Abstract
Homologs of aflatoxin biosynthetic genes have been identified in the pine needle pathogen Dothistroma pini. D. pini produces dothistromin, a difuranoanthraquinone toxin with structural similarity to the aflatoxin precursor versicolorin B. Previous studies with purified dothistromin suggest a possible role for this toxin in pathogenicity. By using an aflatoxin gene as a hybridization probe, a genomic D. pini clone was identified that contained four dot genes with similarity to genes in aflatoxin and sterigmatocystin gene clusters with predicted activities of a ketoreductase (dotA), oxidase (dotB), major facilitator superfamily transporter (dotC), and thioesterase (dotD). A D. pini dotA mutant was made by targeted gene replacement and shown to be severely impaired in dothistromin production, confirming that dotA is involved in dothistromin biosynthesis. Accumulation of versicolorin A (a precursor of aflatoxin) by the dotA mutant confirms that the dotA gene product is involved in an aflatoxin-like biosynthetic pathway. Since toxin genes have been found to be clustered in fungi in every case analyzed so far, it is speculated that the four dot genes may comprise part of a dothistromin biosynthetic gene cluster. A fifth gene, ddhA, is not a homolog of aflatoxin genes and could be at one end of the dothistromin cluster. These genes will allow comparative biochemical and genetic studies of the aflatoxin and dothistromin biosynthetic pathways and may also lead to new ways to control Dothistroma needle blight.
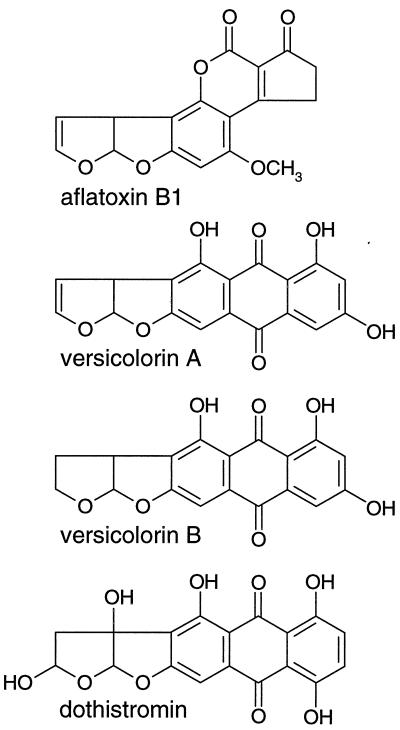
Dothistromin is a difuranoanthraquinone toxin that has remarkable structural similarity to versicolorin B, a precursor of the aflatoxin family of compounds (Fig. 1). Dothistromin is produced by the pine pathogen Dothistroma pini Hulbary (28) as well as by several Cercospora species, including the peanut pathogen C. arachidicola (47). The structure of dothistromin has been confirmed by using spectroscopic and crystallographic methods (4, 27).
FIG. 1.
Chemical structures of aflatoxin B1, versicolorin A, versicolorin B, and dothistromin.
There is evidence that dothistromin shares biosynthetic steps with aflatoxin, which is produced by Aspergillus parasiticus and Aspergillus flavus, and with the aflatoxin precursor sterigmatocystin, which is produced by Aspergillus nidulans. In a 13C nuclear magnetic resonance study of dothistromin biosynthesis, the labeling pattern in the bistetrahydrofurano side chain was identical to those found in aflatoxin and sterigmatocystin (44). Furthermore, aflatoxin precursors, including averantin, averufin, and versicolorins, were detected in culture filtrates of dothistromin-producing species (19, 47).
Although the production of aflatoxin is coordinated with asexual sporulation (1, 29), no clear biological role has been discovered for this complex group of secondary metabolites in the fungi that produce them. Dothistromin is thought to have a role in pathogenicity of the necrotrophic pathogen D. pini, as the injection of purified dothistromin into pine needles results in necrotic lesions and red-band symptoms, the same as those seen in Dothistroma needle blight (43). Benzoic acid is produced in cells adjacent to those killed by dothistromin, leading to extensive needle death (26). This defoliation leads to reduced wood yield and, in extreme cases, death of the tree. Several species of pine are susceptible to D. pini infection, including Pinus radiata Don, which is a major commercial crop of many countries in the southern hemisphere. Aerial spraying of infected forests with copper fungicides is the current method of disease control.
Dothistromin is a potent and broad-spectrum toxin. Toxicity has been demonstrated towards mature pine embryos and leaf callus with only 13 nmol of dothistromin per g of tissue. A 40-kDa dothistromin-binding protein was detected in the embryos, but the mechanism of toxicity is not known (34). As well as being a phytotoxin, dothistromin is toxic to a variety of microbial and animal cells (48). It is also weakly mutagenic and clastogenic, which has raised concerns about the health of forest workers (23).
The work outlined here is part of a program aimed at understanding the genetics, biochemistry, and biology of dothistromin, with a particular interest in comparative studies with aflatoxin biosynthesis. A wealth of knowledge has accumulated on the aflatoxin biosynthetic pathways (6, 11). The genes are clustered, with approximately 25 genes within a 60- to 70-kb region of the genome, although the order of homologous genes differs between the aflatoxin cluster of A. parasiticus (53) and the sterigmatocystin cluster of A. nidulans (10, 14).
The specific aim of this work was to determine whether D. pini has aflatoxin-like genes involved in dothistromin biosynthesis. Aflatoxin genes were used as hybridization probes to recover dothistromin genes from a D. pini genomic library. We report here the characterization of D. pini genes that show homology to aflatoxin pathway genes. Detailed characterization of dotA is reported; the predicted gene product shows 80% amino acid identity to the A. parasiticus aflatoxin Ver-1 protein (45) and is involved in dothistromin biosynthesis.
MATERIALS AND METHODS
Strains and culture conditions.
Escherichia coli XL-1 (12) was used for propagating plasmids. E. coli KW251 (39) was the recipient strain for the phage genomic library. D. pini strains NZE1 (ATCC MYA-605) and NZE5 were isolated from P. radiata trees near Rotorua, New Zealand, and cultured and maintained on Dothistroma medium (DM) as described previously (9). To assess radial growth, D. pini strains were point inoculated onto Aspergillus minimal medium (AMM) (ATCC culture medium 687). For standard dothistromin assays, 25 ml of liquid AMM (with 2% glucose), in 250-ml flasks, was inoculated with 10 to 30 mm3 of mycelium macerated with a pestle (Eppendorf no. 0030120973) and incubated for 7 or 10 days at 23°C with orbital shaking at 220 rpm. For thin-layer chromatography (TLC) and mass spectrometric analysis, the same growth conditions were used but with 200 ml of medium in 2-liter flasks. Dothistromin production was also tested after replacing the N source (NaNO3) in AMM with NH4Cl (3.7 g/liter) or the C source (glucose) with peptone (20 g/liter) or by adding freeze-dried and powdered P. radiata needles (20 g/liter).
Isolation and sequencing of the dotA clone.
Genomic DNA was routinely extracted from D. pini by using the method of Al-Samarrai and Schmid (2). For high-molecular-weight DNA for library construction, a plant tissue method (3) was used, followed by CsCl gradient purification. A genomic library of D. pini NZE1 was prepared in λGEM-12 according to the manufacturer's instructions (Promega, Madison, Wis.). A 1.8-kb EcoRI-HindIII fragment of the Aspergillus parasiticus ver-1 gene (45) was labeled with 32P by using a random primer DNA labeling kit (Roche Molecular Biochemicals, Mannheim, Germany) and hybridized to the library at 55°C in buffer containing 3× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate). Standard methods were used for library screening, for the purification of hybridizing clones, and for DNA extraction and restriction mapping (42).
Clone λCGV1 was subcloned into pUC18 and sequenced on both strands by using an ABI Prism dye terminator cycle sequencing ready reaction kit (Perkin- Elmer, Foster City, Calif.) and an ABI 377 automated sequencer. Overlap sequences were obtained from separate overlapping subclones or by PCR amplification of overlap sections from genomic DNA. Computer analyses of sequence data were performed by using the Wisconsin Genetics Computer Group package. To determine whether predicted open reading frames (ORFs) were expressed and to provide a cDNA template to verify dotA introns, reverse transcriptase PCR (RT-PCR) was carried out by using a SuperScript One-Step RT-PCR system (Life Technologies, Rockville, Md.).
Construction of gene replacement vector and fungal transformation.
The dotA disruption vector pR208 was constructed by using a four-step procedure. In the first step, regions flanking a region of the dotA gene were PCR amplified by using XbaI-tailed primers. A 692-bp region upstream of the dotA coding sequence was amplified by using the forward primer 5′-TCCTGCCCATGGTGCGA-3′ (primer 1) and the reverse primer 5′-GACTCTAGAACGAGTCTCAATGTATC-3′ (primer 2) to which an XbaI site (underlined) had been added. A 670-bp downstream region was amplified by using the forward primer 5′-CGTTCTAGAGTCGCTCGCGAGGCGTA-3′ (primer 3) to which an XbaI site had been added and the reverse primer 5′-TCCTCGCCGTCATGGAGTA-3′ (primer 4). The upstream and downstream PCR products each contained 12 bp of matching sequence at one end due to the common XbaI sites and matching 3-bp extensions present in primers 2 and 3.
In the second step, these two products were combined in a further round of PCR by using primers 1 and 4. In the third step, the combined 1.4-kb product from step 2 was cloned into pGEM-T (Promega) according to the manufacturer's instructions. Finally, DNA containing the selectable marker gene hph was cloned into the XbaI site marking the boundary of the upstream and downstream fragments to make gene replacement construct pR208. The hph gene confers resistance to hygromycin, is under the control of the Aspergillus niger glaA promoter and the A. nidulans trpC terminator, and was obtained from plasmid pCWHyg1 (C. Wasmann, University of Arizona). The resulting dotA disruption construct lacked a 536-bp region including a sequence that encodes a conserved adenine nucleotide binding motif involved in the active site of the putative enzyme.
D. pini strain NZE5 was used as a transformation host for replacement of the dotA gene; NZE5 is indistinguishable from NZE1 by random amplified polymorphic DNA analysis (32) but produces consistently higher levels of dothistromin in culture than NZE1 (9). The gene replacement construct was introduced into D. pini NZE5 by protoplast-mediated transformation by selection with 70 μg of hygromycin per ml as previously described (8).
Characterization of dothistromin mutants.
Transformants were purified by two rounds of growth from single spores on selective medium. Targeted replacement of the dotA gene was assessed by screening of transformants with PCR and then by Southern blotting by standard methods (42) with a digoxigenin-labeled probe and chemiluminescent detection (Roche Molecular Biochemicals). Triplicate cultures were grown in order to assay dothistromin that is secreted into the medium. Cultures were harvested by filtration, the mycelium was freeze-dried for dry weight determination, and the growth medium was analyzed for dothistromin by a competitive enzyme-linked immunosorbent assay (ELISA) as previously described (9, 33).
For TLC and mass spectrophotometric analysis, fungal cultures were extracted with acetone and chloroform (1:1, vol/vol); extracts were condensed and dried over phosphorus pentoxide. Samples, resuspended in chloroform, were applied to silica TLC plates and run alongside standards in an ether-methanol-water (96:3:1, vol/vol/vol) solvent system (15). For characterization of the identity of isolated metabolites in comparison to known standards, additional TLC was done in toluene-ethylacetate-acetic acid (50:30:4, vol/vol/vol) and chloroform-methanol (10:0.5, vol/vol) solvent systems. Mass spectrometry was performed by using a Micromass Autospec EBE magnetic sector instrument (Altrincham, Manchester, United Kingdom) as described (17).
Nucleotide sequence accession number.
The genomic DNA nucleotide sequence from ddhA to dotD, including the spacer regions reported here, has been submitted to GenBank under accession number AF448056.
RESULTS
Identification and characterization of dotA.
A Southern blot of D. pini genomic DNA showed strong hybridization of the A. parasiticus ver-1 probe to a single BamHI fragment of approximately 1.5 kb (data not shown). By using the same hybridization conditions, clone λCGV1 was selected from the genomic library for further analysis on the basis of its strong hybridization to ver-1. This clone contained 13.3 kb of D. pini genomic DNA. Restriction mapping and Southern hybridization showed that a 1.5-kb BamHI fragment of this clone hybridized to ver-1. Partial DNA sequence analysis of this fragment revealed similarity to the ver-1 gene: consequently, the entire 13.3-kb clone was subcloned and sequenced.
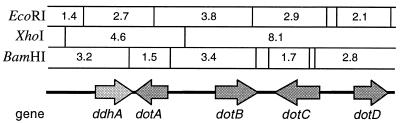
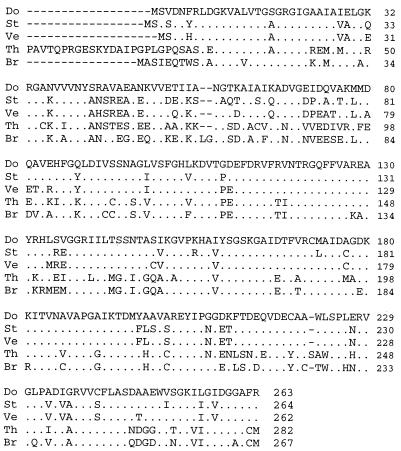
The λCGV1 sequence contained five predicted ORFs (Fig. 2), including one (dotA) that was homologous to the A. parasiticus ver-1 gene. The predicted 263-amino-acid sequence of the dotA product (Fig. 3) contains an adenine nucleotide binding motif found in other fungal ketoreductases, 19GXGIGX24. The proposed amino acid sequence of the dotA product has 80% identity to that of the A. parasiticus ver-1 product (aligned by using the Wisconsin Genetics Computer Group GAP program) and 79% to that of the sterigmatocystin gene stcU (Table 1). There are two introns in the dotA sequence; the first is in the same position as an intron found in the aflatoxin and sterigmatocystin homologs. Intron positions were confirmed by direct sequencing of cDNA obtained by RT-PCR by using primers designed to exons 1 and 3 of the dotA gene. Alignment with other fungal ketoreductases (Clustal W) showed a high level of amino acid identity throughout the predicted amino acid sequence (Fig. 4), particularly with those involved in aflatoxin biosynthesis.
FIG. 2.
Physical restriction map of the 13.3-kb genomic clone λCGV1 containing part of the dothistromin gene cluster (four dot genes) of D. pini. ddhA is not considered a functional part of the cluster (see text). Large arrows indicate the direction of transcription of each gene. Sizes of restriction fragments are indicated (in kilobases).
FIG. 3.
Nucleotide sequence of the dotA gene. The predicted amino acid sequence is shown below the corresponding codon, and the introns are in lowercase letters. Putative aflR-like regulatory binding sites are in bold type, and the positions of gene replacement primers 1 to 4 are underlined. Positions of BamHI and EcoRI sites referred to in Fig. 2 (opposite orientation) are dotted. Nucleotides 1966 to 1964 complement the final stop codon of the ddhA gene.
TABLE 1.
Probable homologs of D. pini dot genesa
| D. pini gene | Putative activity | Pathway | Species | Gene | % aa identity | GenBank accession no. | Reference |
|---|---|---|---|---|---|---|---|
| dotA | Ketoreductase | AF | Aspergillus parasiticus | ver-1 | 80.2 | M91369 | 45 |
| ST | Aspergillus nidulans | stcU | 79.1 | L27825 | 10 | ||
| MEL | Alternaria alternata | BRM2 | 64.6 | AB015743 | 35 | ||
| dotB | Oxidase | ST | Aspergillus nidulans | stcC | 24.0 | U34740 | 10 |
| CH | Caldariomyces fumago | CPO | 24.3 | X04486 | 40 | ||
| dotC | Toxin pump | AF | Aspergillus parasiticus | aftT | 31.2 | AF268071 | |
| HC | Cochliobolus carbonum | toxA | 30.8 | L48797 | 41 | ||
| dotD | Thioesterase | AF | Aspergillus parasiticus | pksL1 | 34.8 | L42766 | 24 |
| ST | Aspergillus nidulans | stcA | 37.9 | U34740 | 10 | ||
| MEL | Aspergillus fumigatus | alb1 | 43.6 | AF025541 | 49 |
Putative peptide activity is based on homology to products of aflatoxin (AF), sterigmatocystin (ST), melanin or other conidial pigment (MEL), CH chloroperoxidase (CH), or HC-toxin efflux pump (HC) filamentous fungal genes. Percent amino acid (aa) identity was calculated with the GAP program (31).
FIG. 4.
Alignment of deduced amino acid sequences: Do, D. pini dotA; St, A. nidulans stcU; Ve, A. parasiticus ver-1; Th, Magnaporthe grisea thnR (GenBank accession no. L22309); Br, Alternaria alternata BRM2. See Table 1 for other accession numbers. BRM2 and thnR are melanin pathway genes. Amino acids identical to those in the D. pini product are shown as dots, and gaps are shown as hyphens.
Identification of other genes clustered with dotA.
Of the four other ORFs in the 13.3-kb clone sequence, three (dotB, dotC, and dotD) show similarity to other aflatoxin genes (Table 1). BlastX matches to other (nonaflatoxin) fungal genes are included for comparison. The proposed dotB gene lies 2.3 kb away from dotA and contains an ORF of 1.2 kb. The 2.3-kb gap does not appear to contain any genes (BlastX and Wisconsin Genetics Computer Group GenScan analysis). The predicted amino acid sequence of the dotB product is similar to those of oxidases and chloroperoxidases of fungi and contains a putative heme-binding site, 49PCPALNALANHG60 (7). Although it shows similarity to the sterigmatocystin gene stcC (Table 1), no equivalent is known for A. parasiticus. The fungus Caldariomyces fumago, whose chloroperoxidase gene product showed the highest amino acid identity to DotB, is a loculoascomycete fungus in the same phylogenetic class as Dothistroma. There are no predicted introns in any of these three genes.
The dotC ORF lies 0.72 kb from dotB and is predicted to encode a hydrophobic 585-amino-acid protein with homology to fungal major facilitator superfamily transporters. DotC shows >30% identity with other fungal major facilitator superfamily proteins, in addition to those shown in Table 1, that have been proposed to export toxins, for example, CFP, a cercosporin transporter of Cercospora kikuchii (13). Like its A. parasiticus and C. kikuchii counterparts, the D. pini dotC gene has three predicted introns, but the positions of these are not conserved.
The dotD ORF lies 1.5 kb from dotC and is predicted to encode a 322-amino-acid polypeptide with homology to the thioesterase domains of polyketide synthase genes involved in aflatoxin, sterigmatocystin, and conidiospore pigment biosynthesis (Table 1). Rather than being part of a larger polyketide synthase gene, dotD is a complete ORF that appears to encode a monofunctional thioesterase enzyme. Since the thioesterase domain of a polyketide synthase is usually at the carboxyl terminus of a polyketide synthase protein, the orientation of dotD within the λCGV1 clone (Fig. 2) further suggests that this is a whole thioesterase gene rather than the tail end of a polyketide synthase gene.
The ORF ddhA encodes a predicted 469-amino-acid protein except for a stop signal at codon 64 that would prevent normal translation. RT-PCR with primers designed to coding regions showed that while the dotA to dotD genes are expressed when cells are grown in DM shake cultures, the ddhA gene is not (results not shown). However, disregarding the nonsense mutation, ddhA has strongest similarities to eubacterial and archaebacterial genes: 34% amino acid identity with a putative glucose/mannose dehydrogenase from Streptomyces coelicolor (AL391754) and 30% identity with a probable polysaccharide biosynthesis protein from Pyrococcus horikoshii (AP000002). There is no evidence to suggest similarity of ddhA with any genes involved in aflatoxin biosynthesis. Clustering of ddhA along with the dot genes in the D. pini genome was verified by Southern blotting. Each of the five ORFs hybridized to a single 21.2-kb XbaI genomic fragment (results not shown).
Utilization of sugar and nitrogen sources in dothistromin biosynthesis.
The production of dothistromin was monitored in shake flask cultures under conditions known to affect aflatoxin biosynthesis. Aflatoxin production is induced when cultures are transferred from peptone to glucose salts medium (46). This was mirrored by a higher level of dothistromin production in glucose-grown than in peptone-grown cultures (Table 2). Similarly, ammonium supports aflatoxin synthesis and nitrate represses it (16), while for sterigmatocystin the reverse has been observed (25). The pattern of dothistromin expression was similar to that of sterigmatocystin, with no dothistromin detectable in ammonium-containing medium despite good growth of the mycelium. However, RT-PCR with primers designed to the coding region of dotA detected a basal level of expression of dotA in ammonium as well as in nitrate and peptone medium (results not shown). Glucose medium supplemented with pine needles supported surprisingly low dothistromin production, although the mycelium dry weight was substantially increased compared to that of the glucose control. The amount of dothistromin in the culture medium declined over time. For example, after 10 days of growth in AMM (with glucose and nitrate), dothistromin levels dropped from 1.13 ± 0.10 μg/ml (7 days) to 0.23 ± 0.06 μg/ml despite an increase in mycelium dry weight from 0.91 ± 0.28 mg/ml to 2.45 ± 0.75 mg/ml over the same time period.
TABLE 2.
Dothistromin production and growth of D. pini NZEI (7-day shake flasks)a
| Carbon and nitrogen sources | Dothistromin (μg/ml) | Mycelium dry wt (mg/ml) |
|---|---|---|
| Glucose + NaNO3 | 1.13 ± 0.10 | 0.91 ± 0.28 |
| Peptone + NaNO3 | 0.09 ± 0.06 | 1.46 ± 0.82 |
| Glucose + NH4Cl | ND | 2.95 ± 0.65 |
| Pine needles + glucose + NaNO3 | 0.61 ± 0.25 | 5.91 ± 0.62 |
Results are means ± SEM (n = 3). ND, not detected. Least significant difference (P = 0.05) = 0.45 μg of dothistromin and 2.03 mg of mycelium per ml.
The nucleotide sequences of the dot genes were analyzed for matches to the AflR regulatory protein-binding motif (TCG SWNNS CGR) found in A. parasiticus aflatoxin genes (22). Two of the five D. pini ORFs have a matching sequence upstream of the predicted translation start site: dotA at −205 and dotD at −267. The dotB ORF has a degenerate form (TCG N5 CGC) at −517. However, all five ORFs have at least one copy of the related sequence TCG N11 CGA within 500 bp upstream of the predicted translation start site. The two versions (N5 and N11) of this putative regulatory motif are highlighted in the upstream region of dotA in Fig. 3. A search for an aflR-like gene in the D. pini genome was made by using degenerate nested PCR with primers designed to conserved motifs of AflR proteins from Aspergillus spp. No aflR-like genes were recovered from D. pini with this screen.
Construction and characterization of D. pini dotA mutants.
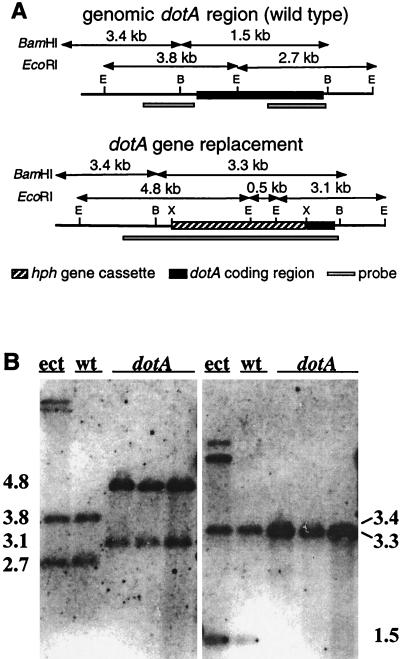
Following transformation of D. pini NZE5 with the gene replacement construct pR208, two dotA mutants (no. 32 and 34) were obtained along with many ectopic transformants (5.4% targeting efficiency). Targeted replacement of the dotA gene was confirmed in D. pini mutants by Southern blotting and hybridization. The results for mutant no. 32 and two replicates (independent single-spore isolates) of mutant no. 34 are shown in Fig. 5A and B. The identity of these mutants as D. pini was confirmed by ribosomal DNA-internal transcribed spacer DNA amplification and DNA sequence comparison to control strains (9). Purified dotA mutants spot inoculated onto AMM plus glucose plates produced a bright yellow pigment after 10 days of incubation at 22°C that was indicative of versicolorin accumulation. Yellow pigment was not evident in the wild-type (untransformed) or ectopic strains.
FIG. 5.
(A) Structure of the D. pini dotA locus before and after gene replacement. Restriction sites: B, BamHI; E, EcoRI. (B) Southern analysis of the wild-type (wt) strain NZE5, ectopic transformant (ect), and dotA mutant transformants (dotA) after digestion with EcoRI (left) or BamHI (right) and hybridization with the hph cassette and dotA flanking regions, as indicated in panel A. Size markers are indicated (in kilobases).
Measurements of radial colony growth on agar plates suggested that the dotA mutants grow more slowly than the wild-type parent strain. After 10 days on AMM plus glucose, the dotA mutant no. 34 had radial growth of 4.2 ± 0.17 mm (mean ± standard error of the mean [SEM]; n = 6), which was significantly less than NZE5, with 5.9 ± 0.08 mm (T = 10.8; df = 10; P < 0.05). After 4 weeks of steady growth, the difference in growth rates was still significant: dotA mutant no. 34, 10.7 ± 0.42 mm; NZE5, 13.2 ± 0.17 mm (T = 5.5; df = 10; P < 0.05). There were no significant differences in growth rates between the two dotA mutant strains.
Identification of metabolites accumulated by dotA mutants.
ELISAs showed that the wild-type isolate produced dothistromin in the range from 225 to 980 ng/ml (n = 3) after 7 days of incubation. Both dotA mutants, however, consistently produced ≤22.5 ng/ml, which was the lower limit of resolution of the ELISA. Mycelium biomass of the mutants was 76 to 95% of the wild-type yield under these growth conditions. Dothistromin production by the mutants did not increase during prolonged incubation for 10 days. Within the limitations of this assay, it is evident that the mutants produced at least 10-fold less dothistromin than the wild-type strain.
Mass spectrometry and TLC in several solvent systems showed the production by the wild-type strain of large amounts of a red compound that was indistinguishable from dothistromin. This compound was not detected in the dotA mutant. Hence, this was further evidence that the dotA mutant is impaired in dothistromin biosynthesis. TLC analysis followed by mass spectrometry indicated the accumulation of versicolorin A in the dotA mutant at much higher levels than those found in the wild type. This is consistent with the expectation that the dotA mutant is blocked in dothistromin production at a step equivalent to that blocked in ver-1 mutants of A. parasiticus (37). Although D. pini makes the aflatoxin precursor versicolorin A, no aflatoxin was detected from either the wild-type or the mutant D. pini strain by either TLC or mass spectrometry.
DISCUSSION
The high level of predicted amino acid identity between DotA and A. parasiticus Ver-1 suggests a ketoreductase function for DotA. Furthermore, the accumulation of versicolorin A by the dotA mutant suggests similar biosynthetic roles for the two enzymes. Although the precise function of the Ver-1 ketoreductase is not known, it is thought to be required, along with a dehydratase enzyme, in a two-step dehydroxylation reaction to convert versicolorin A to 6-deoxyversicolorin A (37).
Dothistromin shares a saturated bisfuran ring with versicolorin B but differs in the arrangement of hydroxyl groups on the anthraquinone rings (Fig. 1). In view of this, it might be expected that versicolorin B is the substrate of DotA rather than the unsaturated bisfuran form, versicolorin A, that accumulated in the dotA mutant. However, both versicolorin A and versicolorin B can serve as substrates for the homologous A. nidulans sterigmatocystin ketoreductase enzyme StcU (36) and the desaturase enzyme (52) in aflatoxigenic fungi. Versicolorin A might be the DotA substrate and the direct precursor of dothistromin. Alternatively, versicolorin B might be the DotA substrate, with versicolorin A produced in the mutant as a by-product when versicolorin B accumulates. Other unsaturated bisfuran structures related to dothistromin have been found in D. pini (19), and hence it is likely that a desaturase ortholog exists in D. pini that could convert versicolorin B to versicolorin A.
The clustering of other aflatoxin-like genes with dotA was expected in view of the gene clustering seen in aflatoxigenic fungi, although no conservation of gene order is apparent. Of the linked dot genes, the lowest amino acid identity to aflatoxin genes was shown by dotB, a possible homolog of the A. nidulans stcC oxidase gene. There is little information on the role of stcC, but the presence of a homolog in the dothistromin cluster would suggest a function before the versicolorin A/B branch point at which the dothistromin and aflatoxin pathways appear to diverge.
The predicted DotC transporter is a member of the major facilitator superfamily proteins. Many fungi have similar proteins that are involved in the efflux of natural toxic compounds and fungicides. Botryotinia fuckeliana has at least three major facilitator superfamily transporter proteins, including Bcmfs1 (Table 1), along with at least 10 ABC (ATP-binding cassette) transporters that are also involved in multidrug resistance (50). It has been proposed that some fungal efflux pumps contribute to self-protection against the toxin. For example, the toxA gene of Cochliobolus carbonum encodes an efflux pump that appears to be essential for the survival of strains producing the phytotoxin HC-toxin (41).
The role of the AflT transporter (P.-K. Chang, J. Yu, D. Bhatnagar, and T. E. Cleveland, paper presented at the USDA-ARS Aflatoxin Elimination Workshop, St. Louis, Mo., 25 to 27 October 1998, p. 501, abstr. no. O-31) in aflatoxin-producing fungi is not clear, particularly since there are no reports of a similar gene in the sterigmatocystin cluster. It is feasible that the DotC transporter is required to transport dothistromin out of the cell so it can reach its target in the host plant, but it is also possible that DotC is necessary to protect the cell against autotoxicity. In either case, it would be a good target for control of the pathogen. However, there may be other mechanisms in place to avoid dothistromin toxicity to the fungal cells. The observation that dothistromin levels in the culture medium were lower at 10 days than at 7 days suggests that dothistromin may be metabolized, conjugated, or otherwise inactivated in culture. Similarly, sterigmatocystin is degraded in cultures of A. nidulans over time (Nancy Keller, personal communication). In plant cells, dothistromin is degraded by photolytic degradation and/or peroxide-catalyzed oxidation with H2O2 to yield the products CO2 and oxalic acid (26).
The discovery of a putative monofunctional thioesterase gene (dotD) was at first surprising because the large type I polyketide synthases that function early in the aflatoxin pathways are multifunctional enzymes with a thioesterase domain as one component. The role of the thioesterase domain is to release the polyketide product from the polyketide synthase complex. Two aflatoxin polyketide synthase gene sequences have been published for different strains of A. parasiticus (pksA [15] and pksL1 [24]). While only one group reported a thioesterase domain, the sequences are very similar and both contain this domain. However, many other fungal polyketide synthase genes, such as the wA spore pigment polyketide synthase gene of A. nidulans (38), do not include a thioesterase domain. Currently under investigation in our laboratory is a D. pini library clone with part of a polyketide synthase gene matching other domains of A. parasiticus aflatoxin polyketide synthase genes.
The ddhA gene was not considered a dot gene on the basis of its similarity to polysaccharide biosynthesis genes and its lack of similarity to fungal aflatoxin/sterigmatocystin genes. It is possible that ddhA marks one end of a cluster of dothistromin genes, although it was considerably closer (0.23 kb) to dotA than the A. parasiticus sugar utilization cluster genes that mark the end of the aflatoxin cluster are to the aflatoxin gene moxY (5 kb) (54). Moreover, while sugar utilization genes in A. parasiticus were speculated to have a role in allowing uptake of sugars for aflatoxin biosynthesis, no functional significance is apparent for polysaccharide biosynthesis in the expression or biological activity of dothistromin.
Dothistromin expression strongly mirrored that of sterigmatocystin production by A. nidulans rather than aflatoxin by A. parasiticus in that ammonium strongly repressed while nitrate supported the production of dothistromin in culture. However, while the growth of A. nidulans was severely impaired in ammonium medium compared to growth in nitrate (25), D. pini grew significantly better in ammonium (Table 2). An inverse relationship between growth rate and secondary metabolite production has been well established for some fungi (1), and in general it does appear that higher growth rates are associated with lower dothistromin levels (Table 2). However, higher rates of dothistromin inactivation or degradation (26) could also account for the lower levels of dothistromin seen in some cases. An apparent contradiction of this inverse relationship was shown by the low growth rates of two independent dotA (dothistromin-deficient) mutants compared to that of the wild-type strain. Although more rigorous growth comparisons under a range of conditions are required, it is feasible that dothistromin production may in fact confer some benefits on the fungus.
It is difficult to predict the biological importance of toxins to the fungi that make them. Laboratory and field tests have indicated that the trichothecene group of mycotoxins play an important role as virulence factors in wheat head blight and maize ear rot caused by Fusarium graminearum (20, 30). However, fumonisins, produced by another Fusarium species (F. verticillioides), are not required for maize ear rot (21). Similarly, although A. parasiticus and A. flavus act as weak pathogens (51), the ability of A. flavus to produce aflatoxins is independent of its ability to infect and multiply in crops (18).
The similarity of the genes within the putative dothistromin and aflatoxin clusters so far leads us to question whether the genes are regulated in the same manner and whether aflatoxin can be produced by dothistromin-producing fungi and vice versa. It is not yet known whether the dothistromin cluster contains an aflR-like regulatory gene, although sequences identical to the TCG N5 CGA aflR binding sites were seen in dotA and dotD. The presence of TCG N11 CGA sequences in all four dot genes and the ddhA gene leads to the speculation that there is a different regulatory gene for dothistromin. However, the expression of dotA in ammonium medium (dothistromin repressing) suggests that dothistromin expression is regulated in a manner different from that for aflatoxin pathway genes.
With regard to metabolite production, no aflatoxin was detected in D. pini cultures. Conversely, in metabolite feeding experiments carried out by the method of Bhatnagar and coworkers (5), dothistromin was not converted to aflatoxin in A. parasiticus. Even though it is possible that dothistromin may not have been taken up by the mycelia, it is also possible that A. parasiticus may lack a set of enzymes that convert dothistromin to versicolorin A for conversion of versicolorin A to aflatoxins.
In summary, the D. pini dotA gene, which is homologous to the ketoreductases of aflatoxin biosynthetic pathways, was shown to be involved in dothistromin biosynthesis. Genes adjacent to dotA also show similarities to genes in the aflatoxin and sterigmatocystin clusters. Further work will allow us to test whether dothistromin is a pathogenicity factor and will provide an interesting comparison of the biochemistry, genetics, and biology between aflatoxin and dothistromin toxins.
Acknowledgments
This work was supported by the NZ Lottery Sciences Board and the Massey University Research Fund. J.M.S. acknowledges the support of the Robert C. Bruce Trust and the NZ Plant Protection Society.
We gratefully acknowledge the help of Stephen Boue (USDA, ARS, New Orleans, La.) for mass spectral analysis. We thank John Linz (Michigan State University) for supplying the ver-1 gene probe and Bill Jones and Dawn Harvey for the dothistromin ELISA.
This work was carried out in compliance with the current laws governing genetic experimentation in New Zealand.
REFERENCES
- 1.Adams, T. H., and J.-H. Yu. 1998. Coordinate control of secondary metabolite production and asexual sporulation in Aspergillus nidulans. Curr. Opin. Microbiol. 1:674-677. [DOI] [PubMed] [Google Scholar]
- 2.Al-Samarrai, T. H., and J. Schmid. 2000. A simple method for extraction of fungal genomic DNA. Lett. Appl. Microbiol. 30:53-56. [DOI] [PubMed] [Google Scholar]
- 3.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. A. Smith, J. G. Seidman, and K. Struhl. 1990. Current protocols in molecular biology. John Wiley and Sons, New York, N.Y.
- 4.Bear, C. A., J. M. Waters, and T. N. Waters. 1972. Crystal structure and absolute configuration of a derivative of dothistromin, a fungal toxin implicated in pine-needle blight. J. Chem. Soc. Perkin Trans. II:2375-2378. [Google Scholar]
- 5.Bhatnagar, D., S. P. McCormick, L. S. Lee, and R. A. Hill. 1987. Identification of O-methylsterigmatocystin as an aflatoxin B1 and G1 precursor in Aspergillus parasiticus. Appl. Environ. Microbiol. 53:1028-1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bhatnagar, D., K. C. Ehrlich, and T. E. Cleveland. Molecular genetic analysis of aflatoxin biosynthesis and its application to understanding the biological role of aflatoxin. Appl. Microbiol. Biotechnol., in press. [DOI] [PubMed]
- 7.Blanke, S. R., and L. P. Hager. 1988. Identification of the fifth axial heme ligand of chloroperoxidase. J. Biol. Chem. 263:18739-18743. [PubMed] [Google Scholar]
- 8.Bradshaw, R. E., A. Bidlake, N. Forester, and D. B. Scott. 1997. Transformation of the fungal forest pathogen Dothistroma pini to hygromycin resistance. Mycol. Res. 101:1247-1250. [Google Scholar]
- 9.Bradshaw, R. E., R. J. Ganley, W. T. Jones, and P. S. Dyer. 2000. High levels of dothstromin toxin produced by the forest pathogen Dothistroma pini. Mycol. Res. 104:325-332. [Google Scholar]
- 10.Brown, D. W., J.-H. Yu, H. S. Kelkar, M. Fernandes, T. C. Nesbitt, N. P. Keller, T. H. Adams, and T. J. Leonard. 1996. Twenty-five coregulated transcripts define a sterigmatocystin gene cluster in Aspergillus nidulans. Proc. Natl. Acad. Sci. USA 93:1418-1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brown, M. P., C. S. Brown-Jenco, and G. A. Payne. 1999. Genetic and molecular analysis of aflatoxin biosynthesis. Fungal Genet. Biol. 26:81-98. [DOI] [PubMed] [Google Scholar]
- 12.Bullock, W. O., J. M. Fernandez, and J. M. Short. 1987. XL-1 blue: a high efficiency plasmid transforming recA Escherichia coli strain with a β-galactosidase selection. BioTechniques 5:376-378. [Google Scholar]
- 13.Callahan, T. M., M. S. Rose, M. J. Meade, M. Ehrenshaft, and R. G. Upchurch. 1999. CFP, the putative cercosporin transporter of Cercospora kikuchii, is required for wild-type cercosporin production, resistance, and virulence on soybean. Mol. Plant-Microbe Interact. 12:901-910. [DOI] [PubMed] [Google Scholar]
- 14.Cary, J. W., P.-K. Chang, and D. Bhatnagar. 2001. Clustered metabolic pathway genes in filamentous fungi. Appl. Mycol. Biotechnol. 1:165-198. [Google Scholar]
- 15.Chang, P.-K., J. W. Cary, J. Yu, D. Bhatnagar, and T. E. Cleveland. 1995. The Aspergillus parasiticus polyketide synthase gene pksA, a homolog of Aspergillus nidulans wA, is required for aflatoxin B1 biosynthesis. Mol. Gen. Genet. 248:270-277. [DOI] [PubMed] [Google Scholar]
- 16.Chang, P.-K., K. C. Ehrlich, J. Yu, D. Bhatnagar, and T. E. Cleveland. 1995. Increased expression of Aspergillus parasiticus aflR, encoding a sequence-specific DNA-binding protein, relieves nitrate inhibition of aflatoxin biosynthesis. Appl. Environ. Microbiol. 61:2372-2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chang, P.-K., J. Yu, K. C. Ehrlich, S. M. Boue, B. G. Montalbano, D. Bhatnagar, and T. E. Cleveland. 2000. adhA in Aspergillus parasiticus is involved in conversion of 5′-hydroxyaverantin to averufin. Appl. Environ. Microbiol. 66:4715-4719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cotty, P. J. 1989. Virulence and cultural characteristics of two Aspergillus flavus strains pathogenic on cotton. Phytopathologia 79:808-814. [Google Scholar]
- 19.Danks, A. V., and R. Hodges. 1974. Polyhydroxyanthraquinones from Dothistroma pini. Aust. J. Chem. 27:1603-1606. [Google Scholar]
- 20.Desjardins, A. E., and T. M. Hohn. 1997. Mycotoxins in plant pathogenesis. Mol. Plant-Microbe Interact. 10:147-152. [Google Scholar]
- 21.Desjardins, A. E., and R. D. Plattner. 2000. Fumonisin B1-nonproducing strains of Fusarium verticillioides cause maize (Zea mays) ear infection and ear rot. J. Agric. Food Chem. 48:5773-5780. [DOI] [PubMed] [Google Scholar]
- 22.Ehrlich, K. C., B. G. Montalbano, and J. W. Cary. 1999. Binding of the C6-zinc cluster protein, AFLR, to the promoters of aflatoxin biosynthetic genes in Aspergillus parasiticus. Gene 230:249-257. [DOI] [PubMed] [Google Scholar]
- 23.Elliott, G. S., R. W. Mason, D. G. Ferry, and I. R. Edwards. 1989. Dothistromin risk assessment for forestry workers. N.Z. J. Forest Sci. 19:163-170. [Google Scholar]
- 24.Feng, G. H., and T. J. Leonard. 1995. Characterization of the polyketide synthase gene (pksL1) required for aflatoxin biosynthesis in Aspergillus parasiticus. J. Bacteriol. 177:6246-6254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Feng, G. H., and T. J. Leonard. 1998. Culture conditions control expression of the genes for aflatoxin and sterigmatocystin biosynthesis in Aspergillus parasiticus and A. nidulans. Appl. Environ. Microbiol. 64:2275-2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Franich, R. A., M. J. Carson, and S. D. Carson. 1986. Synthesis and accumulation of benzoic acid in Pinus radiata needles in response to tissue injury by dothistromin, and correlation with resistance of P. radiata families to Dothistroma pini. Physiol. Mol. Plant Pathol. 28:267-286. [Google Scholar]
- 27.Gallagher, R. T., and R. Hodges. 1972. The chemistry of dothsitromin, a difuroanthraquinone from Dothistroma pini. Aust. J. Chem. 25:2399-2407. [Google Scholar]
- 28.Gibson, I. A. S. 1972. Dothistroma blight of Pinus radiata. Annu. Rev. Phytopathol. 10:51-72. [Google Scholar]
- 29.Guzmán-de-Peña, D., and J. Ruiz-Herrera. 1997. Relationship between aflatoxin biosynthesis and sporulation in Aspergillus parasiticus. Fungal Genet. Biol. 21:198-205. [DOI] [PubMed] [Google Scholar]
- 30.Harris, L. J., A. E. Desjardins, R. D. Plattner, P. Nicholson, G. Butler, J. C. Young, G. Weston, R. H. Proctor, and T. M. Hohn. 1999. Possible role of trichothecene mycotoxins in virulence of Fusarium graminearum on maize. Plant Dis. 83:954-960. [DOI] [PubMed] [Google Scholar]
- 31.Henikoff, S., and J. G. Henikoff. 1992. Amino acid substitution matrices from protein blocks. Proc. Natl. Acad. Sci. USA 89:10915-10919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hirst, P., T. E. Richardson, S. D. Carson, and R. E. Bradshaw. 1999. Dothistroma pini genetic diversity is low in New Zealand. N.Z. J. Forest Sci. 29:459-472. [Google Scholar]
- 33.Jones, W. T., D. Harvey, S. D. Jones, S. Fielder, P. Debnam, and P. H. S. Reynolds. 1993. Competitive ELISA employing monoclonal antibodies specific for dothistromin. Food Agric. Immunol. 5:187-197. [Google Scholar]
- 34.Jones, W. T., D. Harvey, S. D. Jones, P. W. Sutherland, M. J. Nicol, N. Sergejew, P. M. Debnam, N. Cranshaw, and P. H. S. Reynolds. 1995. Interaction between the phytotoxin dothistromin and Pinus radiata embryos. Phytopathologia 85:1099-1104. [Google Scholar]
- 35.Kawamura, C., T. Tsujimoto, and T. Tsuge. 1999. Targeted gene disruption of a melanin biosynthesis gene affects conidial development and UV tolerance in the Japanese pear pathotype of Alternaria alternata. Mol. Plant-Microbe Interact. 12:59-63. [DOI] [PubMed] [Google Scholar]
- 36.Kelkar, H. S., T. W. Skloss, J. F. Haw, N. P. Keller, and T. H. Adams. 1997. Aspergillus nidulans stcL encodes a putative cytochrome P-450 monooxygenase required for bisfuran desaturation during aflatoxin/sterigmatocystin biosynthesis. J. Biol. Chem. 272:1589-1594. [DOI] [PubMed] [Google Scholar]
- 37.Liang, S.-H., C. D. Skory, and J. E. Linz. 1996. Characterization of the function of the ver-1A and ver-1B genes, involved in aflatoxin biosynthesis in Aspergillus parasiticus. Appl. Environ. Microbiol. 62:4568-4575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mayorga, M. E., and W. E. Timberlake. 1992. The developmentally regulated Aspergillus nidulans wA gene encodes a polypeptide homologous to polyketide and fatty acid synthases. Mol. Gen. Genet. 235:205-212. [DOI] [PubMed] [Google Scholar]
- 39.Murray, N. E. W., W. J. Brammar, and K. Murray. 1977. Lambdoid phages that simplify the recovery of in vitro recombinants. Mol. Gen. Genet. 150:53-58. [DOI] [PubMed] [Google Scholar]
- 40.Nuell, M. J., G.-H. Fang, M. J. Axley, P. Kenigsberg, and L. P. Hager. 1988. Isolation and nucleotide sequence of the chloroperoxidase gene from Caldariomyces fumago. J. Bacteriol. 170:1007-1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pitkin, J. W., D. G. Panaccione, and J. D. Walton. 1996. A putative cyclic peptide efflux pump encoded by the TOXA gene of the plant-pathogenic fungus Cochliobolus carbonum. Microbiology 142:1557-1565. [DOI] [PubMed] [Google Scholar]
- 42.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 43.Shain, L., and R. A. Franich. 1981. Induction of Dothistroma blight symptoms with dothistromin. Physiol. Plant Pathol. 19:49-55. [Google Scholar]
- 44.Shaw, G. J., M. Chick, and R. Hodges. 1978. A 13C NMR study of the biosynthesis of the anthraquinone dothistromin by Dothistroma pini. Phytochemistry 17:1743-1745. [Google Scholar]
- 45.Skory, C. D., P.-K. Chang, J. Cary, and J. E. Linz. 1992. Isolation and characterization of a gene from Aspergillus parasiticus associated with the conversion of versicolorin A to sterigmatocystin in aflatoxin biosynthesis. Appl. Environ. Microbiol. 58:3527-3537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Skory, C. D., P.-K. Chang, and J. E. Linz. 1993. Regulated expression of the nor-1 and ver-1 genes associated with aflatoxin biosynthesis. Appl. Environ. Microbiol. 59:1642-1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stoessl, A., and J. B. Stothers. 1985. Minor anthraquinoid metabolites of Cercospora arachidicola. Can. J. Chem. 63:1258-1262. [Google Scholar]
- 48.Stoessl, A., Z. Abramowski, H. H. Lester, G. L. Rock, and G. H. N. Towers. 1990. Further toxic properties of the fungal metabolite dothistromin. Mycopathology 112:179-186. [DOI] [PubMed] [Google Scholar]
- 49.Tsai, H.-F., Y. C. Chang, R. G. Washburn, M. H. Wheeler, and K. J. Kwon-Chung. 1998. The developmentally regulated alb1 gene of Aspergillus fumigatus: its role in modulation of conidial morphology and virulence. J. Bacteriol. 180:3031-3038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vermeulen, T., H. Schoonbeek, and M. A. De Waard. 2001. The ABC transporter BcatrB from Botrytis cinerea is a determinant of the activity of the phenylpyrrole fungicide fludioxonil. Pest Manag. Sci. 57:393-402. [DOI] [PubMed] [Google Scholar]
- 51.Xu, H., A. Annis, J. Linz, and F. Trail. 2000. Infection and colonization of peanut pods by Aspergillus parasiticus and the expression of the aflatoxin biosynthetic gene, nor-1, in infection hyphae. Physiol. Mol. Plant Pathol. 56:185-196. [Google Scholar]
- 52.Yabe, K., Y. Ando, and T. Hamasaki. 1991. Desaturase activity in the branching step between aflatoxins B1 and G1 and aflatoxins B2 and G2. Agric. Biol. Chem. 55:1907-1911. [Google Scholar]
- 53.Yu, J., P.-K. Chang, D. Bhatnagar, and T. E. Cleveland. 2000. Genes encoding cytochrome P450 and monooxygenase enzymes define one end of the aflatoxin pathway gene cluster in Aspergillus parasiticus. Appl. Microbiol. Biotechnol. 53:583-590. [DOI] [PubMed] [Google Scholar]
- 54.Yu, J., P.-K. Chang, D. Bhatnagar, and T. E. Cleveland. 2000. Cloning of a sugar utilisation gene cluster in Aspergillus parasiticus. Biochim. Biophys. Acta 1493:211-214. [DOI] [PubMed] [Google Scholar]