Abstract
Mycobacterium marinum, a natural pathogen of fish and frogs and an occasional pathogen of humans, is capable of inducing actin tail formation within the cytoplasm of macrophages, leading to actin-based motility and intercellular spread. Actin tail formation by M. marinum is markedly reduced in macrophages deficient in the Wiskott-Aldrich syndrome protein (WASP), which still contain the closely related and ubiquitously expressed protein N-WASP (neuronal WASP). In fibroblasts lacking both WASP and N-WASP, M. marinum is incapable of efficient actin polymerization and of intercellular spread. By reconstituting these cells, we find that M. marinum is able to use either WASP or N-WASP to induce actin polymerization. Inhibition or genetic deletion of tyrosine phosphorylation, Nck, WASP-interacting protein, and Cdc42 does not affect M. marinum actin tail formation, excluding the participation of these molecules as upstream activators of N-WASP in the initiation of actin-based motility. In contrast, deletion of the phosphatidylinositol 4,5-bisphosphate-binding basic motif in N-WASP eliminates M. marinum actin tail formation. Together, these data demonstrate that M. marinum subversion of host actin polymerization is most similar to distantly related Gram-negative organisms but that its mechanism for activating WASP family proteins is unique.
Keywords: mycobacteria, neuronal Wiskott-Aldrich syndrome protein, infection, bacterial motility
Ahighly regulated signal transduction cascade initiates Arp2/3 complex-mediated actin reorganization of the cytoskeleton through the members of the Wiskott-Aldrich syndrome protein (WASP) and WAVE families (reviewed in ref. 1). The ubiquitously expressed N-WASP (neuronal WASP) is a modular protein auto-inhibited by an intramolecular interaction in its inactive state. Binding of specific molecules including phosphatidylinositol 4,5-bisphosphate (PIP2), Cdc42, Nck, and Grb2 to N-WASP disrupts this inhibition and unmasks the WASP homology 2 and acidic (WA) domain, allowing the Arp2/3 complex to initiate de novo actin polymerization (2-6). WASP, a protein found only in hematopoietic cells, is closely related to N-WASP, sharing 50% sequence similarity. The two proteins are similarly organized and are controlled by the same host cell inputs. Less related to N-WASP are the WAVE proteins that contain the WA domain output region but have different activating inputs (7).
Diverse pathogens have evolved mechanisms to hijack this Arp2/3-dependent pathway of actin polymerization for their own benefit (reviewed in ref. 8). For vaccinia virus, actin polymerization is required for viral egress from infected cells. Enterohemorrhagic and enteropathogenic Escherichia coli species' actin polymerization leads to pedestal formation, causing attachment and effacement lesions on gut epithelia. For Listeria, Shigella, Burkholderia, Rickettsia, and Mycobacterium marinum, actin polymerization on the surface of cytoplasmic bacteria results in actin tails, intracellular motility, and direct intercellular spread. Interestingly, these pathogens use independently evolved proteins to target different steps leading to Arp2/3 activation. Listeria, Rickettsia, and perhaps Burkholderia mimic host cell N-WASP to activate the Arp2/3 complex directly, and actin tail formation by these species is independent of host cell N-WASP (9-14). The other pathogens depend on the WASP family for actin polymerization: Shigella and enterohemorrhagic E. coli (EHEC) have molecules that directly recruit host cell N-WASP (10, 15-18), whereas vaccinia and enteropathogenic E. coli (EPEC) initiate actin polymerization upstream of N-WASP in a manner similar to host cell receptor tyrosine kinases, involving host cell factors including Nck, Grb2, and WASP-interacting protein (WIP) (19-23). Understanding how pathogens exploit endogenous signal pathways yields insight into basic cell biology as well as mechanisms of microbial pathogenesis.
We recently identified M. marinum as the only mycobacterial species that clearly escapes from phagosomes and subsequently initiates actin tail formation (24). The Arp2/3 complex was found throughout the actin tail behind M. marinum, but WASP localized to the bacterial pole. Here, we determine the host cell requirements for efficient M. marinum actin polymerization and show the necessity for a member of the WASP family. Similar to Shigella and EHEC, M. marinum actin polymerization occurs independently of known N-WASP activators, including tyrosine phosphorylation, Nck, WIP, and Cdc42. However, unlike any other pathogen, work shown here suggests that the mechanism of M. marinum actin polymerization involves the lipid-binding basic motif of N-WASP (hereafter designated by “B”).
Materials and Methods
Host Cells. Macrophages were derived from the bone marrow of 129/Sv or WASP-/- mice (25) and harvested after 7-21 days of culturing as described in ref. 24. Wild-type, N-WASP-/- (10, 16), Nck-/- (26), and WIP-/- (27) embryonic fibroblast cell lines were maintained as previously described. All cells were seeded onto fibronectin-coated coverslips (Becton Dickinson) 1 day before infection.
Bacteria and Infection. Wild-type (strain M), GFP-expressing (24), or red fluorescent protein-expressing (28) M. marinum were cultured in Middlebrook 7H9 (Difco) supplemented with 0.2% glycerol/0.05% Tween 80/10% ADC enrichment (Fisher). Infection of macrophages for microscopy and intracellular growth was carried out as described in refs. 24 and 29. In brief, intracellular growth was assessed by infecting macrophages or fibroblasts with a multiplicity of infection (moi) of 3 and hypotonically lysing the cells 4 and 24 h postinfection for colony-forming unit quantitation. For fibroblasts, a moi of 60 was used, and infected cells were incubated for 48 h before microscopy. For quantification, intracellular bacteria were identified by phase contrast microscopy, and actin tails were visualized by fluorescence. Each experiment was done three times, with 50 cells counted per experiment. Unpaired t tests with Welch corrections were used to generate P values and determine statistical significance. Intercellular spreading assays were performed in confluent macrophage and fibroblast monolayers infected with fluorescent M. marinum as described in refs. 24 and 30. After infection, wells were overlaid with agar, and culture medium contained 40 μg/ml amikacin. Media were changed every 2 days, and macrophage and fibroblast monolayers were examined for pattern of infection 5 or 8 days later, respectively. Quantification of fluorescent foci of infection was performed by using iplab analysis software (Scanalytics, Billerica, MA).
Expression Constructs and Transfection. The WAVE2 and many of the GFP-tagged murine N-WASP expression constructs have been described (16, 31, 32). GFP-tagged full-length and mini human WASP expression constructs were kindly provided by J. E. Dueber (University of California, San Francisco). N-WASP expression constructs (pEGFP-C1-N-WASP Y253F, pEGFP-C1 N-WASP polyProWA, pEGFP-C3 N-WASP WH1+WA, and pEGFP-C1 N-WASP Δ226-267, pEGFP-C1 N-WASP Δ200-226, and pEGFP-C1 N-WASP Δ158-199) were generated by standard methods. All constructs were confirmed by sequencing, and expression of appropriate-size molecules was monitored by immunoblotting. All constructs that were scored here as negative for restoration of M. marinum actin tail formation are functional in other systems (unpublished data). Fibroblast cell lines were transfected 24 h after plating with FuGENE (Roche Diagnostics) according to the manufacturer's specifications. Transfected cells were infected 20 h after transfection.
Pharmacological Inhibitors. Infected macrophages and fibroblasts were treated with inhibitors 30 or 48 h postinfection, respectively. PP2 (4-amino-5-(4-chlorophenyl)-7-(t-butyl)pyrazolo[3,4-d]pyrimi-dine) (100 μM, Calbiochem) or genistein (250 μM) was added to infected cells for 1 h before fixation to study the effect of tyrosine kinase inhibition. Toxin B (1 μg/ml, Calbiochem) and cytochalasin D (1.3 μM, Sigma) were added to infected cells for 1.5 h before fixation.
Microscopy. Infected cells were fixed, permeabilized, and stained with Alexa Fluor phalloidin (Molecular Probes) as described in ref. 24. Indirect immunofluorescence was performed with anti-N-WASP (provided by J. Taunton, University of California, San Francisco), anti-WAVE2 (Santa Cruz Biotechnology), anti-Nck (Upstate Biotechnology, Lake Placid, NY, and BD Transduction Laboratories), anti-WIP (Santa Cruz Biotechnology), anti-phosphotyrosine (PY99, Santa Cruz Biotechnology; 4G10, Upstate Biotechnology), and species-appropriate Alexa Fluor secondary antibodies (Molecular Probes). Coverslips were mounted with Prolong antifade reagent (Molecular Probes). Images were acquired on a Nikon Eclipse TE300 microscope with an Evolution QEi cooled charge-coupled device (Media Cybernetics, Silver Spring, MD) with iplab acquisition software (Scanalytics).
Results
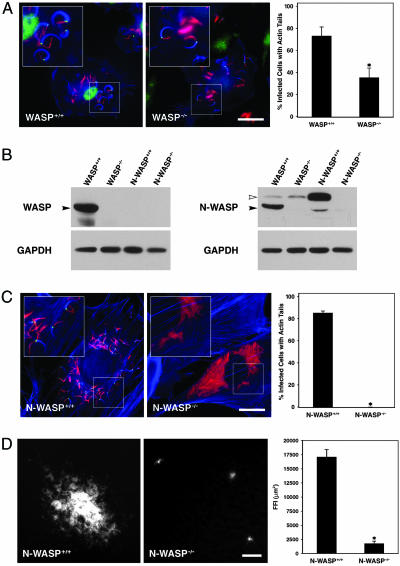
M. marinum Actin Tail Formation and Intercellular Spread in N-WASP-/- Fibroblasts. WASP localized to the pole of M. marinum from which the actin tail formed (Fig. 1A) (24). To determine the requirement for WASP in M. marinum actin tail formation, we infected WASP-/- macrophages and found a significant, ≈2-fold decrease in the number of infected cells in which actin tails were observed (Fig. 1A; WASP+/+, 72.5 ± 8.5%; WASP-/-, 35.10 ± 8.6%; P = 0.04), even though intracellular growth of M. marinum was comparable in WASP+/+ and WASP-/- macrophages by colony-forming unit quantification at 4 and 24 h postinfection (data not shown). Because the deficiency of WASP only partially ablated actin tail formation, we hypothesized that N-WASP might account for the residual actin tail formation. N-WASP was present in WASP-/- macrophages at levels comparable to WASP+/+ macrophages (Fig. 1B). Indeed, we found that N-WASP localized to the pole of M. marinum with actin tails in WASP-/- macrophages (Fig. 1A). M. marinum formed actin tails in N-WASP+/+ fibroblasts that lacked WASP (Fig. 1B), and N-WASP localized at the pole in these cells (Fig. 1C). Most significantly, there was an almost total decrease in actin tail formation in N-WASP-/- fibroblasts that have neither WASP nor N-WASP (Fig. 1C), despite the fact that M. marinum grew similarly in N-WASP+/+ and N-WASP-/- fibroblasts (data not shown). Less than 1% of N-WASP-/- fibroblasts exhibited bacterial actin tails; this small residual bacterial motility was seen in two independently derived N-WASP-/- fibroblast cell lines (data not shown).
Fig. 1.
WASP or N-WASP is required for efficient M. marinum motility. (A) WASP+/+ macrophages (Left) were infected with M. marinum (red) and stained with phalloidin (blue) and with anti-WASP antibodies (green). WASP-/- macrophages (Center) were similarly infected and stained with anti-N-WASP antibodies (green). In Right, the percentage of infected cells with actin tails from three independent experiments is shown (P = 0.037). (Scale bar, 20 μm.) (B) Whole-cell lysates of WASP+/+ and WASP-/- macrophages and N-WASP+/+ and N-WASP-/- fibroblasts were resolved by SDS/PAGE and analyzed for the presence of WASP and N-WASP by Western blotting. Filled arrowheads indicate the WASP bands; open arrowheads indicate the N-WASP bands. The WASP antibody is specific for WASP, but the anti-N-WASP antibody is cross-reactive with WASP, leading to the detection of a WASP band in the WASP+/+ lane. (C) N-WASP+/+ fibroblasts (Left) and N-WASP-/- fibroblasts (Center) were infected with M. marinum (red) and stained with phalloidin (blue) and with anti-N-WASP antibodies (green). In Right, the percentage of infected cells with actin tails was quantified as in A. (Scale bar, 20 μm.) (D) N-WASP+/+ (Left) and N-WASP-/- (Center) fibroblast monolayers were infected with fluorescent M. marinum, and foci of infection were visualized and measured 8 days later (P = 0.002). (Scale bar, 50 μm.) Asterisks in A-D indicate statistically significant differences with P < 0.05.
To determine whether actin tail formation is required for inter-cellular spread of M. marinum, we examined spreading of fluorescent M. marinum within a confluent layer of macrophages or fibroblasts. Fluorescent foci of infection (FFI) in WASP+/+ and WASP-/- macrophages were comparable in size (WASP+/+, 78,726 ± 11,794 μm2; WASP-/-, 94,947 ± 17853 μm2; P = 0.45) and consisted of many cells, representing contiguously infected cells arising from a single originally infected macrophage. This finding suggests that the residual ability of M. marinum to form actin tails in WASP-/- macrophages is sufficient for intercellular spread in macrophages. In contrast, FFI in the N-WASP+/+ monolayer were significantly larger than in the N-WASP-/- monolayer (Fig. 1D; N-WASP+/+, 17,092 ± 1,314 μm2; N-WASP-/-, 1,682 ± 498 μm2; P = 0.002). FFI in the N-WASP-/- monolayer most often involved only a single cell or a few cells.
Transient expression of either N-WASP or WASP restored actin tail formation in M. marinum-infected N-WASP-/- fibroblasts (Fig. 2). In agreement with the immunofluorescence staining of WASP and N-WASP (Fig. 1), the GFP fusion N-WASP and WASP proteins localized to the pole from which the actin tail formed. However, transient expression of GFP-WAVE2 failed to restore actin tail formation by M. marinum in these cells (Fig. 2), and endogenous WAVE2 failed to localize to bacteria assessed by immunofluorescence (data not shown), demonstrating a specificity of M. marinum for WASP and N-WASP among the WASP/WAVE family.
Fig. 2.
M. marinum can use either WASP or N-WASP, but not WAVE2, for intracellular actin tail formation. N-WASP-/- fibroblasts were transiently transfected with GFP-N-WASP (Left), WASP (Center), or WAVE2 (Right) and infected with M. marinum (red). Cells were fixed and stained with phalloidin (blue). (Scale bar, 5 μm.)
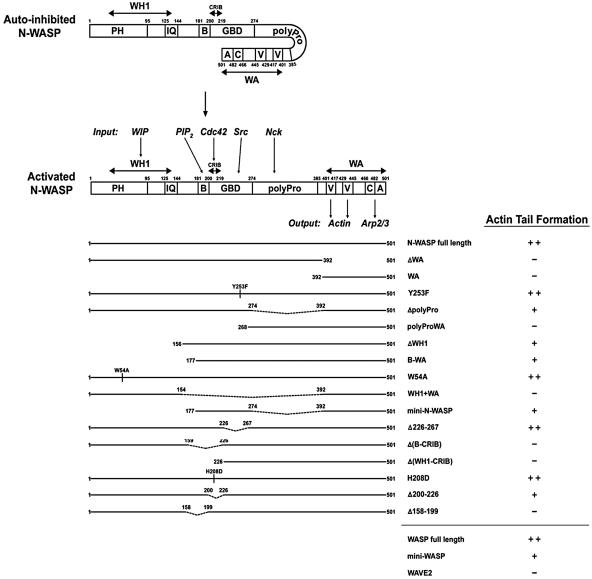
Role of the Arp2/3 Complex, Tyrosine Phosphorylation, Nck, and WIP. To determine how M. marinum recruits WASP and N-WASP, we embarked on a systematic examination of molecules known to interact with the WASP family and analysis of the N-WASP domains required for M. marinum actin polymerization. We verified the necessity of the WA output region of N-WASP that contains the verprolin homology, cofilin homology, and acidic domains by transfecting the N-WASP-/- fibroblasts with an N-WASP construct lacking the WA domain (Fig. 3). Although N-WASP ΔWA associated with the surface of intracellular M. marinum (Fig. 5, which is published as supporting information on the PNAS web site), this construct did not restore actin tail formation (Fig. 3). The WA domain alone of N-WASP did not restore actin tails (Fig. 3). Together, these data demonstrate that the input region of N-WASP is critical for recruitment to M. marinum and that actin and the Arp2/3 complex binding to the WA domain of N-WASP are required for M. marinum actin tail formation.
Fig. 3.
N-WASP domains required for M. marinum actin tail formation. (Upper) Schematic representation of N-WASP in autoinhibited and activated states showing interactions relevant to function is adapted from refs. 16, 31, and 32. WIP, PIP2, Cdc42, Nck, and the Src kinase all bind to the input region, and monomeric actin and the Arp2/3 complex bind to the output region. (Lower) The domain structure of N-WASP constructs used in this study and their ability to restore actin tail formation by M. marinum in N-WASP-/- fibroblasts is shown. -, +, and ++ indicate efficiency of restoration (-, <1 cell with tails per 100 transfected, infected cells; +, 2-20 cells with tails per 100 transfected, infected cells; ++, >20 cells with tails per 100 transfected, infected cells). Also included are results for full-length WASP, mini WASP (equivalent to mini N-WASP), and full-length WAVE2. Each construct was tested two or more times.
To determine whether tyrosine phosphorylation has a role in M. marinum actin polymerization as it does in vaccinia and EPEC (19), we first examined phosphotyrosine staining in M. marinum-infected macrophages and fibroblasts. Anti-phosphotyrosine antibodies showed marked staining of adhesion sites and other cellular structures (Fig. 6, which is published as supporting information on the PNAS web site). In macrophages, ≈25% of all M. marinum with actin tails had phosphotyrosine staining at the pole (Fig. 5A); however, we never detected phosphotyrosine staining associated with bacteria in fibroblasts (Fig. 5B). PP2 (4-amino-5-(4-chlorophenyl)-7-(t-butyl)pyrazolo[3,4-d]pyrimidine), a Src kinase-specific inhibitor, had no effect on actin tail formation in either M. marinum-infected macrophages or fibroblasts, but it did eliminate the occasional phosphotyrosine staining in macrophages (Fig. 6). Genistein, a general tyrosine kinase inhibitor, also had no effect on tail formation (data not shown). Src kinases also can directly activate the WASP family, regulating its activity in hematopoeitic cells and neurons (33, 34). An N-WASP construct with a tyrosineto-phenylalanine mutation (Y253F) that cannot be phosphorylated by Src family kinases in vitro (34) fully restored actin tail formation (Fig. 3). Taken together, these data indicate that tyrosine phosphorylation, either of upstream activators or directly of N-WASP, does not play a significant role in M. marinum actin polymerization.
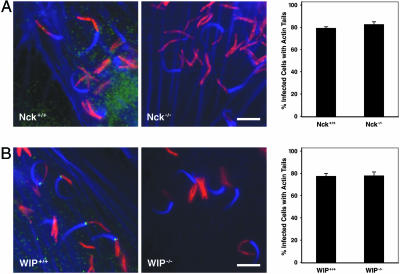
Vaccinia virus and EPEC primarily use Nck to link a phosphorylated microbial molecule to N-WASP, and Nck binding to the polyproline (polyPro) domain of N-WASP is capable of activating the protein in vitro (5). Using two commercially available anti-Nck antibodies, Nck did not localize to the pole of M. marinum actin tails in macrophages or fibroblasts (Fig. 4A; data not shown). Moreover, the percentage of infected cells with actin tails was comparable in Nck+/+ and Nck-/- fibroblasts (Fig. 4A; Nck+/+, 79.3 ± 1.3%; Nck-/-, 82.7 ± 2.4%; P > 0.31), as was the fraction of the total intracellular bacteria that have actin tails (Nck+/+, 20.0 ± 1.0%; Nck-/-, 24.3 ± 2.2%; P > 0.21). Transfection with an N-WASP construct in which the polyPro region was deleted (ΔpolyPro) restored actin tail formation in M. marinum-infected N-WASP-/- fibroblasts, whereas transfection with an N-WASP construct containing the polyPro region and the WA domain alone (polyProWA) did not (Fig. 3).
Fig. 4.
Nck and WIP are not involved in M. marinum actin tail formation. (A) Nck+/+ (Left) and Nck-/- (Center) fibroblasts were infected with M. marinum (red) and stained with phalloidin (blue) and anti-Nck antibodies (green). In Right, the percentage of infected cells with actin tails for Nck+/+ and Nck-/- fibroblasts is shown. (B) WIP+/+ (Left) and WIP-/- (Center) fibroblasts were infected with M. marinum (red) and stained with phalloidin (blue) and anti-WIP antibodies (green). In Right, the percentage of infected cells with actin tails is shown. All quantification is the summary of three independent experiments. (Scale bars, 5 μm.)
The WIP has been implicated in regulation of WASP and N-WASP through eukaryotic signaling cascades and in actin polymerization by vaccinia virus (20). WIP did localize to the pole of M. marinum from which actin tails formed in both macrophages and fibroblasts (data not shown; Fig. 4B). However, M. marinum actin tails in WIP-/- fibroblasts formed at the same frequency (Fig. 4B; fraction of infected cells with tails: WIP+/+, 77.3 ± 2.4%; WIP-/-, 78.0 ± 3.1%; P > 0.88; fraction of intracellular bacteria with tails: WIP+/+, 19.0 ± 0.3%; WIP-/-, 18.9 ± 2.3%; P > 0.99) and had similar morphology to tails in WIP+/+ fibroblasts (Fig. 4B). The members of the WIP family, including WIP, CR16, and WICH, bind to N-WASP by a mechanism requiring tryptophan 54 in the WASP homology 1 (WH1) domain, which includes the pleckstrin homology domain and the calmodulin-binding IQ motif (35). Because some member of this family other than WIP may have a role in activation of N-WASP by M. marinum, N-WASP-/- fibroblasts were transfected with ΔWH1, B-WA, and W54A N-WASP constructs. Actin tail formation by M. marinum in N-WASP-/- fibro-blasts was restored by all constructs, demonstrating that neither WIP nor a WIP-like protein is necessary (Fig. 3). A construct that included only the WH1 domain and the WA domain (WH1+WA) did not restore actin tail formation; thus, the WH1 domain is neither necessary nor sufficient for M. marinum actin polymerization (Fig. 3).
Role of the N-WASP Basic Motif-GTPase-Binding Domain (B-GBD). The mini N-WASP construct containing the basic motif and the GBD (B-GBD) and the WA output domain but lacking the WH1 and the polyPro domains is controlled by PIP2 and Cdc42 in a manner similar to full length N-WASP (3). In our system, mini N-WASP and mini WASP were sufficient to restore actin tail formation in N-WASP-/- fibroblasts, albeit not as efficiently as full-length constructs (Fig. 3). The autoinhibited conformation of the WASP family requires interaction between the carboxyl terminus of the GBD, including amino acids 226-274 of N-WASP, and the WA domain (3, 4). EHEC EspFu binds to this region of N-WASP to initiate pedestal formation (18, 30). However, an N-WASP Δ226-267 construct was fully capable of restoring actin tails in N-WASP-/- fibroblasts, demonstrating that M. marinum does not target this region of the molecule. In contrast, two constructs lacking the N-terminal portion of the B-GBD, including the basic and Cdc42 and Rac interactive-binding (CRIB) motifs (ΔB-CRIB and ΔWH1-CRIB; Fig. 3), were unable to restore actin tail formation in the N-WASP-/- fibroblasts (Fig. 3), focusing our attention on the area to which the host cell factors PIP2 and Cdc42 bind.
Direct binding of GTP-loaded Cdc42 to the CRIB motif is a major mechanism by which the WASP family proteins are activated (2). To determine whether Cdc42 has a role in M. marinum actin polymerization, we transfected N-WASP-/- fibroblasts with an N-WASP H208D construct that abolishes GTPase binding and with an N-WASP Δ200-226 construct that lacks the CRIB motif. Both of these constructs restored actin tail formation by M. marinum (Fig. 3), demonstrating that the bacterium recruits and activates N-WASP independent of GTPases. Furthermore, we treated M. marinum-infected macrophages with Clostridium difficile toxin B, an inhibitor of the entire Rho family of small GTPases (Fig. 7, which is published as supporting information on the PNAS web site). Although toxin B treatment led to dramatic differences in overall actin distribution, M. marinum still formed actin tails at levels similar to untreated cells (Fig. 6; fraction of infected cells with tails: not treated, 84.5 ± 1.7%; toxin B, 81.5 ± 0.1%; P > 0.27; fraction of intracellular bacteria with tails: not treated, 23.1 ± 3.2%; toxin B, 24.4 ± 3.0%; P > 0.85). In contrast, when cytochalasin D was used to inhibit actin polymerization, the percentage of infected cells with actin tails significantly decreased, and the tails that were present were smaller (fraction of infected cells with tails compared with those that were untreated: cytochalasin D, 24 ± 2%; P = 0.0005). Thus, WASP family binding of Cdc42 or a related GTPase is not required for M. marinum actin polymerization.
The host anionic lipid PIP2 is capable of activating N-WASP or WASP by binding to the basic motif, and its main role in this pathway is to synergize with Cdc42 for highly efficient activation of N-WASP (3, 36, 37). Transfection with an N-WASP Δ158-199 construct that lacks the basic motif was unable to be recruited to the bacterial surface (Fig. 5A) or restore actin tail formation in N-WASP-/- fibroblasts (Fig. 3), suggesting a role for this region in M. marinum recruitment of N-WASP and subsequent actin polymerization. This construct is functional; it does support actin tail formation by Shigella and actin pedestal formation by EHEC (Fig. 5B). Thus, in contrast to other pathogens that activate the WASP family, M. marinum actin tail formation requires the basic motif of N-WASP.
Discussion
A diverse group of intracellular pathogens, including M. marinum, enables efficient intercellular spread by activating actin polymerization on their surfaces within host cytoplasm. This motility is used for direct intercellular spread, bypassing host defense mechanisms directed against extracellular pathogens. Pathogens have evolved various methods for subverting the host cytoskeleton to generate Arp2/3-dependent actin polymerization. Unlike Listeria, which directly recruits the Arp2/3 complex (schematically represented in Fig. 8, which is published as supporting information on the PNAS web site), we show here that efficient M. marinum actin polymerization and intercellular spread depends on the WASP family. Unlike Shigella, which exploits only N-WASP, M. marinum can use either family member, depending on availability in accordance with cell type (Figs. 1 and 2). N-WASP-dependent tail formation by M. marinum depends on both the input and output regions, suggesting that this species, like the Gram-negative bacteria, has developed a mechanism of activating N-WASP that depends on the input region in the N-terminal part of the protein, leading to unfolding of the protein and unmasking of the WA output region required for Arp2/3-dependent actin polymerization. The observation that bacterial motility was not completely eliminated by the absence of WASP family proteins suggests the existence of a second, albeit inefficient, mode of actin polymerization by M. marinum.
Because its role was so predominant, we sought to elucidate the mechanism by which M. marinum activates WASP family proteins by using a variety of genetic, biochemical, and pharmacological reagents. M. marinum activation of N-WASP clearly differs from vaccinia and EPEC, which require tyrosine phosphorylation of bacterial molecules A36R and Tir, respectively, to initiate a Nck-dependent cascade for activation of N-WASP (Fig. 8) (19, 23). For both vaccinia and EPEC, the WIP-binding WH1 domain is involved in actin polymerization (16, 20), and WIP is thought to be downstream of Nck in the case of vaccinia but not in EPEC (20, 32). For M. marinum, tyrosine phosphorylation, Nck, and WIP are dispensable for actin tail formation (Figs. 4, 5, and 8). Furthermore, the domains of N-WASP with which they interact, the WH1 and polyPro domains, respectively, can be removed without blocking M. marinum actin polymerization. However, those constructs are less efficient than full-length N-WASP (Fig. 3). It is possible that although not required for N-WASP-dependent tail formation, proteins that interact with the WH1 and polyPro domains contribute to amplification of N-WASP and actin recruitment. Indeed, WIP localizes to the pole behind which M. marinum tails form (Fig. 4). This observation is very similar to what has been shown for Shigella (20), where WIP binding to N-WASP is believed to be downstream of direct bacterial activation of N-WASP.
As illustrated in Fig. 8, M. marinum activation of N-WASP is more similar to the mechanisms described for Shigella and EHEC, which also are independent of tyrosine phosphorylation and bypass Nck or Cdc42 binding (Figs. 4 and 6). Both EHEC and Shigella express proteins that directly bind to N-WASP (18, 38). Shigella IcsA most likely binds to N-WASP at several sites in the input region, including the WH1 and GBD domains (38), whereas EHEC EspFu (16) directly disrupts the autoinhibitory interaction between the GBD and WA domain proteins by binding to the GBD (18, 32). None of the domains required for IcsA or EspFu initiation of actin polymerization is used by M. marinum. Thus, although superficially similar, the three microbes have evolved independent mechanisms for activating N-WASP-dependent actin polymerization.
One way in which the M. marinum mechanism differs from that of all other bacteria is that the basic motif of N-WASP is required for efficient N-WASP recruitment and actin polymerization by M. marinum. This region is known to bind to eukaryotic polyphosphoinositides through an ionic interaction mediated by its high concentration of lysines (3, 37). Unlike other prokaryotes that activate actin polymerization, mycobacterial cell walls are rich in phosphoinositides. One of these, the multiglycosylated lipoarabinomannan, is a potential virulence factor and modulates immune responses (reviewed in refs. 39 and 40). In addition, its precursor, phosphatidylinositol mannoside, also has been found to be associated with the bacterial surface (41). It is intriguing to speculate that mycobacterial phospholipids bind to the basic motif region of N-WASP. We cannot eliminate the possibility that M. marinum may be able to modulate levels of PIP2 on the bacterial surface; however, we have not found any association of the PKCδ pleckstrin homology domain, a PIP2 reporter, with M. marinum (data not shown). PIP2 is not an efficient activator of N-WASP on its own, and it is believed that its major role in host cell actin polymerization is in synergy with Cdc42 or other activators. However, many intracellular pathogens, including Mycobacterium tuberculosis, are known to control levels of host phosphoinositides for their own purposes (reviewed in ref. 42).
In conclusion, M. marinum hijacks eukaryotic signaling for Arp2/3 complex-dependent actin polymerization through activation of the WASP family independent of upstream signaling molecules, resulting in direct intercellular spread. This pathway is similar in principle to the mechanisms evolved by Shigella and EHEC and is another amazing example of convergent evolution by pathogens to exploit a host cell biological pathway for efficient infection. However, data presented here suggest that the M. marinum mechanism is distinct from those thus far described for other pathogens by requiring the basic motif of N-WASP for its activation. It is unknown whether actin-based motility is a characteristic unique to M. marinum among all mycobacteria, although we have not been able to demonstrate this phenomenon with either M. tuberculosis or Mycobacterium ulcerans (43). Further studies of actin polymerization by M. marinum are needed to explore its role in pathogenesis of infection and broaden understanding of actin cytoskeletal dynamics.
Supplementary Material
Acknowledgments
We thank L. Ramakrishnan (University of Washington, Seattle) for the mycobacterial red fluorescent protein expression plasmid, J. Taunton for the anti-N-WASP antibody, J. E. Dueber for the WASP and mini WASP constructs, J. Wehland for critical reading of the manuscript, C. A. Lowell (University of California) for providing WASP-/- mice, S. Benesch (German Research Center for Biotechnology) for the N-WASP-/- fibroblast cell line, T. Pawson (Samuel Lunenfeld Research Institute, Toronto) for the Nck-/- fibroblast cell line, and R. S. Geha (Children's Hospital, Boston) and N. Ramesh (Children's Hospital, Boston) for the WIP-/- fibroblast cell line. This work was supported by Public Health Service Grant AI55614 from the National Institutes of Health (to E.J.B.), Deutsche Forschungsgemeinschaft SBF 621 (K.R. and S.L.), the University of California, San Francisco, Medical Scientist Training Program (L.M.S.), and a fellowship from the ARCS Foundation (to L.M.S.).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: CRIB, Cdc42 and Rac interactive binding; EHEC, enterohemorrhagic E. coli; EPEC, enteropathogenic E. coli; GBD, GTPase-binding domain; PIP2, phosphatidylinositol 4,5-bisphosphate; polyPro, polyproline; WA, WASP homology 2 and acidic; WASP, Wiskott-Aldrich syndrome protein; N-WASP, neuronal WASP; WH1, WASP homology 1 domain; WIP, WASP-interacting protein.
References
- 1.Takenawa, T. & Miki, H. (2001) J. Cell Sci. 114, 1801-1809. [DOI] [PubMed] [Google Scholar]
- 2.Beuzon, C. R., Salcedo, S. P. & Holden, D. W. (2002) Microbiology 148, 2705-2715. [DOI] [PubMed] [Google Scholar]
- 3.Prehoda, K. E., Scott, J. A., Mullins, R. D. & Lim, W. A. (2000) Science 290, 801-806. [DOI] [PubMed] [Google Scholar]
- 4.Kim, A. S., Kakalis, L. T., Abdul-Manan, N., Liu, G. A. & Rosen, M. K. (2000) Nature 404, 151-158. [DOI] [PubMed] [Google Scholar]
- 5.Rohatgi, R., Nollau, P., Ho, H. Y., Kirschner, M. W. & Mayer, B. J. (2001) J. Biol. Chem. 276, 26448-26452. [DOI] [PubMed] [Google Scholar]
- 6.Carlier, M. F., Nioche, P., Broutin-L'Hermite, I., Boujemaa, R., Le Clainche, C., Egile, C., Garbay, C., Ducruix, A., Sansonetti, P. & Pantaloni, D. (2000) J. Biol. Chem. 275, 21946-21952. [DOI] [PubMed] [Google Scholar]
- 7.Miki, H., Suetsugu, S. & Takenawa, T. (1998) EMBO J. 17, 6932-6941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Frischknecht, F. & Way, M. (2001) Trends Cell Biol. 11, 30-38. [DOI] [PubMed] [Google Scholar]
- 9.Welch, M. D., Iwamatsu, A. & Mitchison, T. J. (1997) Nature 385, 265-269. [DOI] [PubMed] [Google Scholar]
- 10.Snapper, S. B., Takeshima, F., Anton, I., Liu, C. H., Thomas, S. M., Nguyen, D., Dudley, D., Fraser, H., Purich, D., Lopez-Ilasaca, et al. (2001) Nat. Cell Biol. 3, 897-904. [DOI] [PubMed] [Google Scholar]
- 11.Breitbach, K., Rottner, K., Klocke, S., Rohde, M., Jenzora, A., Wehland, J. & Steinmetz, I. (2003) Cell. Microbiol. 5, 385-393. [DOI] [PubMed] [Google Scholar]
- 12.Gouin, E., Egile, C., Dehoux, P., Villiers, V., Adams, J., Gertler, F., Li, R. & Cossart, P. (2004) Nature 427, 457-461. [DOI] [PubMed] [Google Scholar]
- 13.Jeng, R. L., Goley, E. D., D'Alessio, J. A., Chaga, O. Y., Svitkina, T. M., Borisy, G. G., Heinzen, R. A. & Welch, M. D. (2004) Cell. Microbiol. 6, 761-769. [DOI] [PubMed] [Google Scholar]
- 14.Stevens, M. P., Stevens, J. M., Jeng, R. L., Taylor, L. A., Wood, M. W., Hawes, P., Monaghan, P., Welch, M. D. & Galyov, E. E. (2005) Mol. Microbiol. 56, 40-53. [DOI] [PubMed] [Google Scholar]
- 15.Egile, C., Loisel, T. P., Laurent, V., Li, R., Pantaloni, D., Sansonetti, P. J. & Carlier, M. F. (1999) J. Cell Biol. 146, 1319-1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lommel, S., Benesch, S., Rottner, K., Franz, T., Wehland, J. & Kuhn, R. (2001) EMBO Rep. 2, 850-857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garmendia, J., Phillips, A. D., Carlier, M. F., Chong, Y., Schuller, S., Marches, O., Dahan, S., Oswald, E., Shaw, R. K., Knutton, S. & Frankel, G. (2004) Cell. Microbiol. 6, 1167-1183. [DOI] [PubMed] [Google Scholar]
- 18.Campellone, K. G., Robbins, D. & Leong, J. M. (2004) Dev. Cell 7, 217-228. [DOI] [PubMed] [Google Scholar]
- 19.Frischknecht, F., Moreau, V., Rottger, S., Gonfloni, S., Reckmann, I., Superti-Furga, G. & Way, M. (1999) Nature 401, 926-929. [DOI] [PubMed] [Google Scholar]
- 20.Moreau, V., Frischknecht, F., Reckmann, I., Vincentelli, R., Rabut, G., Stewart, D. & Way, M. (2000) Nat. Cell Biol. 2, 441-448. [DOI] [PubMed] [Google Scholar]
- 21.Scaplehorn, N., Holmstrom, A., Moreau, V., Frischknecht, F., Reckmann, I. & Way, M. (2002) Curr. Biol. 12, 740-745. [DOI] [PubMed] [Google Scholar]
- 22.Kenny, B. (1999) Mol. Microbiol. 31, 1229-1241. [DOI] [PubMed] [Google Scholar]
- 23.Gruenheid, S., DeVinney, R., Bladt, F., Goosney, D., Gelkop, S., Gish, G. D., Pawson, T. & Finlay, B. B. (2001) Nat. Cell Biol. 3, 856-859. [DOI] [PubMed] [Google Scholar]
- 24.Stamm, L. M., Morisaki, J. H., Gao, L. Y., Jeng, R. L., McDonald, K. L., Roth, R., Takeshita, S., Heuser, J., Welch, M. D. & Brown, E. J. (2003) J. Exp. Med. 198, 1361-1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Snapper, S. B., Rosen, F. S., Mizoguchi, E., Cohen, P., Khan, W., Liu, C. H., Hagemann, T. L., Kwan, S. P., Ferrini, R., Davidson, et al. (1998) Immunity 9, 81-91. [DOI] [PubMed] [Google Scholar]
- 26.Bladt, F., Aippersbach, E., Gelkop, S., Strasser, G. A., Nash, P., Tafuri, A., Gertler, F. B. & Pawson, T. (2003) Mol. Cell. Biol. 23, 4586-4597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Anton, I. M., Saville, S. P., Byrne, M. J., Curcio, C., Ramesh, N., Hartwig, J. H. & Geha, R. S. (2003) J. Cell Sci. 116, 2443-2451. [DOI] [PubMed] [Google Scholar]
- 28.Cosma, C. L., Humbert, O. & Ramakrishnan, L. (2004) Nat. Immunol. 5, 828-835. [DOI] [PubMed] [Google Scholar]
- 29.Gao, L. Y., Groger, R., Cox, J. S., Beverley, S. M., Lawson, E. H. & Brown, E. J. (2003) Infect. Immun. 71, 922-929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gao, L. Y., Laval, F., Lawson, E. H., Groger, R. K., Woodruff, A., Morisaki, J. H., Cox, J. S., Daffe, M. & Brown, E. J. (2003) Mol. Microbiol. 49, 1547-1563. [DOI] [PubMed] [Google Scholar]
- 31.Benesch, S., Lommel, S., Steffen, A., Stradal, T. E., Scaplehorn, N., Way, M., Wehland, J. & Rottner, K. (2002) J. Biol. Chem. 277, 37771-37776. [DOI] [PubMed] [Google Scholar]
- 32.Lommel, S., Benesch, S., Rohde, M., Wehland, J. & Rottner, K. (2004) Cell. Microbiol. 6, 243-254. [DOI] [PubMed] [Google Scholar]
- 33.Cory, G. O., Garg, R., Cramer, R. & Ridley, A. J. (2002) J. Biol. Chem. 277, 45115-45121. [DOI] [PubMed] [Google Scholar]
- 34.Suetsugu, S., Hattori, M., Miki, H., Tezuka, T., Yamamoto, T., Mikoshiba, K. & Takenawa, T. (2002) Dev. Cell 3, 645-658. [DOI] [PubMed] [Google Scholar]
- 35.Zettl, M. & Way, M. (2002) Curr. Biol. 12, 1617-1622. [DOI] [PubMed] [Google Scholar]
- 36.Higgs, H. N. & Pollard, T. D. (2000) J. Cell Biol. 150, 1311-1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rohatgi, R., Ho, H. Y. & Kirschner, M. W. (2000) J. Cell Biol. 150, 1299-1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Suzuki, T., Mimuro, H., Suetsugu, S., Miki, H., Takenawa, T. & Sasakawa, C. (2002) Cell. Microbiol. 4, 223-233. [DOI] [PubMed] [Google Scholar]
- 39.Strohmeier, G. R. & Fenton, M. J. (1999) Microbes Infect. 1, 709-717. [DOI] [PubMed] [Google Scholar]
- 40.Chua, J., Vergne, I., Master, S. & Deretic, V. (2004) Curr. Opin. Microbiol. 7, 71-77. [DOI] [PubMed] [Google Scholar]
- 41.Ortalo-Magne, A., Lemassu, A., Laneelle, M. A., Bardou, F., Silve, G., Gounon, P., Marchal, G. & Daffe, M. (1996) J. Bacteriol. 178, 456-461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pizarro-Cerda, J. & Cossart, P. (2004) Nat. Cell Biol. 6, 1026-1033. [DOI] [PubMed] [Google Scholar]
- 43.Stamm, L. M. & Brown, E. J. (2004) Microbes Infect. 6, 1418-1428. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.