Abstract
A strain that efficiently degraded methyl tert-butyl ether (MTBE) was obtained by initial selection on the recalcitrant compound tert-butyl alcohol (TBA). This strain, a gram-positive methylotrophic bacterium identified as Mycobacterium austroafricanum IFP 2012, was also able to degrade tert-amyl methyl ether and tert-amyl alcohol. Ethyl tert-butyl ether was weakly degraded. tert-Butyl formate and 2-hydroxy isobutyrate (HIBA), two intermediates in the MTBE catabolism pathway, were detected during growth on MTBE. A positive effect of Co2+ during growth of M. austroafricanum IFP 2012 on HIBA was demonstrated. The specific rate of MTBE degradation was 0.6 mmol/h/g (dry weight) of cells, and the biomass yield on MTBE was 0.44 g (dry weight) per g of MTBE. MTBE, TBA, and HIBA degradation activities were induced by MTBE and TBA, and TBA was a good inducer. Involvement of at least one monooxygenase during degradation of MTBE and TBA was shown by (i) the requirement for oxygen, (ii) the production of propylene epoxide from propylene by MTBE- or TBA- grown cells, and (iii) the inhibition of MTBE or TBA degradation and of propylene epoxide production by acetylene. No cytochrome P-450 was detected in MTBE- or TBA-grown cells. Similar protein profiles were obtained after sodium dodecyl sulfate-polyacrylamide gel electrophoresis of crude extracts from MTBE- and TBA-grown cells. Among the polypeptides induced by these substrates, two polypeptides (66 and 27 kDa) exhibited strong similarities with known oxidoreductases.
Methyl tert-butyl ether (MTBE) has been incorporated in reformulated gasoline at concentrations up to 15% (vol/vol) to replace lead tetraethyl in order to comply with the octane index and to reduce the polluting emissions in exhaust gases. Other oxygenates, such as ethyl tert-butyl ether (ETBE) and tert-amyl methyl ether (TAME) and their corresponding alcohols, tert-butyl alcohol (TBA) and tert-amyl alcohol (TAA), can play the same role regarding the octane index. MTBE is the dominant fuel oxygenate (38), with a worldwide production capacity of around 25 million tons. Because of its widespread use and the high frequency of underground tank leakage (35), this compound is now the second most commonly detected contaminant in urban groundwater in the United States (23, 42). In Europe, detectable levels of MTBE in rivers have also been reported (1).
The persistence of MTBE in the environment can be ascribed, on the one hand, to its physicochemical properties (i.e., its low adsorption on organic matter and its high solubility in water) and, on the other hand, to its molecular structure (it has both an ether bond and high steric hindrance, which makes it recalcitrant to microbial degradation).
Early reports based on microcosms having different origins mentioned the recalcitrance of MTBE and TAME (22). Nevertheless, a stable aerobic mixed culture that degraded MTBE via TBA was obtained (31). MTBE biodegradation by microcosms having different origins was reported later (5, 24, 45; K. Park and R. M. Cowan, Preprints Extend. Abstr. Am. Chem. Soc. Div. Environ. Chem. 37:421-424, 1997).
MTBE cometabolism was demonstrated by using propane-grown bacteria (35), an n-butane-grown fungus (17), camphor-grown Pseudomonas putida CAM (36), pentane-grown Pseudomonas aeruginosa (14), ETBE-grown Rhodococcus ruber IFP 2007 (20), and a cyclohexane-grown mixed culture (9). Steffan et al. (36) proposed a pathway for MTBE biodegradation in which MTBE was sequentially degraded to tert-butyl formate (TBF), tert-butyl alcohol, and 2-hydroxy isobutyrate (HIBA). All these systems involved cytochrome P-450 monooxygenases. The genetic organization of the cytochrome P-450 of R. ruber IFP 2007 responsible for ETBE and MTBE oxidation was elucidated by Chauvaux et al. (7).
Pure cultures of only two strains able to use MTBE as a sole carbon and energy source have been isolated previously; these strains are Rubrivivax gelatinosus PM1 (8, 10, 11, 16) and Hydrogenophaga flava ENV 735 (19, 37).
In our laboratory, a strain was isolated from activated sludge based on its ability to grow on TBA and was identified as Mycobacterium austroafricanum IFP 2012 (13). The capacity of this strain to degrade and grow on MTBE as a sole carbon and energy source is described in this paper.
New evidence concerning the MTBE degradation pathway and the nature of the enzymes involved in this pathway is also presented in this paper. Our results also point out differences among the MTBE oxidation systems of M. austroafricanum IFP 2012, R. gelatinosus PM1, and H. flava ENV 735.
MATERIALS AND METHODS
Isolation.
The ability of an activated sludge sample from an urban wastewater treatment plant located near Paris, France, to degrade 200 mg of different fuel oxygenates per liter was evaluated by continuously monitoring oxygen consumption by electrolytic respirometry (Sapromat Apparatus, Voith, Germany). TBA degraders were enriched and isolated from the sludge by five successive additions of TBA and by plating on solid medium containing TBA as the sole carbon and energy source. The pure strain isolated was deposited in the CNCM, Pasteur Institute, Paris, France, as strain CIP I-2562. Stock cultures were kept frozen at −80°C in 20% (vol/vol) glycerol.
16S rDNA and hsp65 analyses.
16S rDNA amplification was performed by using reverse primer 5′-GAGAGTTTGATCCTGGCTCAG -3′ (primer Bott 2) and forward primer 5′-TGCACACAGGCCACAACCCA-3′ (primer Bott 1). The products of amplification were purified with a Qiagen kit, and the sequence was determined by using primer 244 (5′-CCCACTGCTGCCTCCCGTAG-3′).
hsp65 gene amplification was performed by using forward primer Tb11 (5′-ACCAACGATGGTGTGTCCAT-3′) and reverse primer Tb12 (5′-CTTGTCGAACCGCATACCCT-3′) as described by Telenti et al. (39). The products of amplification were purified with a Qiagen kit, and the sequence was determined by using primer Tb11.
In each case, the percentage of identity was determined by using the EMBL/GenBank database with the Blast alignment tool (2).
Growth medium and culture conditions.
M. austroafricanum IFP 2012 was grown in Luria-Bertani medium (LB) (3) or in the defined mineral medium (MM) previously described by Piveteau et al. (29), and the carbon source was added before inoculation. In the latter case, the volume of the headspace was sufficient to prevent any limitation by O2 during growth. When mentioned below, yeast extract (YE) was added to MM to a final concentration of 100 mg/liter. Cultures were grown aerobically in conical flasks and incubated at 30°C with constant agitation.
Growth was monitored by measuring the optical density at 600 nm (OD600) with a UV-1601 spectrophotometer (Shimadzu Corporation, Kyoto, Japan). Unless otherwise stated, tightly closed flasks were inoculated to obtain an initial OD600 of 0.1. From a linear standard curve relating OD600 to cell dry weight, it could be determined that 1 OD600 unit corresponded to a cell dry weight of 0.43 ± 0.01 g/liter (n = 28).
Mineralization yield and carbon recovery.
Cells grown on MM containing TBA (1 g/liter) were harvested by centrifugation, washed twice in phosphate buffer (20 mM, pH 7), and suspended in 80 ml of MM in a 670-ml sidearm flask with a septum. The carbon source was added at the required concentration.
For each experiment four trials were performed. In one of these trials growth was monitored by measuring either CO2 production (mineralization yield) or the OD600 and the residual substrate concentration (carbon recovery). When the maximum growth was reached, 2.4 ml of HNO3 (68%, vol/vol ) was added through the septum in each of the three other sidearm flasks, and the final CO2 concentration in the gaseous phase was measured.
Biomass production based on the difference between the final and initial OD600 values was calculated by using the relationship between optical density and dry weight. The carbon contained in the biomass was evaluated by using C4H8NO2 to determine the composition of the biomass (43).
Degradation assay with resting cells.
M. austroafricanum IFP 2012 was cultivated on MM containing TBA or MTBE or on LB. After growth, cells at an OD600 of 1 were harvested by centrifugation at 23,000 × g for 15 min and washed twice in phosphate buffer (20 mM, pH 7). The cells were resuspended to obtain an initial OD600 of 0.4 in 40 ml of phosphate buffer containing the substrate in 250-ml sealed flasks. After inoculation, the flasks were incubated at 30°C on an orbital shaker. Samples were filtered through 0.22-μm-pore-size filters (Prolabo, Fontenay sous Bois, France) before analysis. Substrate degradation was monitored over a 24-h period by chromatography.
For assays under anaerobic conditions, the cells obtained from aerobic cultures were transferred to an anaerobic chamber under strict anaerobic conditions. Before use, the buffer was boiled with N2 flushing for 1 h and transferred to a glove chamber.
Growth inhibition by acetylene.
Cells were grown on TBA (1 g/liter), collected by centrifugation, washed, resuspended (OD600, 0.2) in 40 ml of MM with the required carbon source, and placed in 250-ml flasks sealed with rubber stoppers. Acetylene (0.4%, vol/vol) was added immediately after inoculation. Growth was monitored by measuring the OD600.
Kinetic analysis of propylene oxidation.
M. austroafricanum IFP 2012 was cultivated on MM supplemented with TBA or MTBE. When the OD600 was 0.5, the cells were centrifuged, washed twice, resuspended in 15 ml of phosphate buffer to obtain an OD600 of 1, and placed in flasks sealed with rubber stoppers. The experiment was started by injecting propylene (40% [vol/vol] in the gas phase). The flasks were incubated at 40°C with agitation. Filtered samples (pore size, 0.22 μm) were analyzed by gas chromatography, and the time course of propylene oxide production was monitored over a 2-h period. To study the effect of acetylene on propylene oxidation, cells resuspended in phosphate buffer were exposed to acetylene (40% [vol/vol] in the gas phase) for 10 min with agitation at 30°C. Acetylene was removed from the flasks by flushing them with air for 15 min. Propylene was then added, and the production of propylene oxide was monitored over time.
Oxygen uptake rate.
TBA-grown cells were collected by centrifugation, washed twice, and resuspended in phosphate buffer (20 mM, pH 7). TBA was added, and the O2 consumption rate was measured with a 12-mm O2 sensor (Ingold, Mettler-Toledo, Paris, France). The measured rates were corrected for endogenous consumption.
Cell extract preparation.
Cells were grown on the appropriate substrate in 300 ml of MM to an OD600 of 1, harvested by centrifugation at 20,000 × g for 15 min, washed twice in phosphate buffer (20 mM, pH 7), and resuspended in 5 ml of phosphate buffer. Cells were broken by three passes through a French press (20,000 lb/in2) and were always maintained on ice. Cell debris was removed by centrifugation twice at 1,000 × g for 2 min. The resulting supernatant was used for an enzymatic activity test and for protein pattern analysis.
Gel electrophoresis.
Soluble proteins from the cell extract were obtained by centrifugation at 15,000 × g for 30 min; the supernatant fraction contained the soluble proteins. Total proteins were assayed by using a Bio-Rad protein assay (Bio-Rad, Munich, Germany) and were analyzed by SDS-7.5 or 15% polyacrylamide gel electrophoresis (PAGE) (26). After electrophoresis, the gels were stained with Coomassie blue.
Analytical assays.
MTBE, ETBE, TAME, TBA, TAA, methanol, ethanol, TBF, isopropanol, acetone, and propylene oxide were quantified by gas chromatography with a Varian 3300 gas chromatograph (Varian, Les Ulis, France) fitted with a flame ionization detector and equipped with a Porabond-Q capillary column (0.32 mm by 25 m; J & W Scientific, Chromoptic, Auxerre, France) by using a two-step temperature gradient in which the temperature increased from 105 to 200°C at a rate of 10°C/min. Helium (1.6 ml/min) was used as the carrier gas. Samples were filtered through 0.22-μm-pore-size filters and were injected without further treatment into the chromatograph.
HIBA, lactate, methacrylate, and formate were quantified by using a high-performance liquid chromatograph (HPLC) (Metrohm) equipped with an anion column (Dual 2; 75 by 4.6 mm) and a conductimetric detector (Metrohm 732). The eluent, a mixture of 90% (vol/vol) 0.05 M sulfuric acid and 10% (vol/vol) acetonitrile, was previously flushed with a Spectra-Physics SCM 400 vacuum flusher and was injected at a rate of 0.5 ml/liter.
The formaldehyde concentration was determined by colorimetry as described by Werringloer (44).
Gaseous carbon dioxide was quantified by thermal conductivity detection with a Varian 3800 gas chromatograph fitted with a Porapak Q column (1,830 by 2 mm). The oven and detector temperatures were 100 and 130°C, respectively. The carrier gas was helium (30 ml/min), and the column was maintained at 50°C.
Chemicals.
ETBE, MTBE, TBA, TAA, TAME, HIBA, methacrylate, and propylene oxide were obtained from Sigma-Aldrich (Saint-Quentin-Fallavier, France). Glucose, methanol, ethanol, isopropanol, formaldehyde, formate, HNO3, toluene, m-xylene, p-xylene, and acetone were supplied by Prolabo. Propylene and acetylene were obtained from Air Liquide (Paris-La Défense, France). All chemicals were the highest purity available.
Nucleotide sequence accession numbers.
The 16S rRNA and hsp65 gene sequences of the new strain M. austroafricanum IFP 2012 have been deposited in the GenBank nucleotide sequence database under accession no. AF487529 and AF487530, respectively.
RESULTS
Isolation and characterization of a TBA-degrading strain.
An activated sludge sample was able to degrade 100% of 200 mg of TBA per liter, 200 mg of TAA per liter, 200 mg of TAME per liter, or 200 mg of ETBE per liter and 41% of 200 mg of MTBE per liter. After the substrate was exhausted, the adaptation phase was extended by periodic additions of substrate over a 2-month period. In the case of TBA, only one strain was isolated after several enrichment steps in MM containing TBA as the sole carbon and energy source and by streaking on solid medium containing TBA. This strain was a gram-positive, strictly aerobic, noncapsulated, nonsporulated rod and formed yellow colonies on solid medium.
Sequence comparison with sequences in the EMBL/GenBank database identified the new isolate as M. austroafricanum, with 97% identity with the hsp65 gene of a strain of M. austroafricanum (accession number AJ310221) (6) and 100% identity with the 16S rDNA of M. austroafricanum IFP 2173 (accession number AF190800) (34). The hsp65 gene, which codes for a 65-kDa heat shock protein, is present in all mycobacteria. It exhibits greater variability than the 16S rDNA gene sequence and is therefore potentially useful for identification of genetically related species. Sequence variations in the hsp65 gene can be exploited to identify mycobacteria to the species level (39).
Degradation capacities of M. austroafricanum IFP 2012.
M. austroafricanum IFP 2012 was tested for its ability to degrade and mineralize fuel oxygenates (MTBE, ETBE, TAME, TBA, and TAA) and C1 compounds (methanol and formate) (Table 1). All of the substrates except ETBE were well degraded. No residual alcohol was detected during growth on TAME and growth on MTBE. TBA was the most readily assimilable substrate. TAME and TAA were both completely degraded within 14 days, whereas about 1 month was necessary for degradation and mineralization of MTBE. Even after 42 days, ETBE was not completely degraded.
TABLE 1.
Degradation and mineralization of gasoline oxygenates and C1 compounds by M. austroafricanum IFP 2012
| Substratea | Incubation time (days) | Degradation (%)b | Mineralization (%)b |
|---|---|---|---|
| MTBE | 30 | 100 | 68.7 (2.4) |
| TAME | 14 | 100 | 54.6 (1.1) |
| ETBE | 42 | 89c | 41.4 (1.9) |
| TBA | 3 | 100 | 54.0 (0.7) |
| TAA | 14 | 100 | 54.7 (0.2) |
| Methanol | 8 | 100 | 50.4 (1.5) |
| Formate | 5 | 100 | 89.1 (0.8) |
Substrates were added at final concentrations of 300 mg/liter for MTBE, TAME, ETBE, methanol, and formate, 250 mg/liter for TBA and TAA, and 200 mg/liter for formaldehyde. The substrate losses measured with abiotic controls for MTBE, TAME, ETBE, TBA, TAA, methanol, and formate were 9.9 (2.1), 20.0 (1.3), 32.0 (0.2), 0.8 (0.3), 3.6 (0.9), 13.3 (9.2), and 22.5 (13.2) mg/liter [mean (standard deviation)], respectively.
Mean (standard deviation) for three samples.
The residual TBA concentration was 25.9 mg/liter (standard deviation, 0.6 mg/liter).
M. austroafricanum IFP 2012 was clearly shown to be able to degrade and mineralize methanol and formate with high mineralization yields compared with those of other methylotrophic strains (4).
Growth of M. austroafricanum IFP 2012 on MTBE and other substrates and metabolite identification.
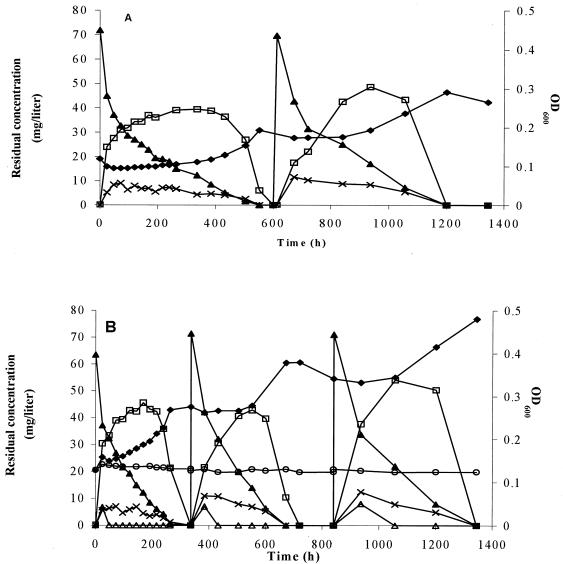
The growth of M. austroafricanum IFP 2012 on MTBE alone was slow and could be divided into two phases (Fig. 1A). In the first phase, MTBE was degraded while TBA accumulated, and this occurred at a nearly constant cell concentration. During the second phase, biomass was readily produced from the TBA accumulated during the first phase. This sequential growth profile could be correlated with the changes in CO2 production observed over time while the organism was growing on MTBE (data not shown). On this substrate, CO2 was produced from TBA after a long lag which corresponded to the conversion of MTBE to TBA. In addition to TBA, another metabolite was detected during growth of M. austroafricanum IFP 2012 on MTBE. This compound was identified as TBF by coupling gas chromatography to mass spectrophotometry. The amounts of TBF and TBA transiently produced after 14 days accounted for 85% (molar) of the amount of MTBE degraded. No other MTBE degradation intermediate could be detected by gas chromatography or HPLC analyses. A second addition of 70 mg of MTBE per liter resulted in similar growth.
FIG. 1.
Growth of M. austroafricanum IFP 2012 on MTBE without (A) and with (B) YE (100 mg/liter). During growth, OD600 (♦) and the concentrations of MTBE (▴), TBF (×), TBA (□), and HIBA (▵) were monitored. In the control (○) growth of M. austroafricanum IFP 2012 on YE (100 mg/liter) as the only carbon source was monitored.
Although 100 mg of YE per liter did not support growth of M. austroafricanum IFP 2012 by itself, it clearly had a stimulatory effect on MTBE consumption (Fig. 1B). With YE only 29 days was required for utilization of 140 mg of MTBE per liter supplied in two successive equivalent additions, compared to the 50 days required for utilization without YE. Analysis of the supernatant revealed transient accumulation of another soluble product of MTBE degradation detected only by HPLC. This compound was identified as HIBA by HPLC-mass spectrophotometry.
A similar effect of YE was observed when glucose was used instead of MTBE as the carbon source (data not shown). Addition of 200 mg of Casamino Acids per liter to MM was more beneficial than addition of the same amount of YE with either MTBE or glucose. This result is different from the results obtained with H. flava ENV 735 (19), which, although requiring YE for good growth, did not require amino acids or vitamins.
Calculation of the carbon balance of M. austroafricanum IFP 2012 growing on MTBE, TBA, or HIBA showed that all of the carbon of each substrate was finally recovered as biomass and CO2 (Table 2). Maximum growth rates were calculated for growth on TBA and HIBA but not for growth on MTBE because no exponential phase was observed with MTBE. The biomass yield (in grams of biomass produced per gram of substrate consumed) was higher with TBA than with MTBE or HIBA.
TABLE 2.
Growth parameters of M. austroafricanum IFP 2012 on MTBE and its metabolites
| Growth substrate | Maximum growth rate (h−1)a,b | Biomass yield (g/g)b | Carbon recovery (%)b,c |
|---|---|---|---|
| MTBE | NDd | 0.436 (0.01) | 99.7 (3.9) |
| TBA | 0.039 (0.001) | 0.609 (0.02) | 97.3 (1.9) |
| HIBA | 0.061 (0.001) | 0.422 (0.01) | 101.6 (1.8) |
Substrates were added at final concentrations of 350 mg/liter for TBA and 250 mg/liter for HIBA.
Mean (standard deviation) for three samples.
Substrates were added at final concentrations of 625 mg/liter for TBA and HIBA and 200 mg/liter−1 for MTBE. The initial OD600 were 0.1 for the experiments with TBA and HIBA and 0.4 for the experiment with MTBE. At the end of growth, the amount of biomass, the amount of CO2 produced, and the residual carbon source amount were determined.
ND, not determined.
The ability of M. austroafricanum IFP 2012 to grow on isopropanol (400 mg/liter), acetone (400 mg/liter), and methacrylate (100 mg/liter), three postulated intermediates of MTBE degradation (36), was demonstrated. Increases in the OD600 of 0.9, 0.7, and 0.3 U were obtained with isopropanol, acetone, and methacrylate, respectively. Growth on C1 compounds was poor. An increase of only 0.15 OD600 unit was obtained with methanol (after three successive additions of 200 mg/liter). Growth on formate was not measurable. Nevertheless, none of the postulated intermediates of the MTBE pathway (isopropanol, acetone, methacrylate, formaldehyde, and formate) was detected in this study.
Moreover, M. austroafricanum IFP 2012 was able to grow on 250 mg of toluene per liter, 250 mg of p-xylene per liter, and 250 mg of m-xylene per liter, with increases in the OD600 of 0.25, 0.3, and 0.3 U, respectively. In each case, with cells precultivated on TBA a lag phase was observed before growth, suggesting that the enzymatic systems responsible for degradation of TBA and for degradation of p-xylene, m-xylene, and toluene were different.
MTBE degradation capacity of M. austroafricanum IFP 2012 resting cells.
MTBE, TBA, and HIBA degradation activities were assessed by using resting cells of M. austroafricanum IFP 2012 grown on MTBE, TBA, and LB (Table 3). MTBE degradation, TBA degradation, and HIBA degradation were monitored as a function of time. No lag phase occurred with any substrate with MTBE- and TBA-grown cells.
TABLE 3.
Degradation activities of M. austroafricanum IFP 2012 resting cells with MTBE and related substrates
| Substrate for resting cell production | Specific degradation activity (μmol/h/g of biomass) witha:
|
||
|---|---|---|---|
| MTBEb | TBA | HIBA | |
| MTBE | 263 (21)c | 248 (10) | 498 (22) |
| TBA | 597 (47) | 315 (11) | 540 (33) |
| LB | 11 (4) | 65 (3) | 41 (4) |
Substrates were added at final concentrations of 340 μM for MTBE and HIBA and 470 μM for TBA. Mean (standard deviation) for three samples unless indicated otherwise.
The product of MTBE degradation was TBA.
Mean (standard deviation) for two samples.
MTBE degradation occurred after growth on MTBE or TBA. Moreover, the MTBE degradation activity was twice as high when TBA-grown cells were used as when MTBE-grown cells were used, suggesting that TBA could be a good inducer of the MTBE degradation pathway. When 30 mg of MTBE per liter was used, the TBA produced from MTBE was not degraded. The molar ratios of the amount of TBA plus TBF produced to the amount of MTBE utilized were 0.68 ± 0.08 and 0.69 ± 0.01 for MTBE- and TBA- grown resting cells, respectively. When a lower initial concentration of MTBE (20 mg/liter) was used, the TBA produced from MTBE was totally degraded. However, TBA consumption took place when the MTBE concentration was less than 3 mg/liter. Besides, when TBA was used as a substrate, it was degraded immediately (results not shown). The activities of resting cells with TBA and HIBA were unchanged when MTBE or TBA was used as the growth substrate.
After growth on LB, a noninducing medium, a lag phase occurred before degradation of each of the three substrates began. LB-grown cells were finally able to degrade MTBE, TBA, and HIBA, but at different levels; TBA and HIBA were completely degraded after 100 h, whereas only around 57% of MTBE was degraded during the same period. This could mean that LB-grown cells required a longer adaptation time on MTBE than on TBA or HIBA.
Involvement of a nonhemic monooxygenase system in MTBE and TBA degradation.
No MTBE and TBA degradation activities could be observed in cell extracts from MTBE- and TBA-grown cells. The nature of the systems involved in the degradation of these compounds was investigated.
First, oxygen was necessary for MTBE or TBA degradation by resting cells of M. austroafricanum IFP 2012. The average residual concentrations of MTBE and TBA after 24 h of incubation under aerobic conditions were 10 and 23%, respectively, while they were 45 and 75%, respectively, under anaerobic conditions.
When crude extracts of MTBE- or TBA-grown cells were used, no cytochrome P-450 was detected by observation of spectra obtained after reaction with carbon monoxide (33). In addition, no staining of the soluble protein from TBA-grown cells separated by gel electrophoresis was observed when a stain specific for cytochromeP-450 (3,3′,5,5′-tetramethylbenzidine) was used (40). The absence of cytochrome P-450 in cell extracts was consistent with production of propylene oxide from propylene by resting cells grown on MTBE (179 ± 4 μmol of propylene oxide/h/g of biomass) or on TBA (303 ± 14 μmol of propylene oxide/h/g of biomass) as alkenes are known inhibitors of cytochrome P-450 (21).
In the presence of 10 mM methimazole, a specific inhibitor of flavin-containing monooxygenases (41), no growth of M. austroafricanum IFP 2012 was detected when TBA (800 mg/liter) was used as a carbon source. A similar inhibitory effect was detected (to a lesser extent) when HIBA (800 mg/liter) was used as the growth substrate; the maximum OD600 in the presence of the same concentration of methimazole was one-half the maximum OD600 in the absence of the inhibitor. This inhibitor had no effect on the growth of M. austroafricanum IFP 2012 on 1 g of glucose per liter.
Growth of M. austroafricanum IFP 2012 on MTBE and TBA was completely inhibited in the presence of acetylene at a concentration as low as 0.4% (vol/vol) in the gas phase. In contrast, growth on HIBA or LB was not affected whether acetylene was added or not. Moreover, exposure of TBA-grown cells to a higher concentration of acetylene (40% [vol/vol] in the gas phase) resulted in loss of 86 and 99% of the MTBE- and TBA-specific degradation activities of resting cells, respectively. Propylene oxide production by resting cells was also affected by exposure to this alkyne. The production of propylene oxide by TBA-grown cells dropped from 386 ± 23 to 40 ± 14 μmol of propylene oxide/h/g of biomass when these cells were previously exposed to acetylene. In these experiments, acetylene was flushed away by air before propylene was added to the assay mixture, suggesting that acetylene acted as an irreversible inactivator rather than as a competitive inhibitor. All these results confirmed that monooxygenase-type enzymes were involved in the oxidation of MTBE and TBA by M. austroafricanum IFP 2012. In addition, the monooxygenase involved in oxidation of MTBE has a behavior similar to that of the monooxygenase involved in oxidation of TBA.
The apparent half-saturation constant (Ks) towards TBA of TBA-grown whole cells was estimated by monitoring O2 consumption and was found to be 1.1 mM. The Ks for MTBE could not be measured by the same method, probably because of a higher affinity of the system for MTBE.
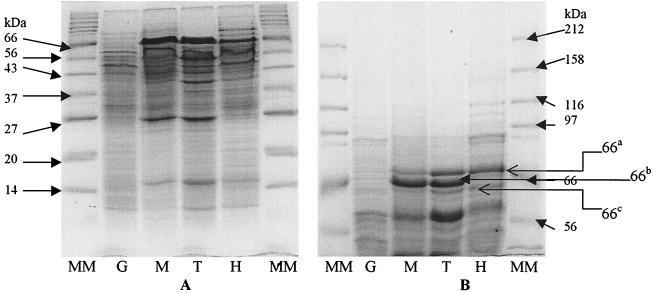
The protein patterns of M. austroafricanum IFP 2012 cellular extracts after growth on MTBE and after growth on TBA were quite similar (Fig. 2). Compared with the protein pattern obtained with glucose-grown cells, several polypeptides were specifically induced with MTBE or TBA, as shown by the bands around 66 kDa (bands 66a, 66b, and 66c) and the bands at 40, 27, and 15 kDa. These induced polypeptides were purified by collecting the bands, subsequently digested with trypsin, and microsequenced by the Edman reaction, giving 20-amino-acid peptides. The 40-and 15-kDa bands, which were also present in isopropanol- and acetone-grown cell extracts, were not taken into account. Analysis of the resulting sequences by using the Blast (2) and Fasta (27) alignment tools revealed that the band 66c and 66b and 27-kDa proteins had high levels of similarity with known proteins (90% similarity with a 68-kDa mycobacterial antigen, 92% similarity with a GMP family oxidoreductase of Caulobacter crescentus, and 57% similarity with a eucaryotic dehydrogenase of Rattus norvegicus, respectively). Band 66a was also present in the pattern of HIBA-grown cells, and the corresponding polypeptide was probably involved in degradation of HIBA. The polypeptides corresponding to band 66b and the 27-kDa band seem to correspond to oxidoreductases involved in MTBE and TBA degradation. Characterization of these bands is being investigated. Moreover, a comparison of the protein patterns in membrane fractions from glucose- and TBA-grown cells did not reveal any difference (data not shown).
FIG. 2.
SDS-PAGE of M. austroafricanum IFP 2012 grown on various substrates. Portions (25 μg) of the total proteins of crude extracts of cells grown on glucose (lane G), MTBE (lane M), TBA (lane T), and HIBA (lane H) were analyzed by SDS-15% PAGE (A) or SDS-7.5% PAGE (B). Lanes MM contained protein standards (7702S; BioLabs).
Selective effect of Co2+ during growth on MTBE of M. austroafricanum IFP 2012.
It was previously shown that Co2+ had a positive effect on the growth of Burkholderia cepacia IFP 2003 on MM containing TBA and cobalt in two forms, CoCl2 at a concentration of 1 mg/liter and cyanocobalamine at a concentration of 1.5 μg/liter (29). The effect of Co2+ on the growth of M. austroafricanum IFP 2012 on several substrates was also investigated (Table 4). The presence of Co2+ in the mineral culture medium had no effect on the growth of M. austroafricanum IFP 2012 on ethanol, isopropanol, and acetone. In contrast, growth on TBA and growth on HIBA were increased 5.5- and 2-fold, respectively, by the presence of this cation.
TABLE 4.
Effect of cobalt on growth of M. austroafricanum IFP 2012
| Substratea | Growth rate (mg of biomass/h/liter)
|
|
|---|---|---|
| Without CoCl2 | With 1 mg of CoCl2 liter−1 | |
| Ethanol | 4.9 | 4.7 |
| Acetone | 2.2 | 2.2 |
| Isopropanol | 6.7 | 7.6 |
| HIBA | 4.9 | 10.3 |
| TBA | 2.6 | 14.4 |
Substrates were added at final concentrations of 5, 2, 3, 3, and 7 g/liter for ethanol, acetone, isopropanol, HIBA, and TBA, respectively, by using three successive equivalent additions. The initial OD600 was 0.05 in each trial, and growth was monitored by measuring the OD600 for 240 h.
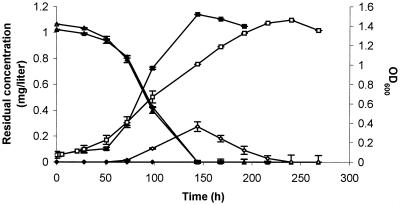
In addition, when TBA was used as a carbon source (Fig 3) in the absence of Co2+, significant transient accumulation of HIBA was detected at 20°C (but not at 30°C), which was not the case when Co2+ cations were supplied in the culture medium. Co2+ ions have no effect on TBA degradation but a strong positive effect on growth, which could be ascribed to their effect on HIBA degradation. Indeed, during growth on HIBA in absence of Co2+, the degradation of HIBA was about one-half that in the presence of the cation (79 ± 11 versus 152 ± 5 mg of HIBA degraded/h/g of biomass). This suggested that no HIBA was detected during growth on TBA in the presence of Co2+ because the HIBA degradation rate was at least equal to the TBA degradation rate in this case.
FIG. 3.
Growth of M. austroafricanum IFP 2012 on TBA in the presence and in the absence of CoCl2. The strain was cultivated on MM containing 1 g of TBA per liter and 1 mg of CoCl2 per liter. Cells were harvested and washed twice in MM without CoCl2. The final cell suspension was used to inoculate MM containing 1 g of TBA per liter in the presence (triplicates) and in the absence (triplicates) of 1 mg of CoCl2 per liter, which was incubated at 20°C with agitation. During growth, OD600 was monitored in cultures with (▪) and without (□) CoCl2. The concentrations of the following residual substrates were measured in cultures: TBA with (▴) and without (▵) CoCl2 and HIBA with (♦) and without (⋄) CoCl2.
Adding 1 mg of Co2+ per liter to Co2+-starved cells led to a strong and immediate increase in growth on HIBA, whereas no effect was observed when the same amount of cyanocobalamine was added (results not shown).
DISCUSSION
M. austroafricanum IFP 2012 is the third pure bacterial strain which is able to grow on MTBE as a sole carbon and energy source and to mineralize it. In contrast to R. gelatinosus PM1 and H. flava ENV 735, M. austroafricanum IFP 2012 is gram positive, and it was isolated by using enrichment medium containing TBA, not MTBE. An MTBE degradation rate of 20 nmol/min/mg of cell protein (calculated by assuming a cell protein content of 50%) was obtained for M. austroafricanum IFP 2012, and this rate is very similar to the rate reported for R. gelatinosus PM1 (19 nmol/min/mg of cell protein). The MTBE degradation rate for H. flava ENV 735 is higher, 86 nmol/min/mg of cell protein.
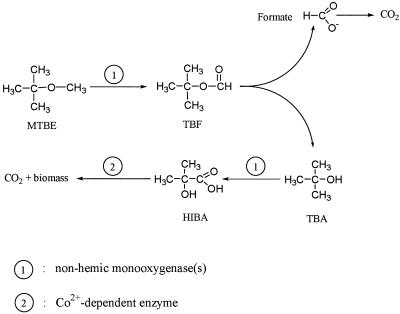
The pathway for cometabolic degradation of MTBE was previously proposed to occur sequentially via TBF (17) to TBA to 2-methyl-2-hydroxy-1-propanol and then to HIBA (36). A similar pathway was possibly used by strains using MTBE as a carbon and energy source since HIBA and TBA were detected in studies in which either H. flava ENV 735 (19) or M. austroafricanum IFP 2012 (this study) was used. Moreover, we also detected TBF and identified it as an intermediate of MTBE degradation. Oxidation of MTBE probably produced the hemiacetal intermediate tert-butoxy methanol (12). The latter compound can produce either formaldehyde plus TBA or TBF, which could be further degraded into formate and TBA. The fact that TBF was identified during growth of M. austroafricanum IFP 2012 on MTBE supports the latter pathway, thereby excluding any formation of formaldehyde (Fig. 4). The fate of HIBA was not elucidated since no degradation compounds were detected when HIBA was used as a growth substrate. Nevertheless, it should be mentioned that M. austroafricanum IFP 2012 was able to grow on isopropanol and acetone, and moreover, the protein patterns obtained with isopropanol- and acetone-grown cells contained the same strongly expressed 40- and 27-kDa polypeptides as the patterns obtained with cells grown on MTBE, TBA, and HIBA; these results are consistent with the HIBA degradation pathway proposed by Steffan et al. (36).
FIG. 4.
Pathway for MTBE degradation in M. austroafricanum IFP 2012.
Understanding the physiology of strains growing on MTBE is important for elucidating the limitations on MTBE degradation in the environment. In fact, such strains are difficult to isolate because of their very slow growth on MTBE. The reasons for this slow growth are still unclear, as discussed by Salanitro (32) and Hatzinger et al. (19). In the case of M. austroafricanum IFP 2012, we observed at least two reasons for its slow growth on MTBE: (i) the cleavage of the ether bond leading to conversion of MTBE to TBA at a nearly stoichiometric ratio before TBA was utilized for growth and (ii) the requirement of the strain for an essential oligo element, Co2+, for good assimilation of accumulated TBA.
To address the first reason, investigations were done to determine (i) the nature of the enzymatic systems involved in MTBE and TBA degradation by M. austroafricanum IFP 2012 and (ii) the specificity of M. austroafricanum IFP 2012 with other ethers (TAME and ETBE), which could also provide interesting data about structural limitations during the initial attack on the ether bond. With respect to the enzymatic systems involved, all of the results confirmed that monooxygenase-type enzymes were involved in the oxidation of MTBE and TBA: (i) oxygen was required for MTBE and TBA degradation, (ii) propylene oxide was produced from propylene by MTBE- and TBA-grown cells, (iii) propylene oxide production and MTBE and TBA degradation were inactivated by acetylene, a mechanism-based inactivator of several moonoxygenases (15), and (iv) growth on TBA was inhibited by addition of 10 mM methimazole, an inhibitor of flavin-containing monooxygenases (41). All these observations show that at least one monooxygenase is involved in MTBE and TBA degradation. Since no cytochrome P-450 could be detected and since propylene oxide was produced, the monooxygenase was likely to be nonhemic; however, no activity could be measured in crude cell extracts. Characterization of the gene(s) involved in MTBE and/or TBA degradation is being investigated to address this point. In H. flava ENV 735, the enzymatic system responsible for MTBE degradation was different from that responsible for TBA degradation, as shown by the responses of the systems to oxygenase inhibitors (19). If only one monooxygenase were involved in degradation of both MTBE and TBA, MTBE catabolism could be slowed down. In M. austroafricanum IFP 2012, the monooxygenase involved in oxidation of MTBE was strongly related to that involved in oxidation of TBA, as shown by the following findings: (i) MTBE and TBA degradation activities were both inducible; (ii) TBA was a better inducer of MTBE degradation activity than MTBE itself; (iii) degradation of MTBE and TBA were both inactivated by the addition of acetylene (which also stopped growth); (iv) during growth on MTBE, TBA was first accumulated and was consumed only when MTBE was totally degraded; and, finally, (v) the protein patterns of MTBE- and TBA-grown cells of M. austroafricanum IFP 2012 were identical. The latter point suggests that the same oxygenase catalyses both MTBE oxidation and TBA oxidation. If this is the case, the oxygenase has a higher affinity for MTBE than for TBA, which results in accumulation of TBA from MTBE, followed by consumption of TBA when the concentration of MTBE is low. However, the resolution of SDS-PAGE may not be sufficient to separate all the polypeptides expressed. Therefore, we cannot rule out the possibility that two different oxygenases, both induced by TBA, are involved and that the TBA-oxidizing enzyme is inhibited by high concentrations of MTBE. In the case of H. flava ENV 735, MTBE degradation activity was expressed constitutively after growth on LB (19), whereas TBA degradation was induced by TBA and acetylene only inhibited degradation of TBA. In R. gelatinosus PM1, the MTBE degradation pathway was inducible, but two different enzymatic systems were required since TBA-grown cells did not consume O2 when they were exposed to MTBE (11).
The specificity of MTBE degraders regarding the utilization of other ethers could be very useful for understanding the mechanism of the initial attack on the ether bond. Like R. gelatinosus PM1, M. austroafricanum IFP 2012 was able to grow on TAME. In addition, M. austroafricanum IFP 2012 grew slowly on ETBE. Both TAME and MTBE contain a methoxy group, whereas ETBE contains an ethoxy group; these groups release C1 and C2 compounds, respectively. The alcohols TBA and TAA produced from the ethers are well utilized by the strain. These results confirm that the initial steps, from the ether to the tertiary alcohol, are crucial and are highly dependent upon the structure of the ether. Additionally, it is interesting that methylotrophy is a common trait of the three strains that degrade MTBE; M. austroafricanum IFP 2012 is able to grow on methanol and to mineralize methanol and formate, R. gelatinosus PM1 is able to grow on formate and formaldehyde (11), and H. flava ENV 735 is able to grow on formaldehyde (19). This explains why no compounds arising from the release of the methoxy group are detected during growth on MTBE. The involvement of methylotrophs in MTBE biodegradation should be addressed in more detail.
Concerning the second possible limiting point during MTBE degradation, Hatzinger et al. (19) recently suggested that certain cofactors or inducers could be necessary for efficient growth of H. flava ENV 735 on downstream metabolites of MTBE. The positive effect of CoCl2 on TBA degradation by B. cepacia IFP 2003 was shown previously (29). The present study showed that M. austroafricanum IFP 2012 required addition of Co2+ specifically during HIBA catabolism. The possible involvement of cobalt in a cobalamin-dependent enzymatic system was not considered for M. austroafricanum IFP 2012 because (i) the growth medium already contained cyanocobalamin and (ii) addition of cyanocobalamin had no effect. Kobayashi and Shimizu (25) reviewed all known non-corrin-cobalt-containing enzymes. To date, cobalt is known to be involved in the structure of eight different enzymes, including a carboxyltransferase (18) and a mutase (28, 30). It plays a role in stabilization of the three-dimensional structure of proteins by binding with specific amino acids. It is worth mentioning that a decarboxylation step was required for HIBA catabolism.
The use of M. austroafricanum IFP 2012 in bioremediation processes should be facilitated by the facts that (i) TBA-grown cells are able to degrade MTBE and (ii) biomass production on TBA is good (0.61 g·g−1). Thus, biomass that efficiently degrades MTBE should be easily produced. For use in contaminated aquifers, M. austroafricanum IFP 2012 has additional interesting capacities as it is able to grow on toluene , p-xylene, and m-xylene.
The present results highlight the biodiversity of strains that degrade MTBE, both taxonomically and enzymatically. Such diversity justifies investigations of the genes involved in fuel oxygenate degradation (7). Detection of these genes in the environment would be a suitable approach for evaluating the existing potential for natural attenuation of MTBE.
Acknowledgments
We thank P. Beguin, S. Chauvaux, C. Gaillardin, R. Marchal, and J. P. Vandecasteele for helpful discussions. We also thank D. Lyew for linguistic advice. We are indebted to A. Fafet and J. O. Païssé for mass spectrophotometry analyses and to C. Le Dantec for 16S rDNA and hsp65 analyses. We are grateful to J. d'Alayer for sequencing peptides from TBA-induced proteins.
REFERENCES
- 1.Achten, C., and W. Puttmann. 2000. Determination of methyl tert-butyl ether in surface water by use of solid-phase microextraction. Environ. Sci. Technol. 34:1359-1364. [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ausebel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1990. Current protocols in molecular biology. Greene Publishing and Wiley Interscience, New York, N.Y.
- 4.Babel, W. 1984. Utilization of C1 compunds by acidophiles, p. 141-146. In R. L. Crawford and R. S. Hanson (ed.), Microbial growth on C1 compounds. American Society for Microbiology, Washington, D.C.
- 5.Bradley, P. M., F. H. Chappelle, and J. E. Landmeyer. 2001. Methyl t-butyl ether mineralization in surface-water sediment microcosms under denitrifying conditions. Appl. Environ. Microbial. 67:1975-1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brunello, F., M. Ligozzi, E. Cristelli, S. Bonora, E. Tortoli, and R. Fontana. 2001. Identification of 54 mycobacterial species by PCR-restriction fragment length polymorphism analysis of the hsp65 gene. J. Clin. Microbiol. 39:2799-2806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chauvaux, S., F. Chevalier, C. Le Dantec, F. Fayolle, I. Miras, F. Kunst, and P. Béguin. 2001. Cloning of a genetically unstable cytochrome P-450 gene cluster involved in the degradation of the pollutant ethyl tert-butyl ether by Rhodococcus ruber. J. Bacteriol. 183:6551-6557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen, C.-I., H. Hohnbaum, A. Smith, K. Scow, D. Chang, and K. Jackson. 2000. Scale-up of PM-1 MTBE-degrading culture production for field application, p. 282-283. In D. L. Drogos and A. L. Diaz (ed.), Exploring the environmental issues of mobile, recalcitrant compounds in gasoline. ACS, San Francisco, Calif.
- 9.Corcho, D., R. J. Watkinson, and D. N. Lerner. 2000. Cometabolic degradation of MTBE by a cyclohexane-oxidising culture, p. 183-189. In G. B. Wickramanayake, A. R. Gavaskar, B. C. Alleman, and V. S. Magar (ed.), Bioremediation and phytoremediation of chlorinated and recalcitrant compounds. Batelle Press, Columbus, Ohio.
- 10.Deeb, R. A., H.-Y. Hu, J. R. Hanson, K. M. Scow, and L. Alvarez-Cohen. 2001. Substrate interactions in BTEX and MTBE mixtures by an MTBE-degrading isolate. Environ. Sci. Technol. 35:312-317. [DOI] [PubMed] [Google Scholar]
- 11.Deeb, R. A., S. Nishino, J. Spain, H.-Y. Hu, K. Scow, and L. Alvarez-Cohen. 2000. MTBE and benzene biodegradation by a bacterial isolate via two independent monooxygenase-initiated pathways, p. 280-282. In D. L. Drogos and A. L. Diaz (ed.), Exploring the environmental issues of mobile, recalcitrant compounds in gasoline. ACS, San Francisco, Calif.
- 12.Fayolle, F., J.-P. Vandecasteele, and F. Monot. 2001. Microbial degradation and fate in the environment of methyl tert-butyl ether and related fuel oxygenates. Appl. Microbiol. Biotechnol. 56:339-349. [DOI] [PubMed] [Google Scholar]
- 13.François, A., P. Piveteau, F. Fayolle, R. Marchal, P. Béguin, and F. Monot. 2001. Selection of a defined mixed culture for MTBE mineralization, p. 153-160. In V. S. Magar, J. T. Gibbs, K. T. O’Reilly, M. R. Hyman, and A. Leesou (ed.), Bioremediation of MTBE, alcohols, and ethers. Battelle Press, Columbus, Ohio.
- 14.Garnier, P. M., R. Auria, C. Augur, and S. Revah. 1999. Cometabolic biodegradation of methyl t-butyl ether by Pseudomonas aeruginosa grown on pentane. Appl. Microbiol. Biotechnol. 51:498-503. [DOI] [PubMed] [Google Scholar]
- 15.Hamamura, N., R. T. Storfa, L. Semprini, and D. J. Arp. 1999. Diversity in butane monooxygenases among butane-grown bacteria. Appl. Environ. Microbiol. 65:4586-4592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hanson, J. R., C. E. Ackerman, and K. M. Scow. 1999. Biodegradation of methyl tert-butyl ether by a bacterial pure culture. Appl. Environ. Microbiol. 65:4788-4792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hardison, L. K., S. Currys, L. M. Ciuffetti, and M. R. Hyman. 1997. Metabolism of diethyl ether and cometabolism of methyl tert-butyl ether by a filamentous fungus, a Graphium sp. Appl. Environ. Microbiol. 63:3059-3067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harmon, F. R., N. H. Goss, and H. G. Wood. 1982. Stabilization of the quaternary structure of transcarboxylase by cobalt(II) ions. Biochemistry 21:2847-2852. [DOI] [PubMed] [Google Scholar]
- 19.Hatzinger, P. B., K. McClay, S. Vainberg, M. Tugusheva, C. W. Condee, and R. J. Steffan. 2001. Biodegradation of methyl tert-butyl ether by a pure bacterial culture. Appl. Environ. Microbiol. 67:5601-5607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hernandez-Perez, G., F. Fayolle, and J.-P. Vandecasteele. 2001. Biodegradation of ethyl t-butyl ether (ETBE), methyl t-butyl ether (MTBE) and t-amyl methyl ether (TAME) by Gordonia terrae. Appl. Microbiol. Biotechnol. 55:117-121. [DOI] [PubMed] [Google Scholar]
- 21.Hyman, M., P. Kwon, K. Williamson, and K. O'Reilly. 1998. Cometabolism of MTBE by alkane-utilizing micoorganisms, p. 321-326. In G. B. Wickramanayake and R. E. Hinchee (ed.), Natural attenuation of chlorinated and recalcitrant compounds. Batelle Press, Columbus, Ohio.
- 22.Jensen, H. M., and E. Arvin. 1990. Solubility and degradability of the gasoline additive MTBE, methyl tert-butyl ether and gasoline compounds in water, p. 445-448. In F. Arendt, M. Hinsenveld, and W. J. Van den Brink (ed.), Contaminated soil '90. Kluwer, Dordrecht, The Netherlands.
- 23.Johnson, R., J. Pankow, D. Bender, C. Price, and J. Zogorsky. 2000. MTBE. To what extent will past releases contaminate community water supply wells? Environ. Sci. Technol. 34:210A-217A. [DOI] [PubMed] [Google Scholar]
- 24.Kane, S. R., H. R. Beller, T. C. Legler, C. J. Koester, H. C. Pinkart, R. U. Halden, and A. M. Happel. 2001. Aerobic degradation of methyl tert-butyl ether by aquifer bacteria from leaking underground storage tank sites. Appl. Environ. Microbiol. 67:5824-5829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kobayashi, M., and S. Shimizu. 1999. Cobalt proteins. Eur. J. Biochem. 261:1-9. [DOI] [PubMed] [Google Scholar]
- 26.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 27.Pearson, W. R. 1996. Effective protein sequence comparison. Methods Enzymol. 266:227-258 [DOI] [PubMed] [Google Scholar]
- 28.Petrovitch, R. M., F. J. Ruzicka, G. H. Reed, and P. A. Frey. 1991. Metal cofactors of lysine 2,3-aminimutase. J. Biol. Chem. 266:7656-7660. [PubMed] [Google Scholar]
- 29.Piveteau, P., F. Fayolle, J.-P. Vandecasteele, and F. Monot. 2001. Biodegradation of t- butyl alcohol and related xenobiotics by a methylotrophic bacterial isolate. Appl. Microbiol. Biotechnol. 55:369-373. [DOI] [PubMed] [Google Scholar]
- 30.Reed, G. H., and M. D. Ballinger. 1995. Characterization of a radical intermediate in the lysine 1,3-aminomutase reaction. Methods Enzymol. 258:362-379. [DOI] [PubMed] [Google Scholar]
- 31.Salanitro, J. P., L. A. Diaz, M. P. Williams, and H. L. Wisniewski. 1994. Isolation of a bacterial culture that degrades methyl t-butyl ether. Appl. Environ. Microbiol. 60:2593-2596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Salanitro, J. P. 1995. Understanding the limitations of microbial metabolism of ethers used as fuel octane enhancers. Curr. Opin. Biotechnol. 6:337-340. [Google Scholar]
- 33.Schoene, B., R. A. Fleischmann, H. Remmer, and H. F.Von Oldershausen. 1972. Determination of drug metabolizing enzymes in needle biopsies of human liver. Eur. J. Clin. Pharmacol. 4:65-73. [DOI] [PubMed] [Google Scholar]
- 34.Solano-Serena, F., R. Marchal, S. Casarégola, C. Vasnier, J.-M. Lebeault, and J.-P. Vandecasteele. 2000. A new Mycobacterium strain with extended degradation capacities for gasoline hydrocarbons. Appl. Environ. Microbiol. 66:2392-2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Squillace, P. J., J. S. Zogorski, W. G. Wilber, and C. V. Price. 1996. Preliminary assessment of the occurrence and possible sources of MTBE in groundwater in the United States, 1993-1994. Environ. Sci. Technol. 30:1721-1730. [Google Scholar]
- 36.Steffan, R. J., K. McClay, S. Vainberg, C. W. Condee, and D. Zhang. 1997. Biodegradation of the gasoline oxygenates methyl tert-butyl ether, ethyl tert-butyl ether, and tert-amyl methyl ether by propane-oxidizing bacteria. Appl. Environ. Microbiol. 63:4216-4222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Steffan, R. J., S. Vainberg, C. Condee, K. McClay, and P. Hatzinger. 2000. Biotreatment of MTBE with a new bacterial isolate, p. 165-173. In G. B. Wickramanayake, A. R. Gavaskar, B. C. Alleman, and V. S. Magar (ed.), Bioremediation and phytoremediation of chlorinated and recalcitrant compounds. Batelle Press, Columbus, Ohio.
- 38.Swain, E. J. 1999. U.S. MTBE production at a record high in 1998. Oil Gas J. 14(June):99-101. [Google Scholar]
- 39.Telenti, A., F. Marchesi, M. Balz, F. Bally, E. C. Bottger, and T. Bodmer. 1993. Rapid identification of mycobacteria to the species level by polymerase chain reaction and restriction enzyme analysis. J. Clin. Microbiol. 31:175-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thomas, P. E., D. Ryan, and W. Levin. 1976. An improved staining procedure for the detection of the peroxidase activity of cytochrome P-450 on sodium dodecyl sulfate polyacrylamide gels. Anal. Biochem. 75:168-176. [DOI] [PubMed] [Google Scholar]
- 41.Tomasi, I., I. Artaud, Y. Bertheau, and D. Mansuy. 1995. Metabolism of polychlorinated phenols by Pseudomonas cepacia AC1100: determination of the first two steps and specific inhibitory effect of methimazole. J. Bacteriol. 177:307-311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.United States Environmental Protection Agency. 1999. Achieving clean air and clean water: the report of the blue ribbon panel on oxygenates in gasoline. Publication EPA420-R-99-021. U.S. Government Printing Office, Washington, D.C.
- 43.Van Dijken, J. P., and W. Harder. 1975. Growth yields of microorganisms on methanol and methane. Theoretical study. Biotechnol. Bioeng. 17:15. [Google Scholar]
- 44.Werringloer, J. 1978. Assay of formaldehyde generated during microsomal oxidation reactions. Methods Enzymol. 52:297-302. [DOI] [PubMed] [Google Scholar]
- 45.Yeh, C. K., and J. T. Novak. 1994. Anaerobic biodegradation of gasoline oxygenates in soils. Water Environ. Res. 66:744-752. [Google Scholar]