Abstract
The cbnA gene encoding the chlorocatechol dioxygenase gene from Ralstonia eutropha NH9 was introduced into rice plants. The cbnA gene was expressed in transgenic rice plants under the control of a modified cauliflower mosaic virus 35S promoter. Western blot analysis using anti-CbnA protein indicated that the cbnA gene was expressed in leaf tissue, roots, culms, and seeds. Transgenic rice calluses expressing the cbnA gene converted 3-chlorocatechol to 2-chloromucote efficiently. Growth and morphology of the transgenic rice plants expressing the cbnA gene were not distinguished from those of control rice plants harboring only a Ti binary vector. It is thus possible to breed transgenic plants that degrade chloroaromatic compounds in soil and surface water.
Chlorinated aromatic compounds are used in large quantities as herbicides, pesticides, and solvents (9). Contamination of the soil by these compounds is a very serious environmental problem because they are toxic and resistant to degradation. Some microorganisms degrade these compounds aerobically (1). In microbial degradation of these chlorinated aromatic compounds, the chlorinated aromatic compounds are converted to respective chlorocatechols and then the chlorocatechols are degraded by the modified ortho-cleavage pathway (12). Therefore, in most common examples of bacterial aerobic degradation of chlorinated aromatics, chlorocatechols generated from various chlorinated aromatics through convergent pathways are key intermediates in the degradative pathway of chlorinated aromatic compounds and the degradation pathway of chlorocatechol has a significant role in the complete degradation of chlorinated aromatic compounds.
A gram-negative bacterium, Ralstonia eutropha (Alcaligenes eutrophus) NH9, isolated in Japan grows on a medium containing 3-chlorobenzoate as the sole carbon source and degrades 3-chlorobenzoate via 3-chlorocatechol via the modified ortho-cleavage pathway (8). A gene cluster cbnR-ABCD encodes the enzymes responsible for the degradation of chlorocatechol and is encoded on a large plasmid, pENH91. Chlorocatechols are converted to the 3-oxoadipate by the action of four enzymes and are funneled to the 3-oxoadipate pathway. The first enzyme, chlorocatechol 1,2-dioxygenase (EC 1.13.11), encoded by the cbnA gene, cleaves the aromatic ring of chlorocatechol with consumption of molecular oxygen. This enzyme reaction is one of the important steps for the degradation of chlorocatechols because cleavage of the aromatic ring reduces its toxic activity.
Bioremediation is a low-cost treatment alternative for the cleanup of chloroaromatic compound-contaminated soils and surface water. Microorganisms have been used to degrade organic substances in the biologic treatment of wastewater (10). In biodegradation of chlorinated aromatic compounds released in the environment, treatment with microorganisms that degrade these compounds have been extensively studied. Degradation by microorganisms can only occur, however, when environmental conditions are suitable. It is also difficult to maintain growth of these microorganisms in the polluted soil and surface water. Phytoremediation using plants to remove or inactivate pollutants from soil and surface water has received increasing attention in recent years (2). The use of plants for bioremediation is cost-effective, less disruptive to the environment, and is sustainable technology because plants use photosynthetic energy to degrade compounds and can be eventually used as biomass. To date, metal-accumulating plants that can clean up metals in soils were developed by genetic engineering techniques (5). Molecular breeding of transgenic plants that can degrade chloroaromatic compounds, however, has not yet been reported. This is the first report, to our knowledge, that describes the successful expression of a chlorocatechol-degrading gene in plants.
MATERIALS AND METHODS
Construction of a binary vector plasmids for rice transformation.
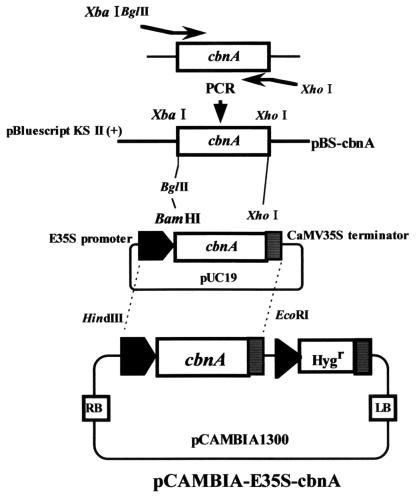
To express the cbnA gene in rice, pCAMBIA-E35S-cbnA was constructed (Fig. 1). The cbnA gene encoding chlorocatechol dioxygenase (765 bp) was amplified by PCR using primers cbnA-F (5′-TCTAGAAGATCTATGAACGAACGAGTGAAGCAGGTTGC-3′) and cbnA-R (5′-CTCGAGTCATGCGTGCTCCCGGGG-3′) and pBLcbn1 as a template (7). PCR was performed for 30 cycles (30 s at 94°C, 30 s at 60°C, 60 s at 72°C), using KOD-Dash polymerase (Toyobo, Osaka, Japan). The amplified cbnA was subcloned into pBluescript KSII (+) between the XbaI and XhoI sites and its sequence was confirmed. The resultant plasmid was designated pBS-cbnA. The cbnA gene subcloned in pBS-cbnA was excised with BglII and XhoI, and then subcloned into pE7131 between BamHI and XhoI sites. The cbnA gene was placed under the control of the enhanced CaMV35S promoter (E35S) and CaMV35S poly(A) signal (6). The promoter-cbnA-terminator cassette was excised from the plasmid using HindIII and EcoRI, and subcloned into a Ti binary vector pCAMBIA1300 (CAMBIA, Canberra, Australia). The constructed plasmid, pCAMBIA-E35S-cbnA, carrying the hygromycin phosphotransferase gene (hpt) as a selectable marker, was transformed into A. tumefaciens EHA101 by electroporation. Agrobacterium cultures were grown for 10 to 12 h in LB medium containing hygromycin (50 mg/liter) and used for Agrobacterium-mediated transformation of rice.
FIG. 1.
Plasmid construction used for transformation of rice plants. For transformation of rice plants, the cbnA gene was cloned between the E7131 promoter and CaMV35S terminator in pCAMBIA1300. E35S, E7131 promoter; hpt, the hygromycin phosphotransferase gene.
Transformation of rice plants.
Agrobacterium-mediated transformation of rice plants (Oryza sativa L. subsp. japonica cultivar Notohikari) was performed as described previously (4).
PCR analysis of transgenic rice callus.
To analyze the integration of the cbnA gene in transgenic rice plants, PCR method was used. PCR was performed for 30 cycles (30 s at 94°C, 30 s at 60°C, 60 s at 72°C), using KOD-Dash polymerase using primers cbnA-F and cbnA-R, and genomic DNAs from the transformants as template
Antibody production.
Polyclonal anti-CbnA antibodies were generated in rabbits against a synthetic antigen peptide (Sawady Technologies, Tokyo, JAPAN) with the 14-amino-acid sequence of CbnA (NH2-WHSTPDGKYSGFHD-COOH) that corresponds to the 122nd through 135th amino acid residues. Rabbits were initially injected with 0.2 mg of the peptide with adjuvant, followed by two booster injections with equivalent amounts of peptide in adjuvant at 2-week intervals. Immunoaffinity antibody purification was performed by coupling 5.0 mg of antigen to 5 ml of agarose gel. The crude serum was applied to the gel-containing column. After the washes, the adsorbed protein was eluted at low pH and collected in Tris buffer to neutralize the elution buffer. The purified antibody was concentrated and dialyzed against phosphate buffered saline using an Amicon stirred cell concentrator.
Western blot analysis.
Leaf tissue (100 mg) of plants was harvested and ground in 1 ml of ice-cold extraction buffer (phosphate buffer ]pH 6.8], 50 mmol/liter; EDTA, 10 mmol/liter; 0.1% Triton; 0.1% Sarkosyl; dithiothreitol, 1 mmol/liter). The homogenate was centrifuged at 13,000 × g for 10 min at 4°C. The supernatant was used as a crude enzyme sample. The protein amount was determined using a DC protein assay kit II (Bio-Rad Laboratories, Inc.). The crude enzyme samples were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Proteins blotted onto Hybond P membrane (Amersham Pharmacia Biotech) from SDS-PAGE gel were probed with antibody against CbnA at a 1:1,000 dilution. Proteins recognized by the CbnA antibody were visualized by chemiluminescence Plus Western blotting detection kit (Amersham Pharmacia Biotech) as described in manufacturer's protocol.
Enzyme assay of chlorocatechol dioxygenase activity.
Quantitative enzyme assay of the transgenic calluses and leaf tissues was performed by incubating 1 ml of 2-mmol/liter 3-chlorocatechol in phosphate buffer (50 mmol/liter; pH 7.4) with transgenic calluses or leaf tissues. For transgenic rice calluses, 50 mg of the transgenic calluses grown on the solid medium was added to the 3-chlorocatechol solutions and incubated with gentle shaking. Leaf tissues (50 mg) from the transgenic rice plants were incubated with the 3-chlorocatechol solutions. After incubation, transformation of 3-chlorocatechol to 2-chlormuconate was analyzed using high performance liquid chromatography (HPLC). After incubation of transgenic callus or leaf tissues with 3-chlorocatechol as described above, substrate consumption in the supernatant was measured by HPLC (model CL 6A; Shimazu, Kyoto, Japan) on a unisil Q C18 (GL Sciences, Inc.) reverse-phase column, using acetonitrile:10-mmol/liter H3PO4 (pH 2.5) (50:50, vol/vol) as the solvent at a flow rate of 1 ml min−1. After injection of 20 μl of the sample, absorbance at 230 nm was measured. E. coli cells expressing the cbnA gene were incubated with 3-chlorocatechol and 3-chlorocatechol was converted to 2-chloromuconate. This was used as a standard of 2-chloromuconate to verify its retention time on HPLC analysis.
Other methods.
Genomic DNAs from rice callus and plants were isolated as described elsewhere (13). Other recombinant DNA methods were performed as described previously (10).
RESULTS
Expression of the cbnA gene in rice calluses.
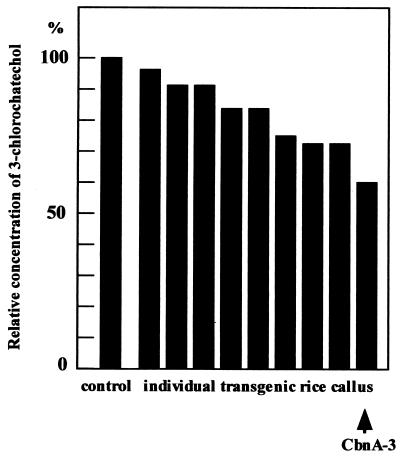
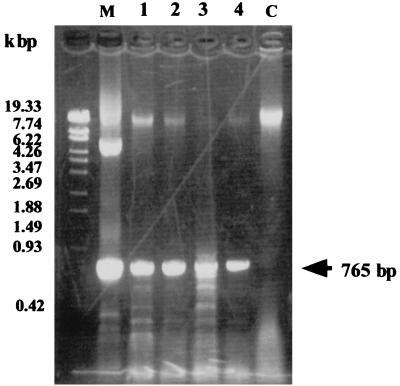
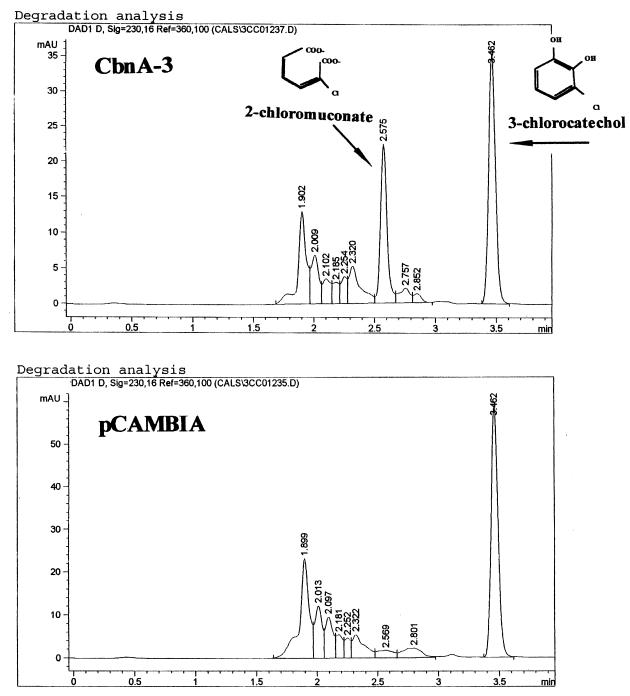
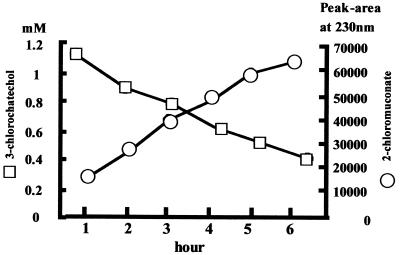
Transgenic rice calluses with the hygromycin-resistant phenotype were isolated. Chlorocatechol dioxygenase activity was quantitatively analyzed to determine the expression of the cbnA gene in the rice transgenic calluses. Consumption of 3-chlorcatechol in the assay solution by each transformants was compared to that of callus harboring Ti vector without the cbnA gene (Fig. 2). Among 16 transformants tested, nine calluses showed clear activity. Integration of the cbnA gene into their chromosomes was assessed with the PCR method. All of the calluses that showed dioxygenase activity had the cbnA gene in their genomes (Fig. 3). One of the clones, CbnA-3, was used for further quantitative analysis of chlorocatechol dioxygenase activity by HPLC. After incubation of the transgenic rice calluses with 3-chlorocatechol for 3 h, 3-chlorocatechol was effectively converted to 2-chloromuconate (Fig. 4). Conversion of 3-chlorocatechol to 2-chloromuconate by the transgenic rice calluses was time-dependent (Fig. 5) and after 16 h incubation, 3-chlorocatechol was completely converted to 2-chloromuconate. These results strongly indicated that the cbnA gene was successfully expressed in rice cells.
FIG. 2.
Dioxygenase activity in callus of different transgenic rice. Relative consumption of 3-chlorocatechol in the reaction mixture was shown. The concentration of 3-chlorocatechol in the reaction mixture was analyzed by HPLC. 3-chlorocatechol concentration in the reaction mixture with the control callus harboring only a Ti binary vector was shown as 100%.
FIG. 3.
PCR amplification of a 675-bp fragment of the cbnA gene integrated into transgenic rice calluses. The transgenic line number is indicated at the top of each lane. M, pCAMBIA-E35S-cbnA plasmid as a positive control for PCR; C, a control transgenic plant harboring a pCAMBIA vector.
FIG. 4.
Oxidation of 3-chlorocatechol to 2-chloromuconate by transgenic rice cell line CbnA-3. Transgenic rice cell line CbnA-3 expressing the cbnA gene and the control cell line harboring pCAMBIA were incubated in 3-chlorocatechol for 3 h. After incubation, supernatants of the solutions were analyzed by HPLC.
FIG. 5.
Time-dependent oxidation of 3-chlorocatechol to 2-chloromuconate by the transgenic cell line CbnA-3. The transgenic rice cell line CbnA-3 was incubated in 3-chlorocatechol as described in Fig. 4. Concentration of 3-chlorocatechol at each time point was analyzed by HPLC. Levels of 2-chloromuconate are given as peak areas.
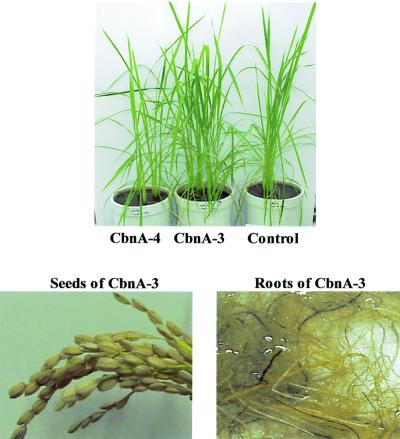
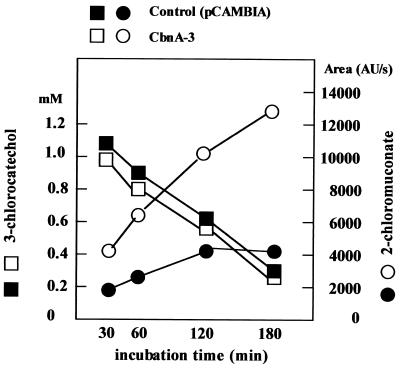
Rice plants were regenerated from the transgenic callus expressing the cbnA gene. Transgenic rice plants could not be distinguished morphologically from the control rice plants harboring the pCAMBIA vector (Fig. 6). To confirm the expression of the cbnA gene in transgenic rice plants, leaf tissues of transgenic plants were cut into small pieces with a razor blade and used for quantitative assay of chlorocatechol dioxygenase activity. In the quantitative assay by HPLC, conversion of 3-chlorcatechol to 2-chloromuconate by transgenic leaf tissues was confirmed and the transformation was time dependent, suggesting that 3-chlorocatechol was degraded by the chlorocatechol dioxygenase produced in the transgenic rice plants (Fig. 7). Decrease of 3-chlorocatechol was also observed when control leaf tissues harboring Ti vector without the cbnA gene was used. This may due to the adsorption of 3-chlorocatechol to leaf tissues, since 2-chlormuconate was not detected after a 6-h incubation.
FIG. 6.
Morphology of the transgenic rice plants. Each tissue of the transgenic rice plants is compared with a control plant harboring pCAMBIA1300. Leaves and roots were morphologically normal and the transgenic rice plants were fully fertile and grains were morphologically indistinguishable.
FIG. 7.
Time-dependent oxidation of 3-chlorcatechol to 2-chloromuconate by leaf tissues of transgenic rice plant. Leaf tissue of transgenic rice plant CbnA-3 was incubated in 3-chlorcatechol and concentration of 3-chlorocatechol and 2-chloromuconate were analyzed by HPLC as described in Materials and Methods.
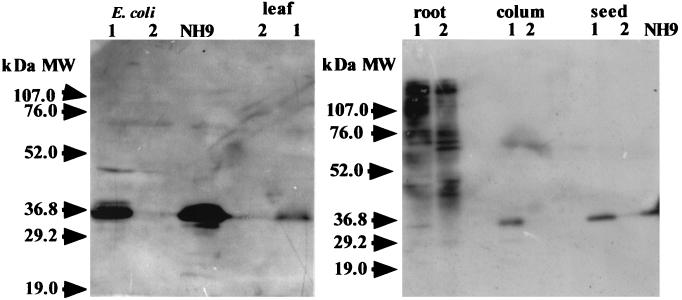
Antibody raised against the CbnA peptide specifically reacted with CbnA produced in R. eutropha NH9 and E. coli (Fig. 8). Predicted molecular weight of CbnA calculated from the deduced amino acid sequence is 27522. The molecular weight of CbnA on SDS-PAGE was higher than the predicted molecular weight. Although we did not the exact reason, this may due to the amino acid composition of CbnA or posttranslational modification. Western blot analysis using anti-CbnA antibody indicates that the cbnA gene was expressed in leaves, culms, roots, and seeds of the transgenic rice plants (Fig. 8). Expression levels of the cbnA gene in above ground organisms were similar. However, expression of the cbnA gene in roots was very low. Although many protein bands were observed in the root cell extracts, the antibody bound nonspecifically to these products because the same bands were also detected in cell extracts from control plants harboring the pCAMBIA vector. Genomic Southern blot analysis indicated that the cbnA gene was integrated into chromosomes of the transgenic rice plants (Fig. 9).
FIG. 8.
Western blot analysis of extracts from transgenic rice plant line CbnA-3. Cell extract from tissues of each transgenic rice plant line CbnA-3 was analyzed by Western blot analysis with anti-CbnA antibody as described in MATERIALS AND METHODS. NH9, R. eutropha NH9; E. coli lane1, E. coli harboring pUC19-cbnA; lane 2, E. coli harboring pUC19; transgenic rice plant lane 1, control rice plant harboring pCAMBIA1300; Lane 2, transgenic rice plant line CbnA-3.
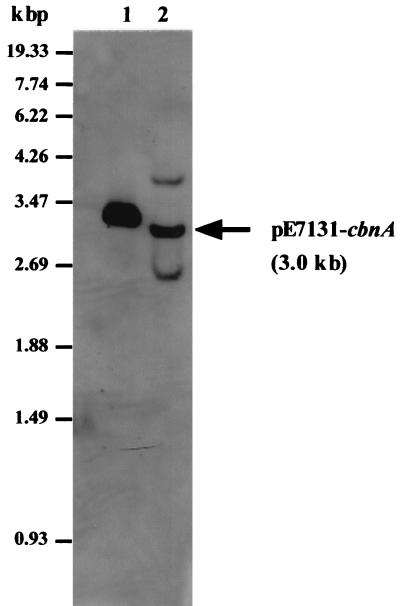
FIG. 9.
Southern blot analysis of the transgenic rice plant line CbnA-3. Genomic DNA from the transgenic rice plant line CbnA-3 was digested with HindIII and EcoRI, which cut out the promoter-cbnA-terminator cassette (see Fig. 1) and Southern blot was probed with a 765-bp fragment of the cbnA gene labeled with digoxigenin. Hybridized bands were detected with antidigoxigenin antibody. Lane 1, pCAMBIA-E35S-cbnA; lane 2, Genomic DNA from the transgenic rice plant line CbnA-3.
Generation of T2 plants.
Seeds were harvested from the T0 transgenic rice plants and germinated on the hygromycin-containing plates. T1 plants harboring the cbnA gene were selected. All of the T1 plants that grew on the hygromycin-containing plate had chlorocatechol dioxygenase activity and Western blot analysis indicated that they expressed the cbnA gene. Further, T2 plants were grown and confirmed the expression of the cbnA gene. These results suggest that expression of the cbnA gene did not have toxic effects on rice plants. Molecular breeding of transgenic crops or other plants expressing the cbnA gene is therefore possible.
DISCUSSION
Rice is a very important crop in Asia. Several herbicides and pesticides containing chlorinated compounds have been used and have spread in the environment. Therefore, phytoremediation of these chemical compounds will be a powerful technique to degrade chlorinated pollutants in soil. Molecular breeding of transgenic rice plants that express genes encoding enzymes to degrade chlorinated chemical compounds would enable us to remove these chemical compounds from soil and surface water in fields. Chlorocatechols are intermediate products of the degradation of polychlorinated biphenyls via chlorobenzoates, and conversion of chlorocatechols to chloromuconates is a rate-limiting step for complete degradation of polychlorinated biphenyls (3). Therefore, we introduced the cbnA gene encoding chlorocatechol dioxygenase from R. eutropha NH9 into rice plants. The cbnA gene under the control of the E7131 promoter, which has seven tandemly repeated core enhancer sequences of the CaMV35S promoter, was successfully expressed in rice plants. In contrast to rice plant, the cbnA gene was not expressed in tobacco BY-2 cells under the control of the CaMV35S promoter. This might be due to the high GC content (65%) of the cbnA gene, because GC contents of the genes from dicot plants such as tobacco (lower than 50%) are know to be lower than those of monocot plants such as rice (based on gene sequence deposited in GenBank). Therefore, codon usage of the cbnA gene would be preferable for its expression in rice plants.
3-Chlorocatechol was converted to 2-chloromuconate when incubated with the entire transgenic callus, suggesting that 3-chlorocatechol and 2-chloromuconate can pass through the cell wall and cell membrane because chlorocatechol dioxygenase produced by the rice cells localizes in the cytoplasmic space. Also, leaf tissues of the transgenic rice plants transformed 3-chlorcatechol to 2-chloromuconate. However, we could not show that roots of the transgenic rice plant transform 3-chlorcatechol to 2-chloromuconate after a 16-h incubation. This may due to the lower expression of the cbnA gene in root tissue and/or lower penetration efficiency of 3-chlorocatechol into root tissue. It may require longer incubation time to degrade 3-chlorocatechol by root. Rapid degradation of 3-chlorocatechol by root tissue would be achieved by high level expression of the cbnA gene in surface cells of roots using a root specific promoter to drive the cbnA gene.
We were able to breed transgenic rice plants that express the bacterial chlorocatechol dioxygenase gene. The transgenic rice plants will aid in the microbial degradation of chlorinated compounds in soil and surface water. In the future, phytoremediation using this type of transgenic plant producing several kinds of enzymes that degrade chlorinated compounds will be a cost-effective and sustainable technology for removing chlorinated pollutants from the environment.
Acknowledgments
This work was supported by the Ministry of Education, Culture, Sports, Science, and Technology of Japan grant 411 (The environmental risk of endocrine disruptor) and a Ministry of Agriculture, Forestry and Fisheries of Japan (Development of innovative plants and animals using transformation and cloning).
We thank M. Ugaki of National Institute of Agrobiological Science for providing pE7131. We acknowledge the Center for Genetics and Molecular Biology, Mie University, for allowing growth of transgenic rice plants to be conducted in one of their greenhouses.
M. Shimizu and T. Kimura contributed equally to this work.
REFERENCES
- 1.Alexander, M. 1981. Biodegradation of chemicals of environmental concern. Science 211:132-138. [DOI] [PubMed] [Google Scholar]
- 2.Cunningham, S. D., W. R. Berti, and J. W. Huang. 1995. Phytoremediation of contaminated soils. Trends Biotechnol. 13:393-397. [Google Scholar]
- 3.Furukawa, K. 1982. Microbial degradation of polychlorinated biphenyls, p. 33-57. In A. M. Chakrabarty (ed.), Biodegradation and detoxification of environmental pollutants. CRC Press, Boca Raton, Fla.
- 4.Hiei, Y., S. Ohta, T. Komari, and T. Kumashiro. 1994. Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J. 62:271-282. [DOI] [PubMed] [Google Scholar]
- 5.Kramer, U., and A. N. Chardonnens. 2001. The use of transgenic plants in the bioremediation of soils contaminated with trace elements. Appl. Microbiol. Biotechnol. 55:661-672. [DOI] [PubMed] [Google Scholar]
- 6.Mitsuhara, I., M. Ugaki, H. Hirochika, M. Ohshima, T. Murakami, Y. Gotoh, Y. Katayose, S. Nakamura, R. Honkura, S. Nishimiya, K. Ueno, A. Mochizuki, H. Tanimoto, H. Tsugawa, Y. Otsuki, and Y. Ohashi. 1996. Efficient promoter cassettes for enhanced expression of foreign genes in dicotyledonous and monocotyledonous plants. Plant Cell Physiol. 371:49-59. [DOI] [PubMed] [Google Scholar]
- 7.Ogawa, N., S. M. McFall, T. J. Klem, K. Miyashita, and A. M. Chakrabarty. 1999. Transcriptional activation of the chlorocatechol degradative genes of Ralstonia eutropha NH9. J. Bacteriol. 181:6697-6705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ogawa, N., and K. Miyashita. 1995. Recombination of a 3-chlorobenzoate catabolic plasmid from Alcaligenes eutrophus NH9 mediated by direct repeat elements. Appl. Environ. Microbiol. 61:3788-3795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pfister, R. M. 1973. Interaction of halogenated pesticides and microorganisms: a review, p. 1-33. In A. I. Laskin and H. Lechevalier (ed.), Handbook of microbiology. CRC Press, Boca Raton, Fla. [DOI] [PubMed]
- 10.Reineke, W. 1998. Development of hybrid strains for the mineralization of chloriaromatics by patchwork assembly. Annu. Rev. Microbiol. 52:287-331. [DOI] [PubMed] [Google Scholar]
- 11.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 12.van der Meer, J. R., W. M. de Vos, S. Harayama, and A. J. B. Zehnder. 1992. Molecular mechanisms of genetic adaptation to xenobiotic compounds. Microbiol. Rev. 56:677-694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wagner, D. B., G. R. Furnier, M. A. Saghai-Maroof, S. M. Williams, B. P. Dancik, and R. W. Allard. 1987. Chloroplast DNA polymorphisms in lodgepole and jack pines and their hybrids. Proc. Natl. Acad. Sci. USA 84:2097-2100. [DOI] [PMC free article] [PubMed] [Google Scholar]