Abstract
Bacillus cereus, a dairy-associated toxigenic bacterium, readily forms biofilms on various surfaces and was used to gain a better understanding of biofilm development by gram-positive aerobic rods. B. cereus DL5 was shown to readily adapt to an attached mode of growth, with dense biofilm structures developing within 18 h after inoculation when glass wool was used as a surface. Two-dimensional gel electrophoresis (2DE) revealed distinct and reproducible phenotypic differences between 2- and 18-h-old biofilm and planktonic cells (grown both in the presence and in the absence of glass wool). Whereas the 2-h-old biofilm proteome indicated expression of 15 unique proteins, the 18-h-old biofilm proteome contained 7 uniquely expressed proteins. Differences between the microcolony (2-h) proteome and the more developed biofilm (18-h) proteome were largely due to up- and down-regulation of the expression of a multitude of proteins. Selected protein spots excised from 2DE gels were subjected to N-terminal sequencing and identified with high confidence. Among the proteins were catabolic ornithine carbamoyltransferase and l-lactate dehydrogenase. Interestingly, increased levels of YhbH, a member of the sigma 54 modulation protein family which is strongly induced in response to environmental stresses and energy depletion via both σB and σH, could be observed within 2 h in both attached cells and planktonic cultures growing in the presence of glass wool, indicating that this protein plays an important role in regulation of the biofilm phenotype. Distinct band differences were also found between the extracellular proteins of 18-h-old cultures grown in the presence and in the absence of glass wool.
The development of multicellular aggregates, also known as biofilms, is a common phenomenon in aqueous environments and occurs through bacterial adhesion at solid-liquid interfaces (43, 44). Biofilm development is widely believed to be initiated by bacteria sensing certain surface-associated parameters that trigger the transition from a planktonic to a biofilm mode of growth (6, 36). This involves a number of changes in gene regulation that cause the adhering cells to become phenotypically (3, 7) and metabolically (6) distinct from their planktonic counterparts. The altered phenotype is believed to be responsible for the distinct properties of bacteria in biofilms, most notably their increased resistance to antimicrobial agents (38). The complex biofilm architecture also provides an opportunity for metabolic cooperation, and niches are formed within the spatially well-organized systems. Consequently, the bacteria are exposed to an array of distinct physicochemical conditions within a biofilm that can result in differential gene expression (6, 27, 28). In this regard, important factors include cell density, as well as the extent and duration of cell-to-cell contact, the concentrations of diffusible substances and/or the ability to establish concentration gradients of diffusible substances, and oxygen availability.
Although the initial stages of biofilm development have been studied thoroughly for gram-negative rods (28), for gram-positive cocci (20, 21), and also for Candida (1), many of the underlying regulatory processes are still not clearly understood. Furthermore, a well-described model of biofilm development for gram-positive rods is lacking. To establish such a model, we have focused our attention on Bacillus cereus due to its distinct ability to adhere to and form biofilms on stainless steel (31) and glass (19). B. cereus is a well-known enterotoxin-producing food-poisoning organism (12, 18) and is regarded as one of the most important organisms impairing the keeping quality of pasteurized milk and milk products (2, 4, 31). Biofilms of this bacterium may serve as a chronic source of microbial contamination, thereby compromising food quality.
Biofilms have been studied predominantly in stagnant batch culture by using microtiter plates (29) or under flow conditions by using various flow cells (48). Whereas these systems are suited for genetic and microscopic studies, they do not yield sufficient biomass for proteomic studies. Alternative systems have been proposed to increase the yield of biofilm biomass; these systems include silicon tubing (37), gravel chips in a chemostat (46), and glass wool (26, 39). Glass wool provides a large surface-to-volume ratio (39) and allows separation of the biofilm biomass from the surrounding planktonic cells for further characterization.
In this paper, we describe the phenotypic changes that take place when planktonic cells of B. cereus DL5 make the transition to the biofilm mode of growth. High-resolution two-dimensional gel electrophoresis (2DE) was used to demonstrate phenotypic differences between 2- and 18-h-old biofilm B. cereus DL5 cells and planktonic cells grown in the presence and absence of glass wool. Planktonic cells grown in the presence of glass wool are referred to below as PGW cells. Comparative analysis of the proteomes indicated that there were distinct differences between the protein profiles. Eight protein spots which varied reproducibly in cellular concentration were selected and subjected to N-terminal protein sequencing.
MATERIALS AND METHODS
Bacterial strain and culture medium.
All experiments were performed by using B. cereus DL5, which was isolated previously from the alkaline wash solution in a dairy factory (19). B. cereus DL5 was routinely cultivated in Standard One Nutrient-like broth (SONLB) (18).
Microscopy of biofilm development and calculation of cell volumes.
To monitor cell attachment and biofilm development, 100 ml of SONLB containing 0.5 g of glass wool (mean diameter, ca. 15 μm; total surface area, 650 cm2; Merck, Darmstadt, Germany) was inoculated with 1 ml of an overnight culture of B. cereus DL5 (final inoculum density, 106 CFU · ml−1) and grown with agitation (200 rpm) at 37°C. Glass wool was removed after 2, 18, 24, and 42 h of incubation, stained with 0.02% (wt/vol) toluidine blue, and then viewed by bright-field microscopy with a Zeiss Axioscope light microscope (Carl Zeiss, Oberkochen, Germany). Images were captured with a COHU monochrome charge-coupled device camera (RS-170; Cohn Inc., San Diego, Calif.). The cell volumes (V) of 2- and 18-h-old planktonic, PGW, and biofilm cells were calculated by using the following formula: V = 4/3Πr3 + Πr2(l − 2r) where r is the radius and l is the cell length. The lengths and widths of at least 50 individual B. cereus DL5 cells were measured in each instance. The cell volumes were subsequently compared to each other by multifactor analysis of variance (Statgraphics 7.0) at the 95% confidence level.
SDS-PAGE analysis of extracellular proteins.
Flasks containing 100 ml of SONLB with and without glass wool were inoculated with 1 ml of an overnight culture of B. cereus DL5 (final inoculum density, 106 CFU · ml−1) and grown with agitation (200 rpm) at 37°C. Following incubation for 2 and 18 h, respectively, bacterial cells were removed from each culture supernatant by centrifugation at 12,000 × g for 10 min. Extracellular proteins were precipitated by mixing the culture supernatant with 4 ml of 50% trichloroacetic acid (Merck) and incubating the preparations on ice for 30 min. The precipitate was collected by centrifugation, washed three times with ice-cold 70% ethanol, dried, and dissolved in an appropriate volume of sample solution (0.5 M Tris-HCl [pH 6.8], 5% 2-mercaptoethanol, 10% glycerol, 2% sodium dodecyl sulfate [SDS]). SDS-polyacrylamide gel electrophoresis (PAGE) was performed with 12% polyacrylamide gels by the method of Laemmli (17). The BENCHMARK prestained protein ladder (GIBCO BRL, Vienna, Austria) was used as a molecular mass marker covering the 220- to 15-kDa range. Following electrophoresis, proteins were visualized by silver diamine staining (9), and the gels were scanned with an Agfa T1200 scanner.
Whole-cell protein extraction for proteome analysis.
Flasks containing 100 ml of SONLB with and without glass wool were inoculated with an overnight culture of B. cereus DL5 and cultured for 2 and 18 h, respectively, as described above. Planktonic cells were concentrated from cultures grown in the absence of glass wool by centrifugation at 12,000 × g for 10 min. Flasks containing glass wool were used to concentrate biofilm and PGW cells as follow. Following aseptic removal of the glass wool, PGW cells were recovered from the culture medium by centrifugation at 12,000 × g for 10 min. The recovered glass wool was washed twice in 0.2 M sodium phosphate buffer (pH 6.9) before it was transferred to a sterile flask containing 45 g of glass beads (diameter, 6 mm) and 5 ml of 10 mM Tris-HCl (pH 7.4). The attached cells were released from the glass wool into the liquid phase by vigorous shaking for 10 min. Complete removal of biofilm cells was verified by microscopic inspection of the glass wool. The liquid phase was recovered, and the biofilm cells were concentrated by centrifugation as described above. Whole-cell proteins were extracted from each growth phase as previously described (26). Prior to electrophoresis, the protein extracts were concentrated (45) and solubilized in 20 μl of lysis buffer consisting of 9 M urea, 65 mM dithioerythritol, 65 mM 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate (CHAPS), and 5% ampholytes (pH 3 to 10; Amersham-Pharmacia Biotech, Uppsala, Sweden).
2DE.
For 2DE nonlinear isoelectric focusing tube gels containing 6.7% polyacrylamide, 15 M urea, and 2% ampholytes (Ampholine pH 5 to 7 and Pharmalyte pH 3 to 10 at a 4:1 ratio; Amersham-Pharmacia Biotech) were cast in 15-cm-long glass tubes (diameter, 1 mm). Gels were preelectrophoresed at 200 V for 15 min, at 300 V for 30 min, and at 400 V for 30 min. The protein content of each extract was determined with a Coomassie Plus protein assay reagent (Pierce) and standardized to ca. 10 μg·μl−1. After prefocusing, 20 μl of protein extract suspended in 5 μl of sample buffer (9.5 M urea, 2% Nonidet P-40, 2% ampholytes, 5% 2-mercaptoethanol) was loaded per gel. Gels were electrophoresed at 400 V for 16 h and then for an additional 1 h at 800 V. After equilibration (62.5 mM Tris-HCl, 2% SDS, 10% glycerol, 5% 2-mercaptoethanol; pH 6.8) for 20 min, the isoelectric focusing gels were embedded in a uniform SDS-10% polyacrylamide separating gel. Electrophoresis was performed at 5 W for 15 min and then at 10 W for 4.75 h. A constant temperature of 18°C was maintained during electrophoresis. After electrophoresis, proteins were visualized by silver diamine staining (9) to allow comparative analysis. For N-terminal sequencing of selected protein spots, gels were stained with a 0.3% Coomassie blue R250 solution for 2 h and destained overnight with a destaining solution (25% methanol, 10% acetic acid). The pH gradient (pH 4 to 7) was determined experimentally by using the 2-D SDS-PAGE standard from Bio-Rad (catalog no. 161-0320). The molecular masses were determined by electrophoresis of a premixed molecular mass marker covering the 14- to 98-kDa range (Roche Diagnostics GmbH, Mannheim, Germany) in the second dimension.
Analysis of proteome patterns.
Whole-cell protein extracts prepared from 2- and 18-h-old planktonic, PGW, and biofilm cells were analyzed on at least three separate occasions, and the corresponding gels were electrophoresed in triplicate. Gel images were obtained with an Agfa T1200 scanner, resized, and matched on a grid similar to the system of Pederson et al. (30). The number of protein spots per gel was determined, and distinct differences between the patterns were noted. The protein spot pattern of the 2-h planktonic proteome was used as the standard for spot matching. Thus, the intensities of the 2-h planktonic spots were considered the reference intensities (0) and were compared to the intensities of the spots of the other proteomes to obtain information regarding up- and down-regulation of specific protein spots (39).
N-terminal amino acid sequencing and protein identification.
The regions of the Coomassie blue-stained gels containing protein spots of interest were excised and electroblotted onto Immobilon-P polyvinylidene difluoride membranes (Millipore, Freehold, N.J.) by using a 0.12 M Tris-0.04 M glycine buffer. The N-terminal sequence was determined by automated Edman degradation with a Procise 492 automatic sequencer (Applied Biosystems, Courtabeuf, France). Searches for homologous amino acid sequences were performed by screening the B. cereus database (http://wit.Integratedgenomics.com/Public/IG_Release.html) (last accessed in September 2001), the SubtiList database (http://genolist.pasteur.fr/SubtiList/genome.cgi), and the nonredundant database at The National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/) with the BLASTP and TBLASTN programs.
RESULTS
Biofilm development on glass wool.
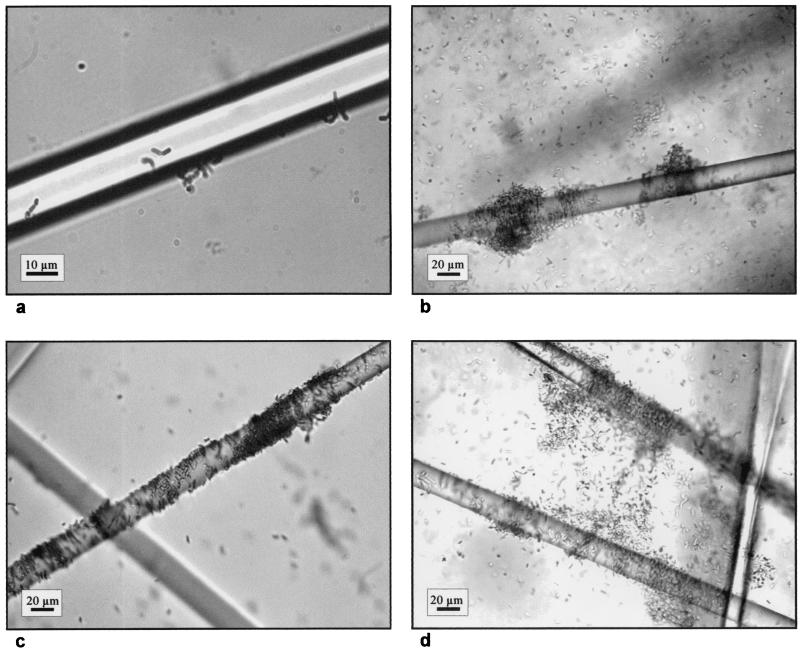
Biofilm development by B. cereus DL5 cells on glass wool in SONLB was monitored at various times by bright-field microscopy. Cells attached to glass wool were observed immediately after inoculation, and microcolonies were observed after 2 h (Fig. 1a). Dense localized biofilm structures formed within 18 h (Fig. 1b and c) and became more dense after 24 h, and there were thick, complex biofilm structures after 42 h of incubation (Fig. 1d). These results confirmed and extended our previous findings by indicating that copious amounts of biofilm could be obtained after 18 h of growth in the presence of glass wool (26).
FIG. 1.
Photomicrographs illustrating the development of B. cereus DL5 biofilms on glass wool in SONLB at 2 h (a), 18 h (b and c), and 42 h (d) after inoculation.
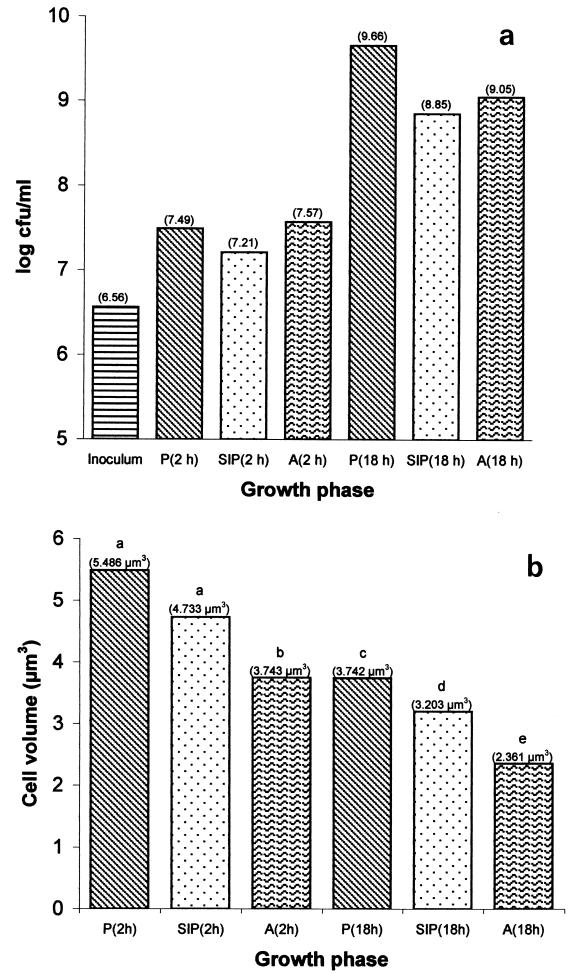
The combined yield of PGW cells and biofilm biomass after 2 h was slightly higher than the yield for a parallel planktonic culture inoculated at the same cell density. After 18 h of culture, the PGW population was much smaller than the planktonic population. The planktonic population was 2.5-fold larger than the combined PGW-biofilm population (Fig. 2a). Since the initial inocula were identical, the decrease in the yield of PGW biomass could have been due either to an adaptational event following reversible attachment or to impairment of the growth of the suspended cells by a biofilm exudate. Interestingly, very few spores were detected in any of the 18-h populations; the highest proportion (0.006%) was observed in the biofilm (data not shown). The dissolved oxygen present in the planktonic and PGW phases was measured immediately after inoculation and after 2 and 18 h of culturing by using a YSI model 85 oxygen, conductivity, salinity, and temperature system equipped with a gold cathode (YSI Inc., Yellow Springs, Ohio). After 2 h of culturing, the dissolved oxygen concentrations in the cultures were nearly identical (3.63 mg · liter−1 without glass wool and 3.73 mg · liter−1 with glass wool), as were the pH values (6.94 and 6.99, respectively). After 18 h, however, the dissolved oxygen concentrations were very low in both cultures (0.03 mg · liter−1 without glass wool and 0.02 mg · liter−1 with glass wool), and the pH values were still nearly identical (6.09 and 6.15, respectively).
FIG. 2.
Culturable counts (expressed as log10 CFU per milliliter of culture) (a) and cell volumes (expressed as cubic micrometers) (b) of B. cereus DL5 planktonic cells (P), PGW cells (SIP), and biofilm cells (A) grown for 2 and 18 h in SONLB at 37°C. Different letters indicate that differences are statistically significant (P < 0.05).
The cell volume of bacteria is known to increase exponentially with the growth rate, so a decrease in the cell volume indicates that there is a decrease in the growth rate (24). This relationship between growth rate and cell size also is true for biofilms of Acinetobacter (15). Consequently, the cell volumes of 2- and 18-h-old planktonic, PGW, and biofilm cells were calculated and compared to each other by using multifactor analysis of variance at the 95% confidence level (Fig. 2b). After 2 h of culturing, the cell volume of PGW cells was slightly smaller than that of planktonic cells, but the difference was not statistically significant (P > 0.05). The cell volume of biofilm cells was, however, significantly smaller (P < 0.05) than the cell volumes of both planktonic and PGW cells, indicating that the growth rate was lower. After 18 h, the cell volume of PGW cells was significantly smaller (P < 0.05) than that of planktonic cells, and the cell volume of biofilm cells was significantly smaller (P < 0.05) than the cell volumes of both planktonic and PGW cells. The biofilm and PGW cells may have been growing at a lower rate than the planktonic cells without glass wool. However, the biofilm yield was greater than the PGW yield (Fig. 2a), suggesting that the proportion of PGW cells attached was larger than the proportion of biofilm cells detaching. In this respect, B. cereus may be different from Pseudomonas aeruginosa; for the latter organism most of the cells occur in the suspended phase and only ca. 15% of the cells are attached (39).
One-dimensional gel electrophoresis of extracellular proteins.
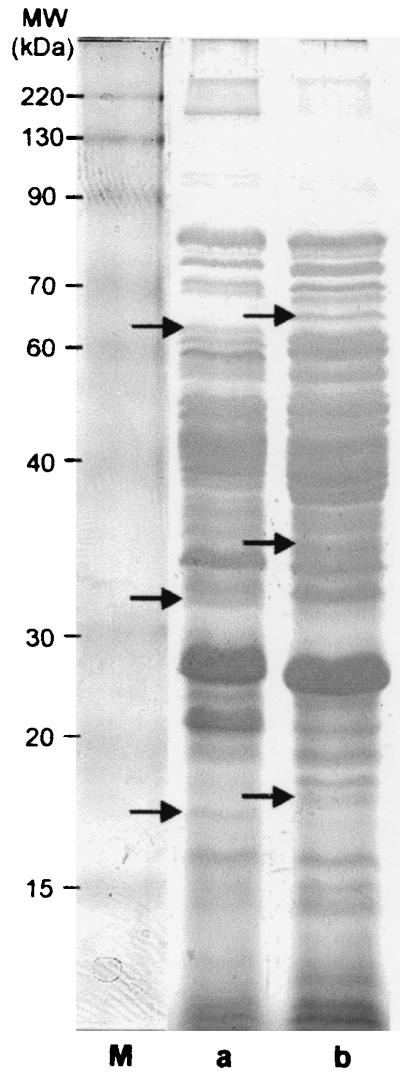
The extracellular proteins of 2- and 18-h-old B. cereus DL5 cultures grown in the presence and absence of glass wool were compared after separation by SDS-PAGE and silver staining. Only a few proteins were excreted after 2 h, and no difference between the protein profiles was observed (data not shown). However, after 18 h of growth, distinct band differences between the extracellular proteins of cultures grown in the presence and in the absence of glass wool could be identified (Fig. 3). This variability in the expression of extracellular proteins may indicate that the secretome of B. cereus is regulated differently in planktonic cells than in PGW and/or biofilm cells.
FIG. 3.
Extracellular protein profiles of 18-h-old planktonic cells grown in the absence of glass wool (a) and combined PGW-biofilm cells (b) of B. cereus DL5 following separation by one-dimensional SDS-12% PAGE and silver staining. A BENCHMARK prestained protein ladder (GIBCO BRL) was used as the molecular weight (MW) marker (lane M). Unique bands are indicated by arrows.
Proteome profile analysis.
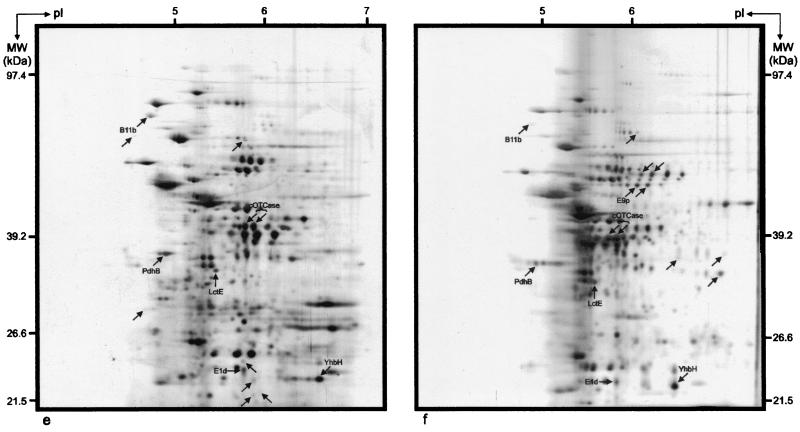
To further characterize protein expression differences among the planktonic, PGW, and biofilm cells, high-resolution 2DE of whole-cell protein extracts was performed. The reproducibility of separation of the total proteins was high, and 345 distinct protein spots were observed in the pH range from 4 to 7 after silver staining. By matching and comparing the 2DE proteomes, proteins were grouped according to their expression characteristics (Table 1). Representative examples of planktonic, PGW, and biofilm proteomes are shown in Fig. 4.
TABLE 1.
Protein expression in B. cereus DL5 planktonic, PGW, and biofilm cells following 2 and 18 h of culturing in the absence and presence of glass wool
| Protein expression | No. of spots | % of total spots (n = 345) |
|---|---|---|
| Proteins unique to a specific growth phase | ||
| Planktonic cells (2 h) | 1 | 0.3 |
| PGW cells (2 h) | 2 | 0.6 |
| Biofilm cells (2 h) | 15 | 4.4 |
| Planktonic cells (18 h) | 1 | 0.3 |
| PGW cells (18 h) | 7 | 2.3 |
| Biofilm cells (18 h) | 7 | 2.0 |
| Proteins up-regulated in a specific phase compared with the levels of the corresponding spot in other phases | ||
| Planktonic cells (2 h) | 3 | 0.9 |
| PGW cells (2 h) | 10 | 2.9 |
| Biofilm cells (2 h) | 4 | 1.2 |
| Planktonic cells (18 h) | 7 | 2.0 |
| PGW cells (18 h) | 30 | 8.7 |
| Biofilm cells (18 h) | 19 | 5.5 |
| Proteins present in a specific phase regardless of time (2 h or 18 h) | ||
| Planktonic cells | 0 | 0 |
| PGW cells | 1 | 0.3 |
| Biofilm cells | 5 | 1.5 |
| Proteins present in all phases and always at the same level (no up- or down-regulation) | 35 | 10.1 |
| Proteins present in planktonic and PGW cells and absent in biofilm cells | 11 | 3.2 |
| Proteins present in PGW and biofilm cells and absent in planktonic cells | 0 | 0 |
| Proteins present in planktonic and biofilm cells and absent in PGW cells | 0 | 0 |
| After 2 h present only in biofilm cells; after 18 h present in PGW and biofilm cells | 2 | 0.6 |
| After 2 h present only in planktonic cells; after 18 h present in planktonic and PGW cells | 1 | 0.4 |
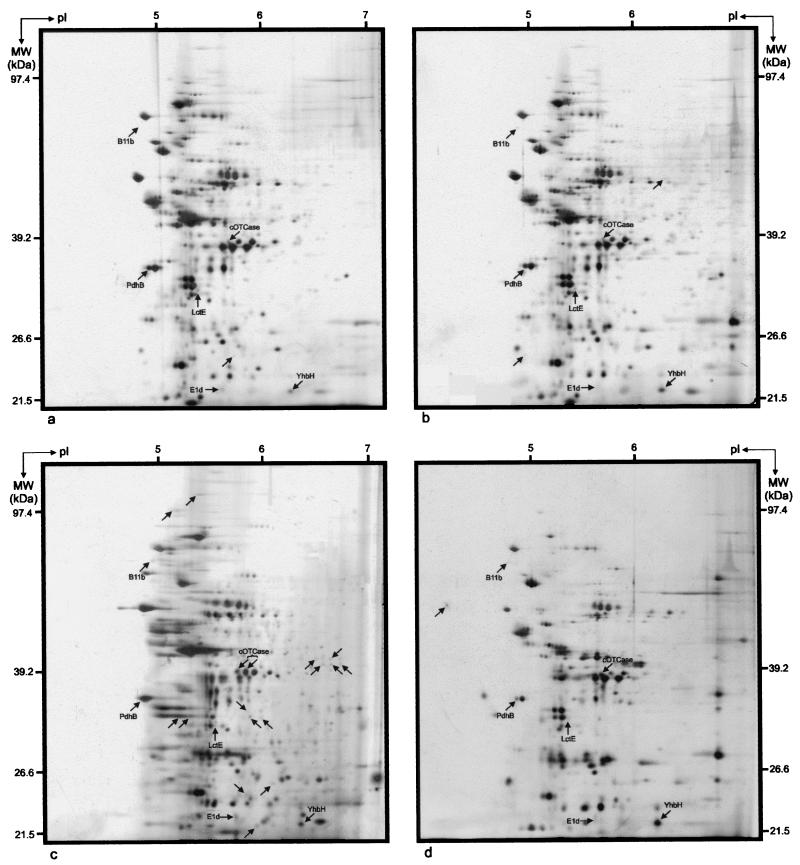
FIG. 4.
2DE proteome patterns for 2-h-old B. cereus DL5 planktonic cells (a), PGW cells (b), and biofilm cells (c) and 18-h-old B. cereus DL5 planktonic cells (d), PGW cells (e), and biofilm cells (f). Unique protein spots are indicated by arrows. The horizontal axes represent pIs of the isoelectric focusing gradients, and the vertical axes represent molecular masses (MW), based on comigration of Bio-Rad 2-D SDS-PAGE standards (Roche Diagnostics GmbH).
Comparison of the 2DE protein patterns obtained after 2 h of growth indicated that 15 protein spots were uniquely expressed in the biofilm mode of growth (i.e., they were not present or could not be detected by silver staining in the other proteomes) (Table 1). This finding supports several recent observations that a number of functions are required de novo in the initial stages of biofilm development (28). The low concentrations of these spots, however, prevented further characterization of them by N-terminal protein sequencing. The planktonic and PGW proteomes produced one and two unique protein spots, respectively (Table 1). In addition to these uniquely expressed proteins, both the attached and PGW populations produced a number of proteins whose levels of expression were up-regulated compared with the planktonic levels. Comparison of the 2DE protein patterns obtained after 18 h of growth indicated the presence of a single unique protein in the planktonic proteome, while the biofilm and PGW proteomes each contained seven unique proteins. Furthermore, 19 proteins were up-regulated in the biofilm proteome, while 30 proteins were found to be up-regulated in the PGW proteome (Table 1). The latter finding is in contrast to data for P. aeruginosa, in which a large number of surface-influenced planktonic proteins are down-regulated compared to planktonically and biofilm-grown cells (39). Following comparison of these 18-h-old protein patterns with those obtained after 2 h of growth, five unique biofilm protein spots and one unique PGW protein spot were identified. No uniquely expressed proteins were observed in the planktonic cells (Table 1). The protein expression profiles also showed that 10% of the proteins were always expressed at the same level in all phases (Table 1). It was also evident that the planktonic and PGW phases had 11 proteins in common which were absent in the biofilm (Table 1). In contrast, neither the PGW and biofilm phases nor the planktonic and biofilm phases displayed uniquely shared proteins (Table 1).
Cumulatively, the results described above indicated that the biofilm proteome differed from the planktonic and PGW proteomes. The microcolony (2-h) proteome differed from the more developed biofilm (18-h) proteome, and the observed difference was not due to a single factor; rather, it was due to a multitude of up- and down-regulated proteins, and posttranslational modification of proteins may also have been involved. Furthermore, the PGW proteome showed more resemblance to the planktonic phenotype than to the biofilm phenotype after 2 h of growth, and the PGW growth phase may be regarded as a transitional state between the true planktonic and biofilm growth phases. However, after 18 h of growth, the PGW phase clearly constitutes a distinct mode of growth.
Identities of differentially expressed proteins.
Eight protein spots were selected for N-terminal protein sequencing. These spots (Table 2) were selected because they varied reproducibly in cellular concentration as a consequence of changes in the growth conditions. The amino acid sequences obtained were subjected to database searches as described in Materials and Methods. The results revealed that five spots displayed 100% homology with previously identified proteins. The molecular mass and pI values obtained by 2DE agreed well with the database values. Protein spot E1d displayed 100% homology with a hypothetical cytosolic protein of B. cereus and 72% amino acid identity with a hypothetical protein (PA2575) of P. aeruginosa. The N-terminal sequence of protein spots B11b and E9p did not display significant similarity to proteins in the databases. The results of this analysis are summarized in Table 3.
TABLE 2.
Growth phase-related differences in the levels of expression of proteins selected for N-terminal protein sequencing
| Protein spot | Level of expression
|
|||||
|---|---|---|---|---|---|---|
| Planktonic cells (2 h) | PGW cells (2 h) | Biofilm cells (2 h) | Planktonic cells (18 h) | PGW cells (18 h) | Biofilm cells (18 h) | |
| B5a | 0a | 0 | 0 | −2 | −1 | −1 |
| B11b | 0 | 1 | 2 | 0 | 2 | 0 |
| D5h | 0 | 0 | 0 | −1 | 3 | 3 |
| E1d | 0 | 0 | 0 | 0 | 1 | 1 |
| E7g | Ab | A | 0 | A | 0 | 0 |
| E7h | 0 | 0 | 3 | 0 | 3 | 3 |
| E9p | A | A | A | A | A | Uc |
| G1b | 0 | 1 | 2 | 2 | 3 | 3 |
A value of 0 indicates a level of synthesis similar to that of the corresponding spot in the 2-h planktonic growth phase (reference spot intensity). A positive value indicates up-regulation, and a negative value indicates repression or down-regulation.
A, protein spot is absent.
U, protein spot is unique to the growth phase.
TABLE 3.
Summary of the proteins identified from 2DE gels of B. cereus DL5 whole-cell protein extracts
| Protein spot | Protein identity | Function | N-terminal sequence | Estimated from gel
|
Calculated from sequence
|
Accession no.
|
|||
|---|---|---|---|---|---|---|---|---|---|
| Size (kDa) | pI | Size (kDa) | pI | B. cereus databasea | SWISS-PROT protein database | ||||
| B5a | Pyruvate dehydrogenase E1 component, beta subunit (PdbB) | Pyruvate + lipoamide → S-acetyl-dihydro-lipoamide + CO2 | QMTMIQAITD | 36 | 4.90 | 35.23 | 4.75 | RZC01921 | P21882 |
| B11b | Unknown | MNVKRHM | 82 | 4.80 | |||||
| D5h | l-Lactate dehydrogenase (LctE) | Pyruvate + NADH → l-lactate + NAD+ | MKKGINRVVL | 34 | 5.50 | 34.79 | 4.99 | RZC05932 | P13714 |
| E1d | Hypothetical cytosolic protein | Unknown | AKDFYSAIEDRRS | 24 | 5.65 | 23.07 | 5.21 | RZC03248 | A83323 |
| E7g | cOTCase | l-Citrulline + Pi → carbamoyl phosphate + ornithine | MLMTRPNLKGRS | 39 | 5.70 | 37.46 | 5.35 | RZC04697 | O86132 |
| E7h | cOTCase | l-Citrulline + Pi → carbamoyl phosphate + ornithine | MLMTRPNLKGRS | 39 | 5.80 | 37.46 | 5.35 | RZC04697 | O86132 |
| E9p | Unknown | MPPVVI | 54 | 6.10 | |||||
| G1b | YhbH light-repressed protein A | Unknown | MKFNI | 23 | 6.20 | 21.08 | 5.63 | RZC04452 | P28368 |
Accession number obtained from the B. cereus database website (http://wit.Integratedgenomics.com/Public/IG_Release.html).
Accession number obtained from the SWISS-PROT protein database website (http://www.expasy.ch/sprot/).
The amino acid sequence derived from protein spot D5h corresponded to the sequence of the l-lactate dehydrogenase (LctE) enzyme. This enzyme catalyzes the reduction of pyruvate to lactate in a single step with simultaneous oxidation of one molecule of NADH to NAD+ (23). Synthesis of this enzyme is drastically increased under anaerobic conditions in the absence of an alternative electron acceptor. LctE was up-regulated in 18-h-old biofilm and PGW cells but down-regulated in 18-h-old planktonic cells (Table 2). The 18-h-old planktonic and PGW phases, however, contained nearly the same levels of dissolved oxygen. It has been reported that oxygen transfer is limited in deeper layers of aerobic biofilms (47) and that gene expression profiles within a biofilm are altered due to oxygen limitation (34). The 18-h-old biofilm could therefore be regarded as anoxic. Biofilm cells experiencing anoxic conditions could be expected to switch to fermentative metabolism, producing more LctE. The high level of LctE expression observed in 18-h-old PGW cells can be attributed either to detached biofilm cells still displaying the biofilm phenotype or to up-regulation by a signal substance excreted by the biofilm.
Protein spots E7g and E7h had the same molecular mass, but their pI values differed. Whereas spot E7h was detected in all of the 2DE protein patterns, spot E7g was identified only in the proteomes of biofilm and 18-h-old PGW cells. The level of expression of spot E7h in these phases was, however, drastically up-regulated compared to that of spot E7g (Table 2). The amino acid sequences derived from both protein gel spots corresponded to a single protein, catabolic ornithine carbamoyltransferase (cOTCase). To determine whether the two isoforms may represent posttranslationally modified versions of the same protein, the amino acid sequence of cOTCase was scanned by using the ScanProsite function available at the ExPASy Molecular Biology server (http://www.expasy.ch/prosite/). The results obtained indicated that the protein contained 12 potential phosphorylation sites for four different kinases. Thus, the spots may represent differentially phosphorylated versions of the same protein. cOTCase is an integral enzyme in the arginine deiminase (ADI) pathway, and Bacillus can grow anaerobically on arginine via the ADI pathway (5, 22). As soon as 2 h after inoculation, biofilm cells strongly expressed cOTCase. This may be indicative of oxygen depletion in microcolonies, or alternatively, it may indicate that the attached cells were preparing for growth within a biofilm before the conditions became anoxic. After 18 h of growth, cOTCase was expressed at very high levels in both PGW and biofilm cells, while the level remained at the early-exponential-phase level in planktonic cells. Similar to LctE, the increased expression of cOTCase in PGW cells can be attributed either to detached biofilm cells still displaying the biofilm phenotype or to up-regulation by a signal substance excreted by the biofilm.
The amino acid sequence derived from protein spot B5a was determined to be the sequence of pyruvate dehydrogenase E1 (beta subunit). This enzyme forms part of the pyruvate dehydrogenase (PDH) complex, which catalyzes the reaction that produces acetyl coenzyme A from pyruvate, yielding 1 mol of NADH. This is a crucial step between glycolysis and the tricarboxylic acid cycle, where acetyl coenzyme A can be further oxidized to generate energy and intermediates for anabolic reactions (40). Under anaerobic conditions, Bacillus subtilis performs mixed acid-butanediol fermentation in which pyruvate is oxidatively decarboxylated by the PDH complex (25). The PDH complex is present at the same level in planktonic, PGW, and biofilm cells after 2 h of growth, but the levels decrease variably in the three phases after 18 h (Table 2). This may indicate that there is a decrease in the synthesis of this housekeeping protein that could form part of a general shutdown of proteins required primarily during the exponential phase.
The amino acid sequence derived from protein spot G1b corresponded to YhbH light-repressed protein A of B. cereus. No information regarding the function of this protein in B. cereus is available yet. YhbH displays 72% amino acid sequence similarity to the general stress protein YvyD of B. subtilis (11) and groups with the sigma 54 modulation protein family. Expression of YvyD is strongly induced in response to different environmental stresses and energy depletion resulting from carbon, phosphate, or oxygen starvation (sigma B dependent), as well as in response to amino acid or nitrogen starvation (sigma H dependent) (10). Sigma H is generally used for transcription of many genes expressed during the transition from exponential growth to the stationary phase, with most of the products functioning in the sporulation process and in competence development (8, 11). YhbH was up-regulated in both biofilm and PGW cells within 2 h, and after 18 h its expression level was higher in these populations than in planktonic cells. This may indicate that the cells were sensing change and consequently preparing for the altered modes of growth.
DISCUSSION
The results obtained in this investigation indicate that B. cereus readily adapts to an attached mode of growth as microcolonies could be observed on glass wool within 2 h after inoculation. The 2DE protein patterns derived from these microcolonies indicated that 15 unique proteins were expressed compared to the patterns for planktonic and PGW proteomes. The smaller cell volume of the biofilm cells may indicate that the growth rate was lower. This may have been due to induction of an initial lag phase during which the cells were adapting to growth on a surface. Similarly, P. aeruginosa enters an extensive lag phase following initial attachment to glass before it resumes growth and cell division (36). It may, however, also be that the cells growing in a biofilm were preparing to switch from aerobic to fermentative growth. This was shown by the up-regulation of expression of cOTCase in the biofilm cells. B. cereus is able to grow aerobically by respiration using oxygen or anoxically by either fermentation of glucose and pyruvate or degradation of l-arginine (22, 42). Therefore, it should be expected that the growth rate would be lower under anoxic conditions, since the ATP yield during fermentative growth is significantly lower than that during respiration with oxygen.
Differences between the microcolony (2-h) proteome and the more developed biofilm (18-h) proteome were largely due to up- and down-regulation of the expression of a multitude of proteins, including cOTCase and LctE. In this respect B. cereus is similar to gram-negative rods such as Escherichia coli (32) and P. aeruginosa (39), both of which display a unique biofilm proteome. For example, transcription of 38% of the genes in E. coli is affected when the bacterium colonizes a surface (34). An explanation for the high levels of the enzymes present in 18-h-old biofilm cells may be that the cells were in the fermentative mode. Cells localized to the interior of the biofilm structure were likely to experience decreased oxygen tensions. The low level of dissolved oxygen measured in the culture medium seems to suggest that these cells were growing anoxically. Fermentative growth and the ADI pathway would enable the biofilm cells to cope with the low oxygen levels and energy depletion. The 18-h-old PGW cells also displayed up-regulation of these enzymes. This apparent switch to fermentative growth was surprising. It may have been due either to a physiological switch induced by temporary association with the surface during reversible attachment or to regulation by a biofilm-excreted signal molecule. The larger cell volume of PGW cells than of biofilm cells suggests that the growth rate was higher. Thus, the lower yield of PGW cells than of biofilm cells may indicate that more cells were attaching than detaching.
The suspended cells growing in the proximity of both a surface and a biofilm displayed a distinct phenotype. Although the PGW population phenotype resembled the planktonic phenotype more closely than the biofilm phenotype, some similarities to attached cells were discernible within 2 h of exposure to the glass wool. Most notably, YhbH was up-regulated in both biofilm and PGW cells compared to the planktonic levels, and cells from both of these growth phases had a smaller cell volume and possibly a lower growth rate. The YhbH protein may play a central role in the switch from the planktonic phenotype to the biofilm phenotype. It displays significant sequence identity to YvyD of B. subtilis, which may indicate its dependence on both σB and σH (8, 10, 11). As σB and σH have central roles in regulating the cell's response to environmental stresses and nutrient starvation, growth at the surface may constitute stress conditions for the cell. Importantly, σB is involved in regulation of biofilm expression and polysaccharide intercellular adhesin synthesis, which is essential for biofilm accumulation in Staphylococcus epidermidis and Staphylococcus aureus (16, 35). Sigma B may also be involved in expression of LctE (33), further highlighting its possible role in regulation of the biofilm phenotype of B. cereus. The up-regulation of YhbH in PGW cells may indicate that these cells sensed the same future stress as the attached cells and started to induce a general stress response. In the case of B. subtilis, the cells have developed a very complex adaptational network in order to deal with stress and starvation. General or nonspecific stress proteins are expressed in a nongrowing cell in order to provide multiple stress resistance in anticipation of future stress (13).
Comparison of the extracellular proteins of 18-h-old cultures grown with and without glass wool by one-dimensional SDS-PAGE analysis showed that the biofilm-associated secretome of B. cereus DL5 is different than that of a planktonic culture. Differences were observed in the expressed proteins, as well as in the up- and down-regulation of protein expression in the different growth phases. The identity of the proteins was not investigated further in this study. A proteomic analysis of B. subtilis 168 extracellular proteins revealed that this organism secretes between 150 and 180 proteins into the culture medium (14). It is also known that among the different classes of extracellular proteins that have been described (41) are degradative enzymes (e.g., proteases, lipases, carbohydratases, DNases, and RNases) which are synthesized as part of an adaptive response to a change in the environment, as well as relatively small proteins (e.g., PhrA and PhrK) that sense the cell density of the population, thereby regulating the onset of post-exponential-phase processes, such as competence development and sporulation. The uniquely secreted PGW-biofilm proteins may therefore include both enzymes and signal molecules.
In conclusion, this study highlighted several changes in the whole-cell protein profiles of B. cereus DL5 growing planktonically and as a biofilm on glass wool. The amounts of several proteins in attached and PGW cells are significantly different from the amounts in planktonic cells. In particular, increased levels of YhbH in both attached and PGW cells were observed within 2 h, indicating that this protein may play an important role in regulation of the biofilm phenotype of B. cereus. Further functional studies are necessary to clarify the roles of this and other proteins in biofilm development.
Acknowledgments
M.C.O. and B.S. were supported by scholarships from the National Research Foundation of South Africa, and M.C.O. received a Claude Harris Leon Foundation Postdoctoral Fellowship. D.L. was supported by the School of Molecular and Cell Biology, University of the Witwatersrand. This research was supported by grants from the National Research Foundation of South Africa to A.V.H. and V.S.B.
We thank J. F. Mostert for supplying B. cereus DL5.
REFERENCES
- 1.Chandra, J., D. M. Kuhn, P. K. Mukherkee, L. L. Hoyer, T. McCormick, and M. Ghannoum. 2001. Biofilm formation by the fungal pathogen Candida albicans: development, architecture, and drug resistance. J. Bacteriol. 183:5385-5394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Christiansson, A., J. Bertilsson, and B. Svensson. 1999. Bacillus cereus spores in raw milk: factors affecting the contamination of milk during the grazing period. J. Dairy Sci. 82:305-314. [DOI] [PubMed] [Google Scholar]
- 3.Costerton, J. W., Z. Lewandowski, D. E. Caldwell, D. R. Korber, and H. M. Lappin-Scott. 1995. Microbial biofilms. Annu. Rev. Microbiol. 49:711-745. [DOI] [PubMed] [Google Scholar]
- 4.Crielly, E. M., N. A. Logan, and A. Anderton. 1994. Studies on the Bacillus flora of milk and milk products. J. Appl. Bacteriol. 77:256-263. [DOI] [PubMed] [Google Scholar]
- 5.Cunin, R., N. Glansdorff, A. Piérard, and V. Stalon. 1986. Biosynthesis and metabolism of arginine in bacteria. Microbiol. Rev. 50:314-352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Davey, M. E., and G. A. O'Toole. 2000. Microbial biofilms: from ecology to molecular genetics. Microbiol. Mol. Biol. Rev. 64:847-867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davies, D. G., and G. G. Geesey. 1995. Regulation of the alginate biosynthesis gene algC in Pseudomonas aeruginosa during biofilm development in continuous culture. Appl. Environ. Microbiol. 61:860-867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Drzewiecki, K., C. Eymann, G. Mittenhuber, and M. Hecker. 1998. The yvyD gene of Bacillus subtilis is under dual control of σB and σH. J. Bacteriol. 180:6674-6680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dunn, M. J. 1996. Detection of proteins in polyacrylamide gels by silver staining, p. 229-233. In J. M. Walker (ed.), The protein protocols handbook. Humana Press Inc., Totowa, N.J.
- 10.Eymann, C., and M. Hecker. 2001. Induction of σB-dependent general stress genes by amino acid starvation in a spoOH mutant of Bacillus subtilis. FEMS Microbiol. Lett. 199:221-227. [DOI] [PubMed] [Google Scholar]
- 11.Eymann, C., G. Mittenhuber, and M. Hecker. 2001. The stringent response, σH-dependent gene expression and sporulation in Bacillus subtilis. Mol. Gen. Genet. 264:913-923. [DOI] [PubMed] [Google Scholar]
- 12.Granum, P. E., and T. Lund. 1997. Bacillus cereus and its food poisoning toxins. FEMS Microbiol. Lett. 157:223-228. [DOI] [PubMed] [Google Scholar]
- 13.Hecker, M., and U. Völker. 1998. Non-specific, general and multiple stress resistance of growth-restricted Bacillus subtilis cells by the expression of the σB regulon. Mol. Microbiol. 29:1129-1136. [DOI] [PubMed] [Google Scholar]
- 14.Hirose, I., K. Sano, I. Shioda, M. Kumano, K. Nakamura, and K. Yamane. 2000. Proteome analysis of Bacillus subtilis extracellular proteins: a two-dimensional protein electrophoretic study. Microbiology 146:65-75. [DOI] [PubMed] [Google Scholar]
- 15.James, G. A., D. R. Korber, D. E. Caldwell, and J. W. Costerton. 1995. Digital image analysis of growth and starvation responses of surface-colonizing Acinetobacter sp. J. Bacteriol. 177:907-915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Knobloch, J. K.-M., K. Bartscht, A. Sabottke, H. Rohde, H.-H. Feucht, and, D. Mack. 2001. Biofilm formation by Staphylococcus epidermidis depends on functional RsbU, an activator of the sigB operon: differential activation mechanisms due to ethanol and salt stress. J. Bacteriol. 183:2624-2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 18.Lindsay, D., V. S. Brözel, J. F. Mostert, and A. von Holy. 2000. Physiology of dairy-associated Bacillus spp. over a wide pH range. Int. J. Food Microbiol. 54:49-62. [DOI] [PubMed] [Google Scholar]
- 19.Lindsay, D., M. C. Oosthuizen, V. S. Brözel, and A. von Holy. 2002. Adaptation of a neutrophilic dairy-associated Bacillis cereus isolate to alkaline pH. J. Appl. Microbiol. 192:81-89. [DOI] [PubMed] [Google Scholar]
- 20.Loo, C. Y., D. A. Corliss, and N. Ganeshkumar. 2000. Streptococcus gordonii biofilm formation: identification of genes that code for biofilm phenotypes. J. Bacteriol. 182:1374-1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mack, D. 1999. Molecular mechanisms of Staphylococcus epidermidis biofilm formation. J. Hosp. Infect. 43:S113-S125. [DOI] [PubMed] [Google Scholar]
- 22.Maghnouj, A., T. F. de Sousa Cabral, V. Stalon, and C. Vander Wauven. 1998. The arcABDC gene cluster, encoding the arginine deiminase pathway of Bacillus licheniformis, and its activation by the arginine repressor ArgR. J. Bacteriol. 180:6468-6475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marino, M., T. Hoffmann, R. Schmid, H. Möbitz, and D. Jahn. 2000. Changes in protein synthesis during the adaption of Bacillus subtilis to anaerobic growth conditions. Microbiology 146:97-105. [DOI] [PubMed] [Google Scholar]
- 24.Møller, S., C. S. Kristensen, L. K. Poulsen, J. M. Carstensen, and S. Molin. 1995. Bacterial growth on surfaces: automated image analysis for quantification of growth rate-related parameters. Appl. Environ. Microbiol. 61:741-748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nakano, M. M., and F. M. Hulett. 1997. Adaptation of Bacillus subtilis to oxygen limitation. FEMS Microbiol. Lett. 157:1-7. [DOI] [PubMed] [Google Scholar]
- 26.Oosthuizen, M. C., B. Steyn, D. Lindsay, V. S. Brözel, and A. von Holy. 2001. Novel method for the proteomic investigation of a dairy-associated Bacillus cereus biofilm. FEMS Microbiol. Lett. 194:47-51. [DOI] [PubMed] [Google Scholar]
- 27.O'Reilly, M., K. Woodson, B. C. A. Dowds, and K. M. Devine. 1994. The citrulline biosynthesis operon, argC-F, and a ribose transport operon, rbs, from Bacillus subtilis are negatively regulated by Spo0A. Mol. Microbiol. 11:87-98. [DOI] [PubMed] [Google Scholar]
- 28.O'Toole, G. A., H. B. Kaplan, and R. Kolter. 2000. Biofilm formation as microbial development. Annu. Rev. Microbiol. 54:49-79. [DOI] [PubMed] [Google Scholar]
- 29.O'Toole, G. A., L. A. Pratt, P. I. Watnick, D. K. Newman, V. B. Weaver, and R. Kolter. 1999. Genetic approaches to study biofilms. Methods Enzymol. 310:91-109. [DOI] [PubMed] [Google Scholar]
- 30.Pederson, S., P. L. Bloch, S. Reeh, and F. C. Neidhardt. 1978. Patterns of protein synthesis in E. coli: a catalogue of the amount of 140 individual proteins at different growth rates. Cell 141:179-190. [DOI] [PubMed] [Google Scholar]
- 31.Peng, J. S., W. C. Tsai, and, C. C. Chou. 2001. Surface characteristics of Bacillus cereus and its adhesion to stainless steel. Int. J. Food Microbiol. 65:105-111. [DOI] [PubMed] [Google Scholar]
- 32.Perrot, F., M. Hebraud, R. Charlionet, G. A. Junter, and T. Jouenne. 2000. Protein patterns of gel-entrapped Escherichia coli cells differ from those of free-floating organisms. Electrophoresis 21:645-653. [DOI] [PubMed] [Google Scholar]
- 33.Petersohn, A., J. Bernhardt, U. Gerth, D. Höper, T. Koburger, U. Völker, and M. Hecker. 1999. Identification of σB-dependent genes in Bacillus subtilis using a promoter consensus-directed search and oligonucleotide hybridization. J. Bacteriol. 181:5718-5724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Prigent-Combaret, C., O. Vidal, C. Dorel, and P. Lejeune. 1999. Abiotic surface sensing and biofilm-dependent regulation of gene expression in Escherichia coli. J. Bacteriol. 181:5993-6002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rachid, S., K. Ohlsen, U. Wallner, J. Hacker, M. Hecker, and W. Ziebuhr. 2000. Alternative transcription factor σB is involved in regulation of biofilm expression in a Staphylococcus aureus mucosal isolate. J. Bacteriol. 182:6824-6826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rice, A. R., M. A. Hamilton, and A. K. Camper. 2000. Apparent surface associated lag time in growth of primary biofilm cells. Microb. Ecol. 41:8-15. [DOI] [PubMed] [Google Scholar]
- 37.Sauer, K., and A. K. Camper. 2001. Characterization of phenotypic changes in Pseudomonas putida in response to surface-associated growth. J. Bacteriol. 183:6579-6589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stewart, P. S. 1996. Theoretical aspects of antibiotic diffusion into microbial biofilms. Antimicrob. Agents Chemother. 40:2517-2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Steyn, B., M. C. Oosthuizen, R. MacDonald, J. Theron, and V. S. Brözel. 2001. The use of glass wool as an attachment surface for studying phenotypic changes in Pseudomonas aeruginosa biofilms by two-dimensional gel electrophoresis. Proteomics 1:871-879. [DOI] [PubMed] [Google Scholar]
- 40.Stülke, J., and W. Hillen. 2000. Regulation of carbon catabolism in Bacillus species. Annu. Rev. Microbiol. 54:849-880. [DOI] [PubMed] [Google Scholar]
- 41.Tjalsma, H., A. Bolhuis, J. D. H. Jongbloed, S. Bron, and J. M. van Dijl. 2000. Signal peptide-dependent protein transport in Bacillus subtilis: a genome-based survey of the secretome. Microbiol. Mol. Biol. Rev. 64:515-547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Unden, G., S. Becker, J. Bongaerts, J. Schirawski, and S. Six. 1994. Oxygen regulated gene expression in facultatively anaerobic bacteria. Antonie Leeuwenhoek 66:2-23. [DOI] [PubMed] [Google Scholar]
- 43.Van Loosdrecht, M. C. M., J. Lyklema, W. Norde, and A. J. B. Zehnder. 1990. Influence of interfaces on microbial activity. Microbiol. Rev. 54:75-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Watnick, P., and R. Kolter. 2000. Biofilm, city of microbes. J. Bacteriol. 182:2675-2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wessel, D., and U. I. Flugge. 1984. A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Anal. Biochem. 138:141-143. [DOI] [PubMed] [Google Scholar]
- 46.Whiteley, M., M. G. Bangera, R. E. Bumgarner, M. R. Parsek, G. M. Teitzel, S. Lory, and E. P. Greenberg. 2001. Gene expression in Pseudomonas aeruginosa biofilms. Nature 413:860-864. [DOI] [PubMed] [Google Scholar]
- 47.Xu, K. D., P. S. Stewart, F. Xia, C.-T. Huang, and G. A. Mcfeters. 1998. Spatial physiological heterogeneity in Pseudomonas aeruginosa biofilm is determined by oxygen availability. Appl. Environ. Microbiol. 64:4035-4039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zinn, M. S., R. D. Kirkegaard, R. J. Palmer, Jr., and D. C. White. 1999. Laminar flow conditions for continuous monitoring of biofilm formation and succession. Methods Enzymol. 310:224-232. [DOI] [PubMed] [Google Scholar]