9). However, omcB is not homologous to known type II secretion genes, and omcB is not part of the multigene cluster encoding the type II secretion system in MR-1.
Complementation of OMCB1 with the various constructs revealed some interesting findings. The amount of MR-1 DNA upstream of omcB in these constructs does not explain the ability or inability to complement OMCB1. For example, pVKomcB had more upstream DNA than pVK21 had, yet pVKomcB did not complement OMCB1, whereas pVK21 fully restored its phenotype. pJQ14 had the most upstream DNA, but it resulted in only a modest increase in Mn(IV) reduction rates and an insignificant increase in OM cytochrome content. While omcB mRNA was not detected in OMCB1/pVKomcB, it was very prominent in OMCB1/pJQ14; however, OmcB was not detected in any subcellular fraction of OMCB1/pJQ14. In addition, OmcA remained mislocalized to the CM of OMCB1/pJQ14. While it is possible that small amounts of OmcB were present in OMCB1/pJQ14, they were below the limit of detection of the antibody used for Western blotting. The lack of detectable OmcB in OMCB1/pJQ14 could result from either a low level of translation or rapid degradation of OmcB apoprotein. The latter possibility is unlikely because anti-OmcB did not detect bands at smaller masses in OMCB1/pJQ14 (data not shown). In total, it appears that despite prominent expression of omcB in OMCB1/pJQ14, OmcB was not detected and there was only a modest increase in the rate of Mn(IV) reduction.
Compared with the presence of pJQ14, the presence of the additional 5.9 kb of downstream DNA in pVK21 allowed for full restoration of the OMCB1 phenotype to that of the wild type. Of the five putative ORFs in this 5.9-kb region, only altA had significant homology to known regulatory genes (specifically, the genes of the AraC family). The AraC family is a family of common prokaryotic transcriptional regulators that positively (for nearly all genes) regulate a wide variety of genes (11, 38). Some members control expression of single genes or operons, whereas others regulate multiple unlinked genes (11). Members of this family exist as either monomers or dimers, and the latter contain dimerization domains (11, 38); the DNA-binding domains of the dimers can bind to two distinct regions of DNA (11). All AraC family members contain helix-turn-helix DNA-binding motifs, and some have additional domains that also bind effector molecules (11). In addition to the BLAST alignments, altA also had other features consistent with an AraC class transcriptional regulator: (i) altA encodes a putative protein of 277 residues, consistent with the 250 to 300 residues expected for this family (11); (ii) altA is in the vicinity of, and downstream of, the ORF(s) that it might regulate, a feature of several other members of the AraC class (36); and (iii) residues 193 to 264 of AltA contain 14 of the 17 most conserved residues (including proper spacing) of the C-terminal region of the AraC-type regulators (11), ATLAGLSPYYLLKQFQHYYGLPPHAYQIQARVRLAKAKIKQGTRLLDVALDCGFHDQSHLNRHFKKTVGVTP (conserved residues are underlined). One hypothesis was that AltA somehow regulates factors that are necessary for the expression, translation, and/or proper localization of OmcB. This hypothesis was refuted because a gene replacement mutant of altA resembled the wild type with respect to Mn(IV) reduction, OM cytochrome content, and localization of OmcA and OmcB to the OM. It is possible that at least one of the other ORFs in this region (orf6 to orf9) is necessary for the synthesis and/or localization of OmcB. However, the fact that this region is intact in the chromosome of OMCB1 argues against a role for a specific gene product. Alternatively, some part of the DNA itself from this region may be important; this is consistent with the requirement that this downstream DNA be present in cis with wild-type omcB. Although many scenarios could be envisioned, one possibility is that this region contains a DNA sequence important for binding one-half of a dimerized transcription factor.
Since the initial characterization of OMCB1, another report has described the isolation of S. putrefaciens strain SR-8, an omcB (mtrC) mutant (3). When examined qualitatively, SR-8 was reported to be deficient in Mn(IV) reduction (3), which is consistent with our quantitative findings for OMCB1. SR-8 was also reported to have markedly lower rates of Fe(III) reduction than MR-1 (3). However, the mutation in SR-8 was polar and blocked expression of the downstream genes mtrA and mtrB (3). Complementation of SR-8 with omcB was not accomplished, but when mtrA and mtrB were expressed from an inducible plasmid, SR-8 remained deficient in Mn(IV) and Fe(III) reduction, which led Beliaev et al. to conclude that OmcB (MtrC) was required for Mn(IV) and Fe(III) reduction (3). While the studies with OMCB1 are consistent with a role for OmcB in Mn(IV) reduction, complementation with pJQ14 suggests that OmcB is not essential for Fe(III) reduction because pJQ14 fully restored Fe(III) citrate reduction and restored 50% of FeOOH reduction (Fig. 11), despite the fact that pJQ14 did not significantly increase the OM cytochrome content (Fig. 5) and did not result in detectable OmcB protein (Fig. 6). One or more of the other ORFs downstream of omcB in pJQ14 may therefore be responsible for restoring Fe(III) reduction. Based on a putative role in metal reduction (2, 3), mtrA and mtrB are possible candidates; however, these genes are expressed at some level in OMCB1 (Fig. 3), and they did not restore Fe(III) reduction to SR-8 (3), so they alone cannot account for the observations regarding Fe(III) reduction. MtrA is a putative periplasmic cytochrome (3) which could potentially serve as an electron carrier, but it is not localized where it could directly transfer electrons to extracellular insoluble metal oxides. MtrB is putatively localized to the OM (2) but does not have any apparent electron carrier moieties, so its direct role in metal reduction is unclear. It is not known if MtrB was properly localized to the OM of SR-8 that was complemented with mtrA and mtrB (3). Nonetheless, the contributions of MtrA and MtrB to metal reduction warrant further examination.
A role in Fe(III) reduction for one of the other downstream ORFs (orf1 to orf5) (Fig. 2) in pJQ14 must also be considered. While BLAST alignments did not reveal any matches of orf1 through orf5 to known electron carriers or redox-active proteins (Table 3), one or more of these ORFs could be responsible for the ability of pJQ14 to fully restore Fe(III) citrate reduction and partially restore FeOOH reduction, either directly or indirectly by facilitating proper localization of Fe(III) reductase components to the OM. Again, however, since these ORFs are intact in OMCB1, it seems that multiple ORFs, or a certain stretch of DNA in this region, must be present in cis to restore Fe(III) reduction.
There are several issues which complicate direct comparisons of OMCB1 and SR-8. First, OMCB1 was isolated by gene replacement, whereas SR-8 was isolated by Tn5 mutagenesis with the insertion site determined by sequencing DNA flanking one end of Tn5. Transposons can mediate polar effects; this was in fact the case for SR-8 (i.e., expression of mtrA and mtrB was also blocked) (3), and the deficiencies in Mn(IV) and Fe(III) reduction were similar to those for a previously reported mtrB mutant (2). In contrast, RT-PCR indicated that mtrA and mtrB are still expressed to some extent in OMCB1 (Fig. 3). Second, while Tn5-based systems are supposed to mediate simple insertions, TnphoA (derived from Tn5) mutagenesis to generate a fumarate reductase mutant of MR-1 resulted in a large (9- to 11-kb) genomic deletion such that the flanking genomic DNA was actually 8 kb away from the fumarate reductase gene (T. M. Mertins, J. M. Myers, and C. R. Myers, Abstr. 100th Gen. Meet. Am. Soc. Microbiol., abstr. H-11, 2000). The reported characterization of SR-8 was not sufficient to rule out such deletions or other unexpected events. Third, while OMCB1 was isolated directly from MR-1, SR-8 was isolated from MR-1R, a rifampin-resistant mutant. Two other independently isolated rifampin-resistant mutants of MR-1, MR-1A and MR-1B, have markedly depressed levels of menaquinone (33), which is required for Mn(IV) and Fe(III) reduction (23); MR-1A and MR-1B are considerably slower than MR-1 at utilizing electron acceptors that require menaquinone (33). It is therefore possible that the differences in Mn(IV) and Fe(III) reduction between SR-8 and MR-1R are smaller than those between SR-8 and MR-1.
The mechanisms of Fe(III) reduction and Mn(IV) reduction in MR-1 have some similarities but are clearly not the same. While OMCB1 is deficient in Fe(III) reduction compared to MR-1, it remains capable of reducing millimolar levels of Fe(III) citrate over a period of days. In contrast, OMCB1 remains a poor reducer of Mn(IV) even after 1 week of incubation. Knockout of omcB therefore compromises Mn(IV) reduction more than it compromises Fe(III) reduction. The results obtained with pJQ14 complementation highlight some other dissimilarities; despite an insignificant increase in OM cytochrome content, continued absence of the OmcB protein, and only a modest increase in Mn(IV) reduction, pJQ14 fully restored Fe(III) citrate reduction. This implies that there are at least some components unique to reduction of Mn(IV) compared to reduction of Fe(III). This is consistent with the previous isolation of mutants of MR-1 and of S. putrefaciens strain 200 that were deficient only in reduction of Mn(IV) or Fe(III) (6, 8, 30).
As for Mn(IV) reduction, it is likely that there are OM components that can mediate Fe(III) reduction (22). The results of studies with polynuclear Fe(III) complexes are consistent with Fe(III) reductase activity in the OM (10). Force microscopy demonstrated that the affinity between MR-1 cells and αFeOOH increased up to fivefold under anaerobic conditions, and force curve signatures were consistent with the presence of an Fe(III) reductase in the OM (17). A mutant of S. putrefaciens strain 200 in which a component needed for type II secretion is blocked is unable to reduce Mn(IV) and Fe(III) oxides (9); this is consistent with a role for OM components in metal reduction since type II secretion systems facilitate translocation of proteins to the outside surface of the OM. Complementation of OMCB1 with pJQ14 fully restored Fe(III) citrate reduction but restored FeOOH reduction to only 50% of the wild-type level (Fig. 11). This may indicate that the organization of OM components in OMCB1/pJQ14 is not optimal for the reduction of insoluble FeOOH but is sufficient for the reduction of soluble Fe(III) citrate. Alternatively, it may indicate that the Fe(III) citrate may have access to a second Fe(III) reductase, possibly in the CM. FeOOH would be unable to make contact with CM components. While some studies have suggested that there is potential for some Fe(III) reductase activity in the CM (10, 22), it is unclear if this activity is associated with anaerobic respiration. If there were both CM and OM Fe(III) reductases, they would both have to be downstream of menaquinone and CymA, since the absence of either results in a nearly total loss of Fe(III) reduction by cells (23, 33). Finding that the type II secretion mutants of strain 200 exhibit profound deficiencies in Fe(III) reduction (9) would suggest that there is a lack of a prominent Fe(III) reductase in the CM because type II secretion would not predictably be necessary for the localization of CM components.
Fe(III) reduction and Mn(IV) reduction in MR-1 are not totally distinct, however; e.g., both require menaquinone and CymA (23, 25, 33). Because Fe(II) is also a reductant of Mn(IV) (29), one cannot consider the two processes in total isolation. For example, the ability of pJQ14 to fully restore Fe(III) citrate reduction to OMCB1 may have contributed to the modest increase in Mn(IV) reduction afforded by pJQ14; i.e., the redox cycling of the 5.4 μM Fe in the defined medium could theoretically have supported a low rate of Mn(IV) reduction.
In summary, complementation of an omcB mutant requires omcB plus 19 kb of downstream DNA to fully restore Mn(IV) reduction and the full complement of OM cytochromes, including proper localization of OmcA and OmcB. Complementation with the most upstream 14.7 kb of this fragment results in restoration of Fe(III) citrate reduction, even though OmcB was not detected. Together, the data imply that OmcB is required for Mn(IV) reduction but not for Fe(III) reduction. Since DNA downstream of omcB in pVK21 and pJQ14 is also intact in the chromosome of the omcB mutant, this DNA must be present on a contiguous fragment (in cis) in order to complement the mutant. The complementation of Mn(IV) and Fe(III) reduction in OMCB1 is clearly complex, and extensive further examination of this region of DNA is required.
Acknowledgments
This work was supported by National Institutes of Health grant R01GM50786 to C.R.M.
The preliminary genomic sequence data was obtained from The Institute for Genomic Research website (http://www.tigr.org). Sequencing of S. putrefaciens MR-1 genomic DNA by The Institute for Genomic Research was accomplished with support from the Department of Energy.
REFERENCES
- 1.Armstrong, P. B., W. B. Lyons, and H. E. Gaudette. 1979. Application of formaldoxime colorimetric method for the determination of manganese in the pore water of anoxic estuarine sediments. Estuaries 2:198-201. [Google Scholar]
- 2.Beliaev, A. S., and D. A. Saffarini. 1998. Shewanella putrefaciens mtrB encodes an outer membrane protein required for Fe(III) and Mn(IV) reduction. J. Bacteriol. 180:6292-6297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beliaev, A. S., D. A. Saffarini, J. L. McLaughlin, and D. Hunnicutt. 2001. MtrC, an outer membrane decahaem c cytochrome required for metal reduction in Shewanella putrefaciens MR-1. Mol. Microbiol. 39:722-730. [DOI] [PubMed] [Google Scholar]
- 4.Brewer, P. G., and D. W. Spencer. 1971. Colorimetric determination of manganese in anoxic waters. Limnol. Oceanogr. 16:107-110. [Google Scholar]
- 5.Burdige, D. J., and K. H. Nealson. 1986. Chemical and microbiological studies of sulfide-mediated manganese reduction. Geomicrobiol. J. 4:361-387. [Google Scholar]
- 6.Burnes, B. S., M. J. Mulberry, and T. J. DiChristina. 1998. Design and application of two rapid screening techniques for isolation of Mn(IV) reduction-deficient mutants of Shewanella putrefaciens. Appl. Environ. Microbiol. 64:2716-2720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Lorenzo, V., M. Herrero, U. Jakubzik, and K. N. Timmis. 1990. Mini-Tn 5 transposon derivatives for insertional mutagenesis, promoter probing, and chromosomal insertion of cloned DNA in gram-negative eubacteria. J. Bacteriol. 172:6568-6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.DiChristina, T. J., and E. F. DeLong. 1994. Isolation of anaerobic respiratory mutants of Shewanella putrefaciens and genetic analysis of mutants deficient in anaerobic growth on Fe3+. J. Bacteriol. 176:1468-1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.DiChristina, T. J., C. M. Moore, and C. A. Haller. 2002. Dissimilatory Fe(III) and Mn(IV) reduction by Shewanella putrefaciens requires ferE, a homolog of the pulE (gspE) type II protein secretion gene. J. Bacteriol. 184:142-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dobbin, P. S., L. M. Requena Burmesiter, S. L. Heath, A. K. Powell, A. G. McEwan, and D. J. Richardson. 1996. The influence of chelating agents upon the dissimilatory reduction of Fe(III) by Shewanella putrefaciens. Part 2. Oxo- and hydroxo-bridged polynuclear Fe(III) complexes. BioMetals 9:291-301. [Google Scholar]
- 11.Gallegos, M.-T., R. Schleif, A. Bairoch, K. Hofmann, and J. L. Ramos. 1997. AraC/XylS family of transcriptional regulators. Microbiol. Mol. Biol. Rev. 61:393-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hopp, T. P., and K. R. Woods. 1981. Prediction of protein antigenic determinants from amino acid sequences. Proc. Natl. Acad. Sci. USA 78:3824-3828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kinder, S. A., J. L. Badger, G. O. Bryant, J. C. Pepe, and V. L. Miller. 1993. Cloning of the YenI restriction endonuclease and methyltransferase from Yersinia enterocolitica serotype O8 and construction of a transformable R− M+ mutant. Gene 136:271-275. [DOI] [PubMed] [Google Scholar]
- 14.Knauf, V. C., and E. W. Nester. 1982. Wide host range cloning vectors: cosmid clone bank of Agrobacterium Ti plasmids. Plasmid 8:45-54. [DOI] [PubMed] [Google Scholar]
- 15.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 16.Lovley, D. R., and E. J. P. Phillips. 1986. Organic matter mineralization with reduction of ferric iron in anaerobic sediments. Appl. Environ. Microbiol. 51:683-689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lower, S. K., M. F. J. Hochella, and T. J. Beveridge. 2001. Bacterial recognition of mineral surfaces: nanoscale interactions between Shewanella and α-FeOOH. Science 292:1360-1363. [DOI] [PubMed] [Google Scholar]
- 18.Maier, T. M., and C. R. Myers. 2001. Isolation and characterization of a Shewanella putrefaciens MR-1 electron transport regulator etrA mutant: reassessment of the role of EtrA. J. Bacteriol. 183:4918-4926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Myers, C. R., and M. L. P. Collins. 1986. Cell-cycle-specific oscillation in the composition of chromatophore membrane in Rhodospirillum rubrum. J. Bacteriol. 166:818-823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Myers, C. R., and J. M. Myers. 1992. Fumarate reductase is a soluble enzyme in anaerobically grown Shewanella putrefaciens MR-1. FEMS Microbiol. Lett. 98:13-20. [Google Scholar]
- 21.Myers, C. R., and J. M. Myers. 1992. Localization of cytochromes to the outer membrane of anaerobically grown Shewanella putrefaciens MR-1. J. Bacteriol. 174:3429-3438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Myers, C. R., and J. M. Myers. 1993. Ferric reductase is associated with the membranes of anaerobically grown Shewanella putrefaciens MR-1. FEMS Microbiol. Lett. 108:15-22. [Google Scholar]
- 23.Myers, C. R., and J. M. Myers. 1993. Role of menaquinone in the reduction of fumarate, nitrate, iron(III) and manganese(IV) by Shewanella putrefaciens MR-1. FEMS Microbiol. Lett. 114:215-222. [Google Scholar]
- 24.Myers, C. R., and J. M. Myers. 1994. Ferric iron reduction-linked growth yields of Shewanella putrefaciens MR-1. J. Appl. Bacteriol. 76:253-258. [DOI] [PubMed] [Google Scholar]
- 25.Myers, C. R., and J. M. Myers. 1997. Cloning and sequence of cymA, a gene encoding a tetraheme cytochrome c required for reduction of iron(III), fumarate, and nitrate by Shewanella putrefaciens MR-1. J. Bacteriol. 179:1143-1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Myers, C. R., and J. M. Myers. 1997. Isolation and characterization of a transposon mutant of Shewanella putrefaciens MR-1 deficient in fumarate reductase. Lett. Appl. Microbiol. 25:162-168. [DOI] [PubMed] [Google Scholar]
- 27.Myers, C. R., and J. M. Myers. 1997. Outer membrane cytochromes of Shewanella putrefaciens MR-1: spectral analysis, and purification of the 83-kDa c-type cytochrome. Biochim. Biophys. Acta 1326:307-318. [DOI] [PubMed] [Google Scholar]
- 28.Myers, C. R., and K. H. Nealson. 1988. Bacterial manganese reduction and growth with manganese oxide as the sole electron acceptor. Science 240:1319-1321. [DOI] [PubMed] [Google Scholar]
- 29.Myers, C. R., and K. H. Nealson. 1988. Microbial reduction of manganese oxides: interactions with iron and sulfur. Geochim. Cosmochim. Acta 52:2727-2732. [Google Scholar]
- 30.Myers, C. R., and K. H. Nealson. 1990. Iron mineralization by bacteria: metabolic coupling of iron reduction to cell metabolism in Alteromonas putrefaciens strain MR-1, p. 131-149. In R. B. Frankel and R. P. Blakemore (ed.), Iron biominerals. Plenum Press, New York, N.Y.
- 31.Myers, C. R., and K. H. Nealson. 1990. Respiration-linked proton translocation coupled to anaerobic reduction of manganese(IV) and iron(III) in Shewanella putrefaciens MR-1. J. Bacteriol. 172:6232-6238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Myers, J. M., and C. R. Myers. 1998. Isolation and sequence of omcA, a gene encoding a decaheme outer membrane cytochrome c of Shewanella putrefaciens MR-1, and detection of omcA homologs in other strains of S. putrefaciens. Biochim. Biophys. Acta 1373:237-251. [DOI] [PubMed] [Google Scholar]
- 33.Myers, J. M., and C. R. Myers. 2000. Role of the tetraheme cytochrome CymA in anaerobic electron transport in cells of Shewanella putrefaciens MR-1 with normal levels of menaquinone. J. Bacteriol. 182:67-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Myers, J. M., and C. R. Myers. 2001. Role for outer membrane cytochromes OmcA and OmcB of Shewanella putrefaciens MR-1 in reduction of manganese dioxide. Appl. Environ. Microbiol. 67:260-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ochman, H., A. S. Gerber, and D. L. Hartl. 1988. Genetic applications of an inverse polymerase chain reaction. Genetics 120:621-623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pradel, E., N. Guiso, and C. Locht. 1998. Identification of AlcR, an AraC-type regulator of alcaligin siderophore synthesis in Bordetella bronchiseptica and Bordetella pertussis. J. Bacteriol. 180:871-880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Quandt, J., and M. F. Hynes. 1993. Versatile suicide vectors which allow direct selection for gene replacement in Gram-negative bacteria. Gene 127:15-21. [DOI] [PubMed] [Google Scholar]
- 38.Rhee, S., R. G. Martin, J. L. Rosner, and D. R. Davies. 1998. A novel DNA-binding motif in MarA: the first structure for an AraC family transcriptional regulator. Proc. Natl. Acad. Sci. USA 95:10413-10418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sambrook, J., E. F. Fritsch, and T. Maniatis, ed. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 40.Simon, R., U. Priefer, and A. Pühler. 1983. A broad host-range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram-negative bacteria. Bio/Technology 1:37-45. [Google Scholar]
- 41.Thomas, P. E., D. Ryan, and W. Levin. 1976. An improved staining procedure for the detection of the peroxidase activity of cytochrome P-450 on sodium dodecyl sulfate polyacrylamide gels. Anal. Biochem. 75:168-176. [DOI] [PubMed] [Google Scholar]