Abstract
Lactic acid bacterial strains were isolated from brines sampled after 7 days of an industrial sauerkraut fermentation, and six strains were selected on the basis of susceptibility to bacteriophages. Bacterial growth in cabbage juice was monitored, and the fermentation end products were identified, quantified, and compared to those of Leuconostoc mesenteroides. Identification by biochemical fingerprinting, endonuclease digestion of the 16S-23S intergenic transcribed spacer region, and sequencing of variable regions V1 and V2 of the 16S rRNA gene indicated that the six selected sauerkraut isolates were Leuconostoc fallax strains. Random amplification of polymorphic DNA fingerprints indicated that the strains were distinct from one another. The growth and fermentation patterns of the L. fallax isolates were highly similar to those of L. mesenteroides. The final pH of cabbage juice fermentation was 3.6, and the main fermentation end products were lactic acid, acetic acid, and mannitol for both species. However, none of the L. fallax strains exhibited the malolactic reaction, which is characteristic of most L. mesenteroides strains. These results indicated that in addition to L. mesenteroides, a variety of L. fallax strains may be present in the heterofermentative stage of sauerkraut fermentation. The microbial ecology of sauerkraut fermentation appears to be more complex than previously indicated, and the prevalence and roles of L. fallax require further investigation.
Sauerkraut fermentation relies on naturally occurring lactic acid bacteria present on the raw cabbage. Several lactic acid bacterial species (mainly Leuconostoc mesenteroides, Lactobacillus brevis, Pediococcus pentosaceus, and Lactobacillus plantarum) are known to contribute to the complex sauerkraut fermentation process (28). L. mesenteroides is thought to be the dominant species in the early heterofermentative stage of this fermentation (13, 14, 28). However, there is little information available regarding the diversity of Leuconostoc species and strains involved in sauerkraut fermentation.
In addition to L. mesenteroides, Leuconostoc strain DSM 20189 was isolated from cabbage fermentation (31); this strain was later identified as Leuconostoc fallax (25). L. fallax strains have been isolated from sauerkraut (18, 31), as well as from fermented rice cake (puto) in the Philippines (20) and from plant exudates of Gerbera jamesonii in The Netherlands (26). Two L. fallax strains have been isolated from exudates of G. jamesonii (26), and five different strains, divided into three pulsed-field gel electrophoresis patterns, have been isolated from fermented rice cake (20). L. fallax was the most prevalent species in puto fermentation, representing more than 20% of all of the isolates screened. Similar to cabbage, puto contains a diverse microflora, including both homo- and heterofermentative lactobacilli, and many different Leuconostoc strains are presumed to be responsible for the initial acid production (20).
Several changes in the taxonomic classification of species within the genus Leuconostoc have been made in the last 10 years. Several new species have been described (3, 10, 12, 20, 21, 25, 31, 34, 38), and three major genera, Leuconostoc, Oenococcus, and Weissella, have been identified (7, 11). Recent improvements in microbial identification and typing provide convenient and accurate methods for classification of environmental and industrial Leuconostoc isolates. Bacteriophages active against L. mesenteroides, L. plantarum, and undefined isolates have been isolated recently from fermenting sauerkraut (42). However, the identity and diversity of the bacterial isolates which were sensitive to bacteriophages were not investigated. The objectives of this study were to identify and characterize the Leuconostoc strains present in sauerkraut fermentation that served as hosts for the propagation of bacteriophages.
MATERIALS AND METHODS
Bacterial strains.
Bacterial isolates were recovered from brines sampled after 7 days of a single industrial sauerkraut fermentation. The brine samples were plated on MRS agar (Difco Laboratories, Detroit, Mich.) and incubated aerobically at 30°C for 20 h. Forty colonies were randomly isolated and screened for bacteriophage sensitivity. Sensitivity to bacteriophages was determined by spotting 5-μl portions of serial dilutions of phage lysates on a lawn of the host strain (42). Phages were isolated from an industrial sauerkraut fermentation (42) and were used in this study only to select phage-sensitive strains. Six isolates were selected on the basis of their sensitivity to different phage lysates. The six bacterial isolates were catalase-negative, gram-positive coccobacilli usually occurring in twisted chains of 4 to 10 smooth lenticular cells. All bacterial strains were grown on MRS agar plates and in MRS broth at 30°C (9). The bacterial strains used in this study are listed in Table 1.
TABLE 1.
Bacterial strains used in this study
| Bacterium | Source or reference |
|---|---|
| Leuconostoc amelibiosum | ATCC 13146a |
| Leuconostoc citreum | ATCC 49370a |
| Leuconostoc fallax | ATCC 700006 |
| Leuconostoc lactis | ATCC 19256 |
| Leuconostoc mesenteroides subsp. cremoris | ATCC 19254 |
| Leuconostoc mesenteroides subsp. dextranicum | ATCC 19255 |
| Leuconostoc mesenteroides subsp. mesenteroides | ATCC 8293 |
| Leuconostoc fallax LA 288b | This study |
| Leuconostoc fallax LA 289b | This study |
| Leuconostoc fallax LA 290b | This study |
| Leuconostoc fallax LA 297b | This study |
| Leuconostoc fallax LA 298b | This study |
| Leuconostoc fallax LA 299b | This study |
| Leuconostoc mesenteroides LA 10c | 35 |
| Weissella paramesenteroides | ATCC 33313 |
ATCC, American Type Culture Collection.
Food Fermentation Laboratory, USDA Agricultural Research Service, Department of Food Science, North Carolina State University.
Strain originally isolated by J. R. Stamer as L. mesenteroides C-33 (35).
Biochemical identification.
Biochemical identification of the bacterial isolates was based on the ability of the isolates to utilize or oxidize different carbon sources, as determined by the Biolog AN MicroPlate method (Biolog, Hayward, Calif.). The selected isolates and Leuconostoc type strains used in this study were initially identified by using this method according to the manufacturer's instructions.
PCR amplification of the ITS region.
Bacterial chromosomal DNA was isolated with a Wizard DNA genomic purification kit (Promega Corp., Madison, Wis.) and was used as the template in a PCR (30) to amplify the intergenic transcribed spacer (ITS) region between the 16S and the 23S rRNA genes. A modification of the procedure of Jensen et al. (17), designed by Breidt and Fleming (5), was used for PCR amplification of the ITS region. The typical 100-μl reaction mixture used for ITS-PCR analysis of Leuconostoc strains contained 70 μl of water, 50 pmol of each primer (Genosys Biotechnologies Inc., The Woodlands, Tex.), 10 μl of 25 mM MgCl2 (Promega), 10 μl of thermophilic DNA polymerase, 10× PCR buffer (Promega), 1 μl of a deoxynucleoside triphosphate mixture (Promega), and 0.2 μg of DNA template. Amplification was carried out by using Taq DNA polymerase (Promega). The primers used were G1-16S (5′GAAGTCGTAACAAGG3′) and L2-23S (5′GGGTTTCCCCATTCGGA3′) (Genosys Biotechnologies Inc.). G1-16S is a primer designed to anneal specifically to a highly conserved region of the 3′ end of the 16S rRNA gene. L2-23S is a primer designed to anneal specifically to a highly conserved region of the 5′ end of the 23S rRNA gene. An initial denaturation step was performed with the reaction mixture prior to addition of Taq polymerase. DNA amplification was performed in a Gradient 96 Robocycler (Stratagene, La Jolla, Calif.) programmed as follows: 10 min at 94°C; 25 cycles of 1 min at 94°C, 5 min at 55°C, and 2 min at 72°C; and 5 min at 72°C. The fragments obtained were subjected to RsaI digestion by following the manufacturer's recommendations (Promega). The DNA band patterns were examined by 5% acrylamide gel electrophoresis, and a 1-kb ladder (Gibco-BRL, Grand Island, N.Y.) was used as a size standard.
RAPD typing.
The method used for random amplification of polymorphic DNA (RAPD) (40, 41) was derived from the method of Johansson et al. (19). The primers used for RAPD analysis of bacterial DNA have been described previously (6, 19, 29). Nine-mers were randomly designed with a G+C content of 80%. The primers used in this study were ED-01 (5′ACGCGCCCT3′) and ED-02 (5′CCGAGTCCA3′) (Genosys Biotechnologies Inc.). The typical 100-μl reaction mixture used for RAPD PCR analysis of L. fallax strains contained 66 μl of water, 100 pmol of primer, 10 μl of thermophilic DNA polymerase, 10× PCR buffer, 10 μl of 25 mM MgCl2, 1 μl of a deoxynucleoside triphosphate mixture, and 0.2 μg of DNA template. An initial denaturation step was performed with the reaction mixture prior to addition of Taq polymerase. The thermal cycler was programmed as follows: 10 min at 94°C; four cycles of 45 s at 94°C, 2 min at 30°C, and 45 s at 72°C; 36 cycles of 15 s at 94°C, 30 s at 36°C, and 45 s at 72°C; and 10 min at 72°C. The DNA amplicons were separated on a 5% acrylamide gel and compared with a 1-kb ladder (Gibco-BRL).
PCR amplification of the 16S ribosomal DNA (rDNA) variable region.
Primers were designed to anneal to highly conserved regions of the 16S rRNA gene and to amplify a 350-bp region of the 16S rRNA gene containing variable regions V1 and V2 (22, 27). The primers used for PCR amplification were 5′AGAGTTTGATCCTGGCTCAG3′ and 5′GTCTCAGTCCCAATGTGGCC3′ (Genosys Biotechnologies Inc.). The thermal cycler was programmed as follows: 10 min at 94°C; 25 cycles of 1 min at 94°C, 2 min at 61°C, and 2 min at 72°C; and 5 min at 72°C. The amplification products were analyzed by electrophoresis in 1% (wt/vol) agarose gels after ethidium bromide (0.5 μg/ml) staining.
16S rDNA sequencing and comparative sequence analysis.
The 350-bp PCR products were purified by using a Wizard PCR Preps DNA purification kit (Promega). DNA samples were sequenced commercially (Davis Sequencing, Davis, Calif.) with a model ABI Prism 277 DNA sequencer (Applied Biosystems, Foster City, Calif.). All sequences were subjected to the BLAST basic local alignment search tool (1) in the GenBank database (2) to determine the most likely identities of the strains. These sequences were also compared to that of the L. fallax type strain, and the percentages of homology were calculated by using BLAST2, taking into account the undetermined nucleotides.
The 16S rDNA sequences of all Leuconostoc species were aligned by using the CLUSTAL W 1.8 program (39), and the longest sequence common to all species was selected to generate a DNA similarity matrix. Percentages of similarity were calculated for the following two different fragments by using BLAST2: (i) the longest sequence available that is common to all Leuconostoc species, and (ii) an ∼300-bp fragment containing variable regions V1 and V2, included in the 350-bp fragment amplified for the sauerkraut isolates. When BLAST2 did not align sequences over the whole length, BestFit (SeqWeb, version 1.1; Wisconsin Package, version 10; Genetics Computer Group Inc., Madison, Wis.) was used. The incompleteness of some of the available sequences containing undetermined nucleotides was taken into account when the percentages of similarity were calculated. Only true mismatches and gaps were discriminative, and an error margin was included to take into account the undetermined nucleotides. The 16S rDNA sequence information is shown in Table 3.
TABLE 3.
16S rRNA gene sequence information
| Species or subspecies | Accession no.a | Total lengthb | Longest common fragment
|
Variable region
|
||||
|---|---|---|---|---|---|---|---|---|
| Lengthb | Position | Nsc | Lengthb | Position | Nsc | |||
| Leuconostoc amelibiosum | S78390 | 1,490 | 1,431 | 44-1474 | 15 | 272 | 44-315 | 6 |
| Leuconostoc argentinum | AF175403 | 1,471 | 1,433 | 17-1449 | 0 | 272 | 17-288 | 0 |
| Leuconostoc carnosum | AB022925 | 1,450 | 1,433 | 7-1439 | 0 | 272 | 7-278 | 0 |
| Leuconostoc citreum | AB022923 | 1,448 | 1,433 | 6-1438 | 0 | 272 | 6-277 | 0 |
| Leuconostoc fallax | S63851 | 1,504 | 1,448 | 42-1489 | 45 | 288 | 42-329 | 5 |
| Leuconostoc gasicomitatum | AF231131 | 1,500 | 1,440 | 18-1457 | 5 | 272 | 18-289 | 0 |
| Leuconostoc gelidum | AB022921 | 1,445 | 1,433 | 1-1433 | 0 | 272 | 1-272 | 0 |
| Leuconostoc kimchii | AF173986 | 1,505 | 1,433 | 17-1449 | 0 | 272 | 17-288 | 0 |
| Leuconostoc lactis | AB023968 | 1,451 | 1,433 | 7-1439 | 0 | 272 | 7-278 | 0 |
| Leuconostoc mesenteroides subsp. cremoris | M23034 | 1,493 | 1,434 | 45-1478 | 6 | 272 | 45-316 | 0 |
| Leuconostoc mesenteroides subsp. mesenteroides | AB023243 | 1,440 | 1,434 | 7-1440 | 0 | 272 | 7-278 | 0 |
| Leuconostoc pseudomesenteroides | AB023237 | 1,448 | 1,433 | 6-1438 | 0 | 272 | 6-277 | 0 |
| Oenococcus oeni | AB022924 | 1,471 | 1,448 | 14-1461 | 1 | 288 | 14-301 | 1 |
| Weissella paramesenteroides | AB023238 | 1,473 | 1,458 | 6-1463 | 3 | 297 | 6-302 | 3 |
The GenBank accession number does not necessarily relate to the American Type Culture Collection type strain. The accession numbers and the corresponding nucleotide sequences can be retrieved from the National Center for Biotechnology Information.
Length in nucleotides.
Ns, nucleotides not defined after nucleic acid sequencing, which usually appear as N in nucleotide sequences.
Cabbage juice preparation.
Filter-sterilized cabbage juice broth (16) was prepared from locally purchased cabbage. After removal of the outer leaves and cores, the cabbage was quartered and heated in an autoclave for 10 min at 121°C to remove growth inhibitors (24). Heated cabbage pieces were processed with a Braun Juicer (Braun Company, Kronberg, Germany). Cabbage juice was extracted from the slurry by centrifugation for 30 min at 11,000 × g. The juice was then centrifuged for 1 h at 20,000 × g and filter sterilized (0.22-μm-pore-size filter; Corning, Corning, N.Y.). The juice was stored either at 4°C or at −20°C and was checked for microbial contamination and inhibition prior to the experiments. Cabbage juice may contain microbial inhibitors (24); therefore, the ability of L. fallax strains to grow in cabbage juice was tested prior to experiments.
Growth in cabbage juice.
The growth of L. fallax strains and the growth of L. mesenteroides strains in cabbage juice were compared. The growth of the type strain and the growth of an experimental strain of each species were monitored by determining changes in pH and optical density at 600 nm (OD600) in triplicate over 90 h of incubation at 18°C. The experimental strains selected for this experiment were L. fallax LA 288 and L. mesenteroides LA 10, a starter culture commonly used for sauerkraut fermentation. For OD600 determination, samples were diluted up to four times to keep the bacterial concentration within the linear range for OD600 measurement with a spectrophotometer.
Cabbage juice fermentation chemistry.
The end products of cabbage juice fermentation by L. fallax and L. mesenteroides strains were determined by high-performance liquid chromatography (HPLC) analysis. Sugars and alcohols were analyzed by HPLC by using an Aminex HPX 87-C column (Bio-Rad, Hercules, Calif.) with a differential refractometer detector. The elution solvent was deionized distilled water at a flow rate fixed at 1 ml/min, and the column temperature was set at 80°C. Acids were analyzed by HPLC by using an Aminex HPX 87-H column (Bio-Rad) associated with a UV detector (λ = 210 nm). The elution solvent was 0.03 N sulfuric acid, the flow rate was 0.8 ml/min, and the column temperature was 60°C.
RESULTS
Biochemical identification.
The six sauerkraut isolates selected were gram-positive heterofermentative cocci occurring in pairs or short chains. The biochemical analysis by the Biolog AN Microplate method (Table 2) revealed that all the sauerkraut isolates selected had a biochemical fingerprint most similar to that of the L. fallax type strain. The levels of similarity of the biochemical fermentation patterns of the sauerkraut isolates to the pattern of the L. fallax type strain ranged from 57 to 89%. The levels of similarity of the fermentation patterns of the other Leuconostoc type strains to the patterns in the database ranged from 53 to 97%.
TABLE 2.
Strain identification
| Strain | Biolog identification
|
ITS patternb | 16S rDNA identificationc,d | |
|---|---|---|---|---|
| Species | % Similaritya | |||
| L. citreum ATCC 49370 | L. citreum | 60 | A | ND |
| L. fallax ATCC 700006 | L. fallax | 97 | B | ND |
| L. lactis ATCC 19256 | L. lactis | 71 | C | ND |
| L. mesenteroides subsp. cremoris ATCC 19254 | L. mesenteroides | 73 | D | ND |
| L. mesenteroides subsp. dextranicum ATCC 19255 | L. mesenteroides | 97 | D | ND |
| L. mesenteroides subsp. mesenteroides ATCC 8293 | L. mesenteroides | 93 | D | ND |
| W. paramesenteroides ATCC 33313 | L. paramesenteroides | 53 | E | ND |
| L. fallax LA 288 | L. fallax | 88 | B | L. fallax (96.7) |
| L. fallax LA 289 | L. fallax | 89 | B | L. fallax (98.1) |
| L. fallax LA 290 | L. fallax | 57 | B | L. fallax (97.7) |
| L. fallax LA 297 | L. fallax | 83 | B | L. fallax (97.8) |
| L. fallax LA 298 | L. fallax | 89 | B | L. fallax (95.2) |
| L. fallax LA 299 | L. fallax | 76 | B | L. fallax (97.8) |
Level of similarity between the biochemical pattern of the strain and the biochemical pattern of the most similar strain present in the Biolog database.
The patterns generated by RsaI digestion of the ITS-PCR-generated fragment are shown in Fig. 1B. Different letters indicate different patterns.
The numbers in parentheses are the percentages of similarity with the ≈300-bp region of the L. fallax type strain containing variable regions V1 and V2.
ND, not determined.
ITS-PCR fragments.
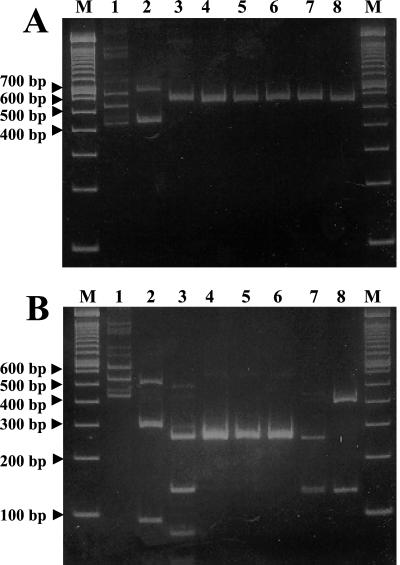
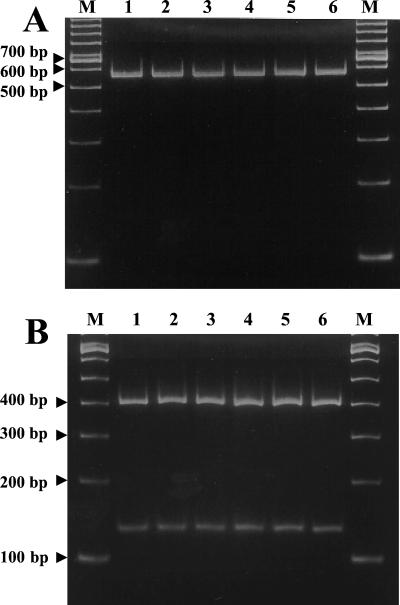
The ITS-PCR fragments of the selected Leuconostoc type strains and their RsaI digests are shown in Fig. 1. Most ITS-PCR fragments were approximately 550 bp long (Fig. 1A); the exceptions were the fragments of Leuconostoc lactis, which produced two bands, and Weissella paramesenteroides, which produced several bands. The RsaI digestion products were different for all the Leuconostoc species included in this experiment. However, the three L. mesenteroides subspecies showed the same patterns (Fig. 1B). The ITS-PCR patterns of the sauerkraut isolates are shown in Fig. 2. The ITS-PCR products of the sauerkraut isolates were all 550 bp long, which is characteristic of the genus Leuconostoc (5). The RsaI digestion products of the sauerkraut isolates were all identical (Fig. 2B), and there were two fragments (400 and 150 bp), which is characteristic of L. fallax (Fig. 1A).
FIG. 1.
(A) ITS-PCR patterns of Leuconostoc species. (B) RsaI digestion of the ITS-PCR fragments of Leuconostoc species. Lane 1, W. paramesenteroides; lane 2, L. citreum; lane 3, L. lactis; lane 4, L. mesenteroides subsp. mesenteroides; lane 5, L. mesenteroides subsp. cremoris; lane 6, L. mesenteroides subsp. dextranicum; lane 7, L. amelibiosum; lane 8, L. fallax; lanes M, molecular weight markers (100-bp DNA ladder).
FIG. 2.
(A) ITS-PCR patterns of the sauerkraut isolates. (B) RsaI digestion of the ITS-PCR products of the sauerkraut isolates. Lane 1, LA 289; lane 2, LA 290; lane 3, LA 288; lane 4, LA 297; lane 5, LA 298; lane 6, LA 299; lanes M, molecular weight markers (100-bp DNA ladder).
16S rDNA variable region sequencing.
DNA sequencing of variable regions V1, V2, and V6 of the 16S rRNA genes has been used previously for identification of lactic acid bacteria (7, 22). Both total 16S rRNA genes and the sequences of the ∼300-bp fragment of the 16S rRNA genes containing variable regions V1 and V2 in Leuconostoc species were compared. Sequence information and similarity data are shown in Tables 3 and 4, respectively. The degrees of similarity between true Leuconostoc species (excluding the species Oenococcus oeni and W. paramesenteroides) ranged from 91.5 to 99.8% for the total 16S rDNA sequence and from 81.3 to 100% for the ∼300-bp fragment containing variable regions. The levels of similarity between the nucleotide sequences of the sauerkraut isolates and the sequence of the L. fallax type strain for this highly variable region ranged from 95.2 to 98.1% (Table 2). In contrast, the levels of similarity between the L. fallax type strain and other Leuconostoc species for this region ranged from 80.1 to 83%.
TABLE 4.
16S rRNA gene similarity
| No. | Species | % Similaritya to:
|
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1b | 2 | 3 | 4 | 5c | 6d | 7 | 8 | 9 | 10e | 11 | 12 | 13 | 14f | ||
| 1 | Leuconostoc amelibiosum | 100 (100) | 98.7 (96.6) | 97.3 (95.5) | 99.3 (97.4) | 94.2 (84.7) | 96.4 (95.5) | 97.2 (94.7) | 96.6 (94.1) | 98.4 (96.6) | 97.6 (94.7) | 97.5 (94.7) | 97.3 (94.0) | 84.9 (72.6) | 89.2 (80.5) |
| 2 | Leuconostoc argentinum | 100 (100) | 97.4 (97.4) | 99.2 (97.8) | 93.4 (83.0) | 96.6 (96.6) | 97.6 (97.0) | 97.3 (97.0) | 99.6 (100) | 97.7 (96.3) | 97.7 (97.0) | 97.6 (96.3) | 85.1 (75.4) | 89.4 (82.0) | |
| 3 | Leuconostoc carnosum | 100 (100) | 97.7 (97.4) | 92.2 (83.0) | 97.6 (98.5) | 98.8 (98.2) | 98.2 (96.0) | 97.1 (97.4) | 97.7 (97.4) | 97.8 (98.1) | 97.6 (97.4) | 85.6 (74.3) | 89.3 (82.0) | ||
| 4 | Leuconostoc citreum | 100 (100) | 93.0 (84.8) | 97.0 (97.4) | 97.7 (96.3) | 98.3 (98.5) | 98.9 (97.8) | 97.7 (95.5) | 97.8 (96.3) | 97.6 (95.6) | 84.3 (74.9) | 89.6 (83.1) | |||
| 5 | Leuconostoc fallax | 100 (100) | 91.5 (81.3) | 92.0 (81.3) | 89.4 (80.1) | 92.2 (83.0) | 92.5 (83.0) | 92.7 (81.8) | 92.6 (82.0) | 85.4 (76.5) | 89.8 (78.1) | ||||
| 6 | Leuconostoc gasicomitatum | 100 (100) | 97.7 (98.9) | 97.2 (96.0) | 97.1 (96.7) | 97.3 (95.9) | 97.4 (96.7) | 97.3 (95.9) | 84.7 (74.0) | 89.3 (82.7) | |||||
| 7 | Leuconostoc gelidum | 100 (100) | 98.7 (95.6) | 97.7 (97.0) | 97.6 (96.3) | 98.1 (97.0) | 98.0 (96.3) | 85.2 (73.6) | 89.2 (82.0) | ||||||
| 8 | Leuconostoc kimchii | 100 (100) | 97.6 (97.1) | 97.2 (94.9) | 97.7 (95.6) | 97.6 (94.9) | 84.7 (72.1) | 88.4 (76.4) | |||||||
| 9 | Leuconostoc lactis | 100 (100) | 97.4 (96.3) | 98.0 (97.0) | 97.8 (96.3) | 84.9 (75.4) | 89.8 (82.0) | ||||||||
| 10 | Leuconostoc mesenteroides subsp. cremoris | 100 (100) | 99.8 (99.3) | 99.0 (98.5) | 84.8 (75.9) | 89.5 (80.7) | |||||||||
| 11 | Leuconostoc mesenteroides subsp. mesenteroides | 100 (100) | 99.6 (99.3) | 84.7 (73.9) | 90.0 (81.3) | ||||||||||
| 12 | Leuconostoc pseudomesenteroides | 100 (100) | 85.0 (76.4) | 90.2 (81.0) | |||||||||||
| 13 | Oenococcus oeni | 100 (100) | 84.5 (76.4) | ||||||||||||
| 14 | Weissella paramesenteroides | 100 (100) | |||||||||||||
Level of similarity for the longest fragment of the 16S rRNA gene common to the Leuconostoc species sequences available (level of similarity for an ≈300-bp fragment of the 16S rRNA gene containing variable regions V1 and V2).
The values are ±1.0% for the longest-fragment values and ±2.2% for the ≈300-bp fragment values due to the incompleteness of the sequence, so that the undetermined nucleotides could be taken into account.
The values are ±3.0% for the longest-fragment values and ±1.7% for the ≈300-bp fragment values due to the incompleteness of the sequence.
The values are ±0.3% for the longest-fragment values due to the incompleteness of the sequence.
The values are ±0.4% for the longest-fragment values due to the incompleteness of the sequence.
The values are ±0.2% for the longest-fragment values and ±1.0% for the ≈300-bp fragment values due to the incompleteness of the sequence.
RAPD typing.
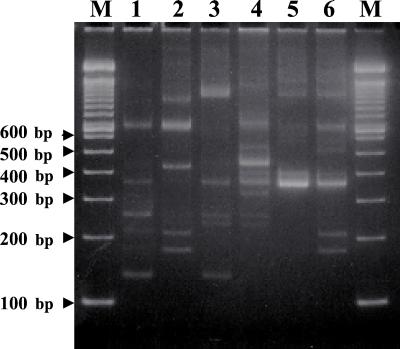
The results of strain typing of the sauerkraut isolates by RAPD fingerprinting are shown in Fig. 3. Most of the RAPD patterns were distinct, with variations in the number of bands, fragment size, and intensity. The number of bands varied between four and eight, and the fragment sizes ranged from 150 to 1,200 bp. Some strains exhibited significant similarity and common bands (Fig. 3, lanes 2 and 6). The patterns were highly reproducible, with variations only in relative band intensities. RAPD typing was capable of producing discriminating DNA fingerprints of the six L. fallax isolates, indicating that there were genetic differences among them.
FIG. 3.
RAPD patterns of the sauerkraut isolates. Lane 1, LA 289; lane 2, LA 290; lane 3, LA 288; lane 4, LA 297; lane 5, LA 298; lane 6, LA 299; lanes M, molecular weight markers (100-bp DNA ladder).
Growth in cabbage juice.
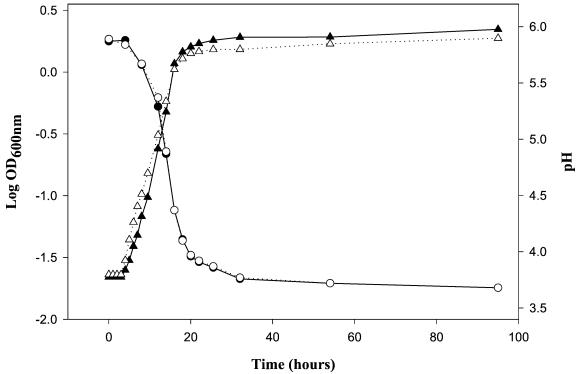
The initial pH of the cabbage juice ranged from 5.98 to 6.03. During fermentation, the pH was reduced to 3.68 by the L. fallax type strain, to 3.69 by L. fallax experimental strain LA 288, to 3.68 by the L. mesenteroides type strain, and to 3.82 by L. mesenteroides experimental strain LA 10. Characteristic growth and acidification patterns are shown in Fig. 4, and the data show that the growth profiles were nearly identical for L. fallax and L. mesenteroides.
FIG. 4.
Cabbage juice fermentation by L. fallax and L. mesenteroides type strains. Symbols: ▴, OD600 of L. fallax; ▵, OD600 of L. mesenteroides; •, pH of L. fallax; ○, pH of L. mesenteroides.
Fermentation end products.
Cabbage juice fermentation by L. fallax and L. mesenteroides experimental and type strains was monitored for end products over a 12-day period by using cabbage juice containing 2% (wt/vol) NaCl. The final pH values ranged from 3.58 to 3.62 for both species. All L. fallax strains produced 40 to 46 mM lactic acid, 53 to 59 mM acetic acid, and 79 to 93 mM mannitol from fructose and glucose, while the malate decarboxylase-positive (MDC+) L. mesenteroides strain produced 58 mM lactic acid, 62 mM acetic acid, and 102 mM mannitol. Carbon dioxide formation was observed but not quantified. The fermentation results are shown in Table 5. These results are consistent with the results of a previous study of cabbage juice fermentation by L. mesenteroides strains (4), in which 40 mM glucose was converted to 40 mM lactic acid and 42 mM acetic acid and 66 mM fructose were converted to 66 mM mannitol. The most significant difference between L. fallax and L. mesenteroides was the inability of the former to carry out the malolactic reaction through the malate decarboxylase. All of the L. fallax strains were unable to use all of the malic acid available, while the MDC+ L. mesenteroides strain exhausted the malic acid. Interestingly, all of the L. fallax strains appeared to ferment the cabbage juice in a pattern similar to that of an MDC− L. mesenteroides strain rather than that of an MDC+ L. mesenteroides strain. However, L. fallax differed in the amount of mannitol produced.
TABLE 5.
Cabbage juice fermentation by L. fallax and L. mesenteroides strains
| Time (days) | Acid concn (mM)
|
Sugar concn (mM)
|
||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Malic
|
Succinic
|
Lactic
|
Acetic
|
Glucose
|
Fructose
|
Mannitol
|
||||||||||||||||||||||
| Aa | Bb | Cc | Dd | A | B | C | D | A | B | C | D | A | B | C | D | A | B | C | D | A | B | C | D | A | B | C | D | |
| 0 | 11.5 | 11.9 | 12.2 | 11.5 | 0.0 | 0.0 | 0.0 | 0.0 | 00.0 | 00.0 | 00.0 | 00.0 | 00.0 | 00.0 | 00.0 | 00.0 | 141.1 | 139.4 | 138.1 | 143.1 | 136.4 | 136.5 | 132.8 | 136.7 | 00.0 | 00.0 | 00.0 | 00.0 |
| 6 | 9.1 | 00.0 | 9.2 | 11.9 | 2.8 | 2.3 | 2.4 | 0.0 | 43.6 | 57.9 | 40.6 | 00.0 | 51.1 | 58.1 | 59.1 | 00.0 | 111.1 | 96.3 | 97.8 | 145.9 | 40.9 | 35.8 | 31.2 | 140.1 | 87.5 | 97.5 | 105.6 | 00.0 |
| 9 | 8.8 | 00.0 | 9.4 | 11.3 | 2.5 | 2.4 | 2.4 | 0.0 | 43.4 | 62.5 | 43.1 | 00.0 | 49.0 | 62.1 | 58.4 | 00.0 | 111.2 | 99.3 | 96.8 | 143.6 | 40.5 | 37.0 | 29.4 | 138.1 | 88.7 | 101.5 | 106.9 | 00.0 |
| 12 | 8.8 | 00.0 | 9.4 | 11.4 | 2.9 | 2.4 | 2.5 | 0.0 | 44.3 | 58.3 | 41.8 | 00.0 | 55.5 | 61.6 | 62.4 | 00.0 | 109.2 | 99.7 | 98.3 | 144.4 | 38.3 | 37.6 | 29.85 | 138.2 | 88.8 | 101.4 | 108.4 | 00.0 |
A, L. fallax (average for seven strains, each examined in duplicate). The coefficients of variation (standard deviation/mean) were within 10.9% of the concentration shown for the seven strains.
B, L. mesenteroides type strain (MDC+), examined in duplicate.
C, L. mesenteroides MDC− strain LA 10, examined in duplicate.
D, cabbage juice, examined in duplicate.
DISCUSSION
This study revealed that a variety of L. fallax strains are present during the heterofermentative stage of sauerkraut fermentation. L. mesenteroides has long been considered to be the preponderant species during the first week of fermentation (14, 28). The presence and diversity of L. fallax strains recovered suggest that our current understanding of the microbial ecology of sauerkraut fermentation is incomplete. Several facts support the possibility that L. fallax strains are dominant late in the heterofermentative stage of sauerkraut fermentation. First, all Leuconostoc strains that were isolated in this study after 7 days of fermentation belonged to the species L. fallax. Second, the frequency of MDC+ strains has been shown to decrease during the first week of sauerkraut fermentation (18), while Leuconostoc strains remain predominant. This observation is consistent with replacement of the mostly MDC+ species L. mesenteroides by the mostly MDC− species L. fallax.
It was shown previously that in glucose broth, L. fallax can grow and lower the pH to 3.9, is resistant to 9% (vol/vol) ethanol, tolerates 5% (wt/vol) salt, and is unable to carry out malolactic fermentation (26). The cabbage juice fermentation end products were virtually equimolar amounts of acetic and lactic acids for MDC+ L. mesenteroides, whereas L. fallax and MDC− L. mesenteroides yielded more acetic acid and were both unable to exhaust malic acid. In this study, it was shown that L. fallax strains behave mostly like MDC−L. mesenteroides strains for cabbage juice fermentation.
Identification of lactic acid bacteria by morphological analysis and biochemical typing is not as reliable or consistent as genotypic characterization (32, 36, 37). As a result, several molecularly based methods have been developed to identify lactic acid bacteria quickly and conveniently. These include ribotyping, pulsed-field gel electrophoresis, 16S rDNA sequencing, RAPD typing, and phage typing (17, 22, 32, 33, 36, 37, 39, 41). ITS-PCR analysis is a rapid and simple way to identify lactic acid bacterial species in vegetable fermentations (5). In this study, RsaI digestion of ITS-PCR products and sequencing of a 350-bp variable region of the 16S rRNA gene provided strong evidence for identification of the species L. fallax. The combination of 16S rRNA gene sequencing with ITS-PCR analysis identified the sauerkraut isolates at the genus and species levels, while RAPD typing differentiated them at the strain level.
The lack of molecular identification methods for L. fallax is likely responsible for the historical failure to distinguish L. fallax from L. mesenteroides. In fermented rice cake, L. mesenteroides has been reported to be the predominant organism (8); however, it was shown later that L. fallax was the prevalent species (20). It seems likely that in the past L. fallax could also have been misidentified as L. mesenteroides in cabbage fermentation. L. fallax is similar to L. mesenteroides from a biochemical and fermentation standpoint, as glucose and fructose are fermented into lactic acid, acetic acid, carbon dioxide, and mannitol. However, one distinctive feature of the two taxa is the ability to ferment malate via the malolactic reaction.
The malolactic reaction is defined as decarboxylation of l-malic acid into l-lactic acid and carbon dioxide by the malolactic enzyme (18, 23). Most L. mesenteroides strains exhibit the malolactic reaction phenotype (15, 18) and can also produce small amounts of succinic acid (around 3 mM). Unlike most leuconostocs, L. fallax does not carry out malolactic fermentation. Therefore, it appears that L. fallax behaves mostly like MDC− L. mesenteroides in terms of cabbage juice fermentation end products. Even though the six L. fallax strains studied here were genetically different, they exhibited very similar biochemical patterns for sauerkraut fermentation and were all MDC−. The malolactic activity of lactic acid bacteria may have important effects on both sensory attributes and chemical properties of fermented cabbage (18). As a result, since L. mesenteroides and L. fallax differ phenotypically in the ability to carry out the malolactic reaction, it is important to determine which Leuconostoc species is predominant in sauerkraut fermentation.
Knowledge of microbial ecology in vegetable fermentations has been improved by the emergence of molecular identification and typing methods. A combination of ITS-PCR analysis and sequencing of a variable region of the 16S rRNA gene can be used to identify L. fallax strains. The discovery of a variety of L. fallax strains in the heterofermentative stage of sauerkraut fermentation encourages further investigation of the prevalence and roles of this species in fermentation of cabbage and perhaps other vegetables.
Acknowledgments
This study was sponsored by the U.S. Department of Agriculture, by Pickle Packers International Inc., and by the NRICGP under project 97-35503-4368.
We acknowledge Janet Hayes for providing bacterial strains and cabbage juice; Roger Thompson for performing the HPLC analysis; Martin Kullen and Eric Altermann for helpful discussions; and Olivia McAuliffe, Michael Callanan, and Sophia Kathariou for reviewing the manuscript.
Footnotes
Paper no. FSR01-26 of the Journal Series of the Department of Food Science, North Carolina State University, Raleigh.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller. E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Benson, D. A., M. S. Boguski, D. J. Lipman, and J. Ostell. 1997. GenBank. Nucleic Acids Res. 25:1-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bjorkroth, K. J., R. Geisen, U. Schillinger, N. Weiss, P. De Vos, W. H. Holzapfel, H. J. Korkeala, and P. Vandamme. 2000. Characterization of Leuconostoc gasicomitatum sp. nov. associated with spoiled raw tomato-marinated broiler meat strips packaged under modified atmosphere. Appl. Environ. Microbiol. 66:3764-3772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Breidt, F., K. A. Crowley, and H. P. Fleming. 1993. Isolation and characterization of nisin-resistant Leuconostoc mesenteroides for use in cabbage fermentations. Appl. Environ. Microbiol. 59:3778-3783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Breidt, F., and H. P. Fleming. 1996. Identification of lactic acid bacteria by ribotyping. J. Rapid Methods Automation Microbiol. 4:219-233. [Google Scholar]
- 6.Brousseau, R., A. Saint-Onge, G. Prefontaine, L. Masson, and J. Cabana. 1993. Arbitrary primer polymerase chain reaction, a powerful method to identify Bacillus thuringiensis serovars and strains. Appl. Environ. Microbiol. 59:114-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Collins, M. D., J. Samelis, J. Metaxopoulos, and S. Wallbanks. 1993. Taxonomic studies on some leuconostoc-like organisms from fermented sausages: description of a new genus Weissella for the Leuconostoc paramesenteroides group of species. J. Appl. Bacteriol. 75:595-603. [DOI] [PubMed] [Google Scholar]
- 8.Cooke, R. D., D. R. Twiddy, and P. J. A. Reilly. 1987. Lactic acid fermentation as a low-cost means of food preservation in tropical countries. FEMS Microbiol. Lett. 46:369-379. [Google Scholar]
- 9.deMan, J. C., M. Rogosa, and M. E. Sharpe. 1960. A medium for the cultivation of lactobacilli. J. Appl. Bacteriol. 23:130-135. [Google Scholar]
- 10.Dicks, L. M. T., L. Fantuzzi, F. C. Gonzalez, M. Du Toit, and F. Dellaglio. 1993. Leuconostoc argentinum sp. nov., isolated from Argentine raw milk. Int. J. Syst. Bacteriol. 43:347-351. [Google Scholar]
- 11.Dicks, L. M. T., F. Dellaglio, and M. D. Collins. 1995. Proposal to reclassify Leuconostoc oenos as Oenococcus oeni [corrig.] gen. nov., comb. nov. Int. J. Syst. Bacteriol. 45:395-397. [DOI] [PubMed] [Google Scholar]
- 12.Farrow, J. A. E., R. F. Facklam, and M. D. Collins. 1989. Nucleic acid homologies of some vancomycin-resistant leuconostocs and description of Leuconostoc citreum sp. nov. and Leuconostoc pseudomesenteroides sp. nov. Int. J. Syst. Bacteriol. 39:279-283. [Google Scholar]
- 13.Fleming, H. P., R. F. McFeeters, and E. G. Humphries. 1987. A fermentor for study of sauerkraut fermentation. Biotechnol. Bioeng. 31:189-197. [DOI] [PubMed] [Google Scholar]
- 14.Fleming, H. P., K. H. Kyung, and F. Breidt. 1995. Vegetable fermentations, p. 631-661. In G. Reed and T. W. Nagodawithana (ed.), Bio/Technology, 2nd ed., vol. 9. VCH Publishing Co., Weinheim, Germany. [Google Scholar]
- 15.Garvie, E. I. 1986. Genus Leuconostoc Van Tieghem 1878, p. 1071-1075. In P. H. A. Sneath, N. S. Mair, M. E. Sharpe, and J. G. Holt (ed.), Bergey's manual of systematic bacteriology, vol. 2. Williams and Wilkins, Baltimore, Md. [Google Scholar]
- 16.Harris, L. J., H. P. Fleming, and T. R. Klaenhammer. 1992. Novel paired starter culture system for sauerkraut, consisting of a nisin-resistant Leuconostoc mesenteroides strain and a nisin-producing Lactococcus lactis strain. Appl. Environ. Microbiol. 58:1484-1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jensen, M. A., J. A. Webster, and N. Straus. 1993. Rapid identification of bacteria on the basis of polymerase chain reaction-amplified ribosomal DNA spacer polymorphisms. Appl. Environ. Microbiol. 59:945-952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johanningsmeier, S. 1999. Malolactic activity of lactic acid bacteria and its effect on sensory and chemical properties of fermented cabbage. M.S. thesis. North Carolina State University, Raleigh.
- 19.Johansson, M. L., M. Quednau, G. Molin, and S. Ahrne. 1995. Randomly amplified polymorphic DNA (RAPD) for rapid typing of Lactobacillus plantarum strains. Lett. Appl. Microbiol. 21:155-159. [DOI] [PubMed] [Google Scholar]
- 20.Kelly, W. J., R. V. Asmundson, G. L. Harrison, and C. M. Huang. 1995. Differentiation of dextran-producing Leuconostoc strains from fermented rice cake (puto) using pulsed field gel electrophoresis. Int. J. Food Microbiol. 26:345-352. [DOI] [PubMed] [Google Scholar]
- 21.Kim, J., J. Chun, and H. U. Han. 2000. Leuconostoc kimchii sp. nov., a new species from kimchi. Int. J. Syst. E vol. Microbiol. 50:1915-1919. [DOI] [PubMed] [Google Scholar]
- 22.Kullen, M. J., R. B. Sanosky-Dawes, D. C. Crowell, and T. R. Klaenhammer. 2000. Use of DNA sequence of variable regions of the 16S rRNA gene for rapid and accurate identification of bacteria in the Lactobacillus acidophilus complex. J. Appl. Microbiol. 89:511-518. [DOI] [PubMed] [Google Scholar]
- 23.Kunkee, R. E. 1967. Malo-lactic fermentation. Adv. Appl. Microbiol. 9:235-279. [DOI] [PubMed] [Google Scholar]
- 24.Kyung, K. H., and H. P. Fleming. 1994. Antibacterial activity of cabbage juice against lactic acid bacteria. J. Food Sci. 59:125-129. [Google Scholar]
- 25.Martinez-Murcia, A. J., and M. D. Collins. 1991. A phylogenetic analysis of an atypical leuconostoc: description of Leuconostoc fallax sp. nov. FEMS Microbiol. Lett. 82:55-60. [DOI] [PubMed] [Google Scholar]
- 26.Middelhoven, W. J., and N. Klijn. 1997. Leuconostoc fallax, an acid and ethanol tolerant lactic acid bacterium. J. Sci. Food. Agric. 75:57-60. [Google Scholar]
- 27.Neefs, J. M., Y. Van de Peer, P. De Rijk, S. Chapelle, and R. De Wachter. 1993. Compilation of small ribosomal subunit RNA structures. Nucleic Acids Res. 21:3025-3049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pederson, C. S., and M. N. Albury. 1969. The sauerkraut fermentation. New York State Agricultural Experiment Station Technical Bulletin no. 824. New York State Agricultural Experiment Station, Geneva, N.Y.
- 29.Plengvidhya, V. 1999. Evaluation of molecular methods to follow the progress of starter cultures in sauerkraut fermentations. M.S. thesis. North Carolina State University, Raleigh.
- 30.Saiki, R. K., D. H. Gelfand, S. Stoffel, S. J. Scharf, R. Higuchi, G. T. Horn, K. B. Mullis, and H. A. Erlich. 1988. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science 239:487-491. [DOI] [PubMed] [Google Scholar]
- 31.Schillinger, U., W. Holzapfel, and O. Kandler. 1989. Nucleic acid hybridization studies on Leuconostoc and heterofermentative lactobacilli and description of Leuconostoc amelibiosum sp. nov. Syst. Appl. Microbiol. 12:48-55. [Google Scholar]
- 32.Schleifer, K. H., M. Ehrmann, C. Beimfohr, E. Brockmann, W. Ludwig, and R. Amann. 1995. Application of molecular methods for the classification and identification of lactic acid bacteria. Int. Dairy J. 5:1081-1094. [Google Scholar]
- 33.Schwartz, D. C., and C. R. Cantor. 1984. Separation of yeast chromosome-sized DNAs by pulsed field gradient gel electrophoresis. Cell 37:67-75. [DOI] [PubMed] [Google Scholar]
- 34.Shaw, B. G., and C. D. Harding. 1989. Leuconostoc gelidum sp. nov. and Leuconostoc carnosum sp. nov. from chill-stored meats. Int. J. Syst. Bacteriol. 39:217-223. [Google Scholar]
- 35.Stamer, J. R., B. O. Stoyla, and B. A. Dunckel. 1971. Growth rates and fermentation patterns of lactic acid bacteria associated with sauerkraut fermentation. J. Milk Food Technol. 34:521-525. [Google Scholar]
- 36.Stiles, M. E., and W. H. Holzapfel. 1997. Lactic acid bacteria of foods and their current taxonomy. Int. J. Food Microbiol. 36:1-29. [DOI] [PubMed] [Google Scholar]
- 37.Stull, T. L., and J. J. LiPuma. 1988. A broad-spectrum probe for molecular epidemiology of bacteria: ribosomal RNA. J. Infect. Dis. 157:280-286. [DOI] [PubMed] [Google Scholar]
- 38.Takahashi, M., S. Okada, T. Uchimura, and M. Kozaki. 1992. Leuconostoc amelibiosum Schillinger, Holzapfel, and Kandler 1989 is a later subjective synonym of Leuconostoc citreum Farrow, Facklam, and Collins 1989. Int. J. Syst. Bacteriol. 42:649-651. [Google Scholar]
- 39.Thompson. J. D., D. G. Higgins, and T. J. Gibson. 1994. Clustal W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Welsh, J., and M. McClelland. 1990. Fingerprinting genomes using PCR with arbitrary primers. Nucleic Acids Res. 18:7213-7218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Williams, J. G. K., A. R. Kubelik, K. J. Livak, J. A. Rafalski, and S. V. Tingey. 1990. DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Res. 18:6531-6535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yoon, S. S., R. Barrangou-Poueys, F. Breidt, T. R. Klaenhammer, and H. P. Fleming. 2002. Isolation and characterization of bacteriophages from fermenting sauerkraut. Appl. Environ. Microbiol. 68:973-976. [DOI] [PMC free article] [PubMed] [Google Scholar]