Abstract
Here, we describe a multigenomic DNA sequence-analysis tool, evoprinter, that facilitates the rapid identification of evolutionary conserved sequences within the context of a single species. The evoprinter output identifies multispecies-conserved DNA sequences as they exist in a reference DNA. This identification is accomplished by superimposing multiple reference DNA vs. test-genome pairwise blat (blast-like alignment tool) readouts of the reference DNA to identify conserved nucleotides that are shared by all orthologous DNAs. evoprinter analysis of well characterized genes reveals that most, if not all, of the conserved sequences are essential for gene function. For example, analysis of orthologous genes that are shared by many vertebrates identifies conserved DNA in both protein-encoding sequences and noncoding cis-regulatory regions, including enhancers and mRNA microRNA binding sites. In Drosophila, the combined mutational histories of five or more species affords near-base pair resolution of conserved transcription factor DNA-binding sites, and essential amino acids are revealed by the nucleotide flexibility of their codon-wobble position(s). Conserved small peptide-encoding genes, which had been undetected by conventional gene-prediction algorithms, are identified by the codon-wobble signatures of invariant amino acids. Also, evoprinter allows one to assess the degree of evolutionary divergence between orthologous DNAs by highlighting differences between a selected species and the other test species.
Keywords: comparative genomics, evolution, gene structure and function
Deciphering the regulatory mechanisms that control coordinate gene expression is a long-standing goal of biology. The comparison of orthologous DNA sequences from multiple vertebrate or invertebrate species promises to identify the cis- regulatory elements that are central to the dynamic interplay between a gene and its transcriptional regulators (1-3). This cross-species comparison, termed phylogenetic footprinting, is based on the hypothesis that functionally important sequences evolve at a significantly slower rate than nonfunctional DNA (1). Phylogenetic footprinting has been used successfully to discover multispecies-conserved sequences (MCSs) that are critical for gene function (reviewed in refs. 2, 4, and 5). An essential first step in this process is the alignment of multiple orthologous DNAs. Multisequence-alignment programs include threaded blockset aligner (6), footprinter (7), conreal (5), and phyme (8). The multiDNA alignments are accomplished either by simultaneous or sequential pairwise alignments of input DNAs, with alignment gaps introduced to optimize the overall homology comparisons.
Individual genome searches have also been commonly used to initiate MCS searches, and two popular whole-genome search algorithms are blast (9) and blat (blast-like alignment tool) (10). One significant difference between the blast and blat algorithms is that blat keeps an index of a species genome in memory and uses this index to scan linearly through the query sequence, whereas blast indexes the query sequence first and then scans linearly along the database. This fundamental difference is the primary reason a blat alignment is significantly faster than other whole-genome alignment algorithms (10). The speed of blat alignment and the current availability of 13 vertebrate and seven Drosophila species blat-formatted genomes (see the Human blat Search database, available at http://genome.ucsc.edu/cgi-bin/hgBlat) enables rapid reference-DNA vs. test-genome pairwise homology searches of related or evolutionary distant species.
Taking advantage of the speed of the blat alignment and the availability of multiple blat-formatted genomes, we developed a simple multigenomic comparative tool that allows one to rapidly identify MCSs as they appear in a species of interest. The evoprinter algorithm superimposes multiple blat readouts of individual reference-DNA vs. test-genome alignments to generate an evolutionary gene print (EvoP) of invariant DNA sequences as they appear in the reference DNA. Unlike most multispecies-alignment programs that display MCSs as consecutive columns of invariant nucleotides interspersed by alignment gaps, the EvoP readout displays only the reference DNA, with no alignment gaps, highlighting a species-centric representation of the conserved sequences. To facilitate the comparative analysis of evolutionary changes between test species, a second algorithm, evodifference (evodif) enables one to identify MCSs that are common to all but one of the test genomes.
To demonstrate the efficacy of evoprinter as a phylogenetic-footprinting tool, we show how EvoPs of well characterized genes (one vertebrate and one Drosophila gene) accurately identify DNA sequences that have been shown to be essential for gene function. Also, we describe how evoprinter can be used to identify genes that had not been noticed by conventional gene-prediction methods.
Materials and Methods
evoprinter is a tool for discovering MCSs that are shared among three or more orthologous DNAs. The program uses the reference DNA outputs of blat alignments and then identifies the sequences within this DNA that are shared by all species. evoprinter is a javascript program that runs on the user's computer. Its algorithm creates an array of strings from the selected blat outputs and then looks for conservation of sequence by looping through the strings one letter at a time (outputting a black capital letter only for the reference DNA nucleotides that are aligned in all test species). Nucleotides within the reference DNA that are not shared are represented by lowercase gray letters. The program requires an up-to-date web browser, and javascript has to be enabled. There is no arbitrary limit on sequence capacity. For example, a 50-kb EvoP can be generated by splicing together two 25-kb blat outputs. The second evodif algorithm reveals what is different in any one species from the EvoP of all other test species (described below).
The first step in generating an EvoP is the curation of the reference DNA (up to 25 kb per alignment) from the University of California, Santa Cruz Genome Browser database (http://genome.ucsc.edu/cgi-bin/hgGateway), the Ensembl database (available at: www.ensembl.org), or the FlyBase database (http://flybase.net). When copied and pasted into the blat engine input window (http://genome.ucsc.edu/cgi-bin/hgBlat), the pairwise alignment is performed between the reference DNA and a selected test species, and the highest-scoring readout alignment is then selected. The readout labeled as “YourSequence” (showing the reference DNA) is then copied and pasted into one of the evoprinter input windows (http://evoprinter.ninds.nih.gov) without removing numbering or spaces. This procedure is repeated with the same reference DNA vs. as many test species as required. evoprinter can also be used to generate a protein EvoP from blat alignments of amino acid sequences.
One important feature of the evoprinter program is its ability to generate EvoPs from subsets of the selected blat readouts by unchecking the species or groups of species to be excluded. This flexibility is particularly useful when assessing whether the loss of an MCS or group of MCSs in one or more blat alignments is caused by (i) small mutational differences; (ii) chromosome rearrangements, including large insertions and/or deletions, resulting in loss of sequence colinearity; (iii) overall sequence divergence being so great that alignment is not achieved for short homologies; or (iv) sequencing gaps in the test genome.
To identify MCSs that are shared by all but one of the test species, deselect all of the test-species readouts that were entered into the evoprinter except for the species in question, and then select the “Highlight Species Differences” button to generate the evodif readout. The lowercase red letters are nucleotides that are lost from the final EvoP if that species is included in the comparison. In addition to assessing the degree of evolutionary divergence, the evodif is particularly useful for identifying chromosome rearrangements (identified by uninterrupted blocks of lost MCSs). Color formatting of the EvoP and evodif readouts can be maintained by dragging the saved HTML output into word (Microsoft).
Identification of potential transcription-factor DNA-binding sites was carried out by using matinspector (11). MicroRNA binding sites in Drosophila were identified as described (12), and human microRNA binding sites were identified by using the Human miRNA Viewer database (www.cbio.mskcc.org/mirnaviewer) as described (13).
Results and Discussion
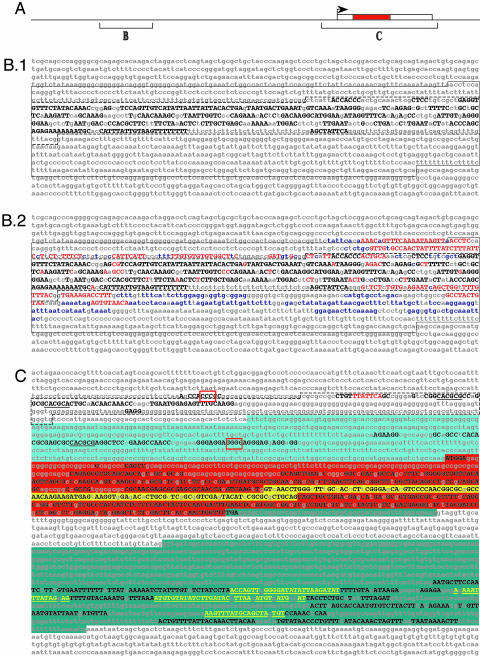
evoprinter Analysis of Vertebrate Achaete-Scute Homologue 1 (Ascl1) Genes Identifies DNA Sequences That Are Essential for Its Expression and Function. The basic helix-loop-helix (bHLH) Ascl1 transcription factor has a critical role in establishing neural cell identities in the developing vertebrate embryo (see refs. 14-16 and references therein). Studies on the mammalian Ascl1 gene (Mash1) demonstrate that it is dynamically expressed in many proliferating CNS and peripheral nervous system neural progenitor cells (NPCs) during murine development (17, 18) and it is also tightly regulated in NPCs that give rise to pulmonary neuroendocrine cells (19). Cis-regulatory elements important for Mash1 expression in mice have been identified in the 5′ flanking intragenic DNA and within the 3′ noncoding region of its transcribed sequence (20, 21). Transgenic studies have localized the Mash1 CNS enhancer to an 1,158-bp region located 7 kb 5′ to its transcription start site (boxed sequence in Fig. 1B) (20). Further dissection of this enhancer has revealed that most of the region- and tissue-specific regulatory elements map to an internal 472-bp region (dashed box in Fig. 1B), and elements that modulate expression levels (enhance or reduce) in flanking sequences map both 5′ and 3′ to the 472-bp region (21).
Fig. 1.
evoprinter analysis of the vertebrate achaete-scute homolog 1 locus. (A) A linear cartoon of the Ascl1 locus 15 kb used in the EvoP analysis indicating the approximate locations of sequences shown in B and C (box represents transcribed region with the red-colored inner box indicating the ORF). (B and C) EvoPs were generated with 15 kb of mouse (B) or human (C) reference-DNA that included the Ascl1-transcribed sequence plus 9 kb of upstream and 3 kb of downstream flanking intragenic sequence. We searched the following test genomes: human, chimpanzee, rhesus monkey, dog, rat, mouse, oppossum, chicken, and X. tropicalis. Invariant MCSs, shared by all test species, are identified with uppercase black letters. (B.1) An EvoP, using all test species, identifies clustered MCSs within the tissue- and region-specific regulatory region of the murine Mash1 CNS enhancer. Shown is the upper DNA strand of 1.9 kb corresponding to nucleotides -8692 to -6784 5′ to the murine Mash1-transcribed region. The solid lined box denotes the 1,158-bp CNS enhancer region, and the dashed-lined inner box identifies the 472-bp domain that contains multiple tissue/region-specific regulatory elements (21). (B.2) The MCSs that are gained when X. tropicalis is excluded from the analysis are shown as uppercase red letters and when both X. tropicalis and chicken genomes are excluded from the EvoP, the additional MCSs are shown as blue lowercase letters. Nonconserved nucleotides are indicated as lowercase gray letters. (C) EvoP analysis of the ash1 proximal promoter region, transcribed sequence, and flanking 3′ intragenic sequence reveals conserved MCSs that contain cis-regulatory and protein-encoding sequences. Shown is 3.9 kb of the human hash1 gene (nucleotides -687 to +3235). The hatched line box denotes a 259-bp region that contains the proximal enhancer and tissue-specific repressor regulatory elements (22). Underlined sequences are HES-1 DNA-binding sites, red-boxed sequences are potential binding sites for IA-1 and a potential FAST-1 binding site is highlighted with red-colored letters (see Results and Discussion). The 5′ UTR of the hash1 transcript is highlighted in light blue, the transcript ORF is shown with red background (the HLH coding sequence is marked with yellow background), and the 3′ untranslated sequence is indicated with a dark blue background. Yellow nucleotides in the 3′ trailer represent potential binding sites for 13 different microRNAs (see Results and Discussion). Note that the 3′ UTR is interrupted by a 359-bp intron (annotation according to the Ensembl sequence data base).
Remarkably, a 15-kb EvoP of this region generated from human, chimpanzee, rhesus monkey, dog, rat, mouse (reference DNA), opossum, chicken, and Xenopus tropicalis DNA identifies a dense cluster of MCSs that are distributed throughout the critical tissue-specific regulatory region (Fig. 1). When the more evolutionarily distant X. tropicalis and chicken species are excluded from the analysis, additional MCSs are identified in enhancer-activator sequences flanking the core tissue-specific regulatory region (Fig. 1B.2). evodif prints of the individual test species revealed also that the opossum has lost MCSs in the 3′ negative-regulatory element (21) that are present in higher vertebrates (data not shown). Outside of the clustered conserved sequences that were detected in the initial EvoP, no MCSs were identified in the flanking 5′ upstream 3.2-kb and 3′ downstream 5.3-kb regions (Fig. 1B and data not shown). The ability of an EvoP to identify biologically significant DNA within the context of reference DNA in excess of 10-kb demonstrates its usefulness as a phylogenetic-footprinting tool.
Transcription-factor DNA-binding site searches have revealed that many of the MCSs have core DNA-binding motifs for different transcription factors, such as homeodomain, bHLH, or Zn-finger proteins, and some have multiple interlocking binding sites for different factors (ref. 20 and data not shown). EvoPs of other characterized vertebrate enhancers have identified clustered MCSs within all cis-regulatory elements examined. For example, within the 90-bp tissue-specific region of the murine anterior neuroectoderm OTX2 enhancer (22), the EvoP reveals that 86% of the nucleotides (77 bp) are part of MCSs (data not shown).
Constitutive expression of human Ascl1 (hash1) gene in lung neoplasms is a feature of one of the most virulent forms of lung cancer, small-cell lung cancer (SCLC) (23, 24). SCLC cell culture studies have demonstrated that hash1 expression in neuroendocrine tumors is controlled in part by a proximal enhancer positioned -234 to -46 from the transcribed sequence and a proximal repressor region located at -308 to -234 (24) (both regions are shown in the hatched box in Fig. 1C). These studies have also indicated that the mammalian homologue of the Drosophila Hairy transcription factor, HES-1 (Hairy Enhancer of Split 1), functions as a direct repressor of hash1 expression by binding to a HES-1 binding site in the proximal promoter region (24). Our EvoP identified a cluster of MCSs within the proximal enhancer/repressor region of the vertebrate ash-1 and two of these MCSs contain HES-1 binding sites (underlined in the dashed box in Fig. 1C). MCSs containing DNA-binding sites for other known transcription factors were also identified. For example, IA-1 (25) and the FAST-1 Smad-interacting protein (26) transcription-factor DNA-binding sites are present in proximal promoter MCSs (Fig. 1C).
In addition to identifying MCSs within the upstream enhancer and proximal promoter, EvoP analysis of the transcribed region revealed multiple MCSs in the 5′ untranslated leader, one of which contains a canonical HES-1 binding site, whereas another harbors a docking site for IA-1 (25) (Fig. 1C, red underline). IA-1 binding sites were also found in proximal and CNS enhancer MCSs (see above). Also suggesting that IA-1 may be a direct regulator of Ascl-1 expression, recent studies (27) have revealed that IA-1 is dynamically expressed in murine CNS neural progenitor cells.
In the Ascl-1 protein-encoding sequence, multiple MCSs are present and most delineate essential amino acid codons as deduced from invariant nucleotides in critical codon positions. A protein EvoP of the different vertebrate Ascl-1 amino acid sequences confirms that many of the conserved nucleotides identified in the genomic EvoP are positioned in invariant codon positions (data not shown).
Within the 3′ UTR of the Ascl1 transcript, the EvoP identifies a dense cluster of MCSs that spans 600 bp of the 1.3-kb trailer (Fig. 1C), and five of the conserved regions (yellow underline in Fig. 1C) harbor potential mRNA binding sites for 13 different human microRNAs (13). In light of studies (21) indicating that the murine Mash1 is under posttranscriptional control, mediated by sequence(s) located 3′ of the protein-coding region (21), it is likely that many or all of these microRNA binding sites are physiologically relevant.
Intragenus evoprinter Analysis of the Drosophila Krüppel (Kr) Gene Loci Identifies Functionally Important DNA. As a second example of the usefulness of evoprinter, we generated an EvoP of the well characterized Drosophila Kr transcription-factor gene. Kr plays multiple roles essential to different phases of Drosophila development. Initially identified as a regulator of thoracic and abdominal segmental identity in the early embryo (ref. 28 and see ref. 29 for review), Kr gene function has been shown to be required for the development of the Malpighian tubule (kidney) (30), muscle (31), and the nervous system (32, 33).
Detailed studies of the cis-regulatory elements that control Kr embryonic expression have identified multiple enhancer regions located upstream of its transcribed sequence (34). The genomic regions that control early blastoderm, muscle precursor, amnioserosa, or CNS expression are shown in Fig. 2. In the early pregastrula embryo, the enhancer regions CD1 and CD2 are required for Kr expression in the central domain of the blastoderm, and the AD1 and AD2 enhancer regions regulate expression in the anterior portion of the late blastoderm (34, 35). During late embryonic development, cis elements that regulate Kr expression in muscle precursor cells and amnioserosa cells have been mapped to CD1 (34). The AD2 region also harbors nervous system-specific (NS2) control elements (34).
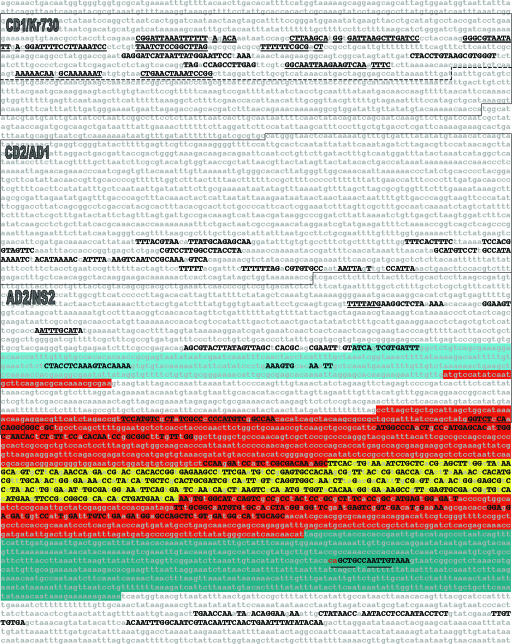
Fig. 2.
evoprinter analysis of the Drosophila Kr gene. The 7.7-kb (upper strand) of a 12-kb genomic EvoP that corresponds to the D. melanogaster reference DNA (nucleotides -4,207 to + 3,531) is shown. The EvoP was generated from blat readouts of the reference DNA aligned with D. simulans, D. yakuba, D. ananassae, D. pseudoobscura, and D. virilis DNAs. MCSs that are shared by all species are shown as uppercase black nucleotides. Boxed sequences represent the cis-regulatory regions described in Results and Discussion. Underlined MCSs within the CD1/Kr730 box contain known transcription-factor binding sites (35, 36). Underlined sequences in the AD2/NS2 box contain potential HB (TTTTAGT) and PDM1 (ATTTGCAT) DNA-binding sites, respectively. The D. melanogaster Kr transcribed sequence is annotated according to FlyBase as follows: 5′ untranslated leader (light blue), protein-encoding sequence (red; Zn finger domain, yellow), and the 3′ untranslated sequence (dark blue). Note that the protein-encoding sequence is interrupted by a 373-bp intron. The underlined nucleotides in the 3′ untranslated transcribed sequence correspond to E-box bHLH binding sites. evodif analysis of the individual test species revealed that the first two nucleotides of the first E-box (red letters) are shared by all tested species except for D. yakuba and D. ananassae.
Further dissection of the 1,159-bp CD1 region revealed that only its first 730 bp (Kr730, dashed line in Fig. 2) contain most, if not all, of the CD1 cis-regulatory elements (36). Cis elements that regulate Kr expression in muscle precursor cells and CNS ventral midline cells also map to the last 3′ 295 bp of Kr730 (34). These studies also demonstrated that Kr730 contains Bicoid (Bcd), Hunchback (Hb), Knirps (Kni), and Tailless (Tll) transcription factor in vivo responsive elements, and in vitro DNA-binding studies have also demonstrated that each of these transcription factors bind directly to different regions of Kr730 (35, 36).
An EvoP of 12 kb spanning the Kr genomic locus using Drosophila melanogaster DNA as the reference DNA and Drosophila simulans, Drosophila yakuba, Drosophila ananassae, Drosophila pseudoobscura, and Drosophila virilis as test genomes has identified multiple MCSs within Kr730 but not in the remaining 420 bp that were found not to be essential for CD1 enhancer activity (36) (Fig. 2). Remarkably, all but three of the MCSs in the Kr730 region are contained in or overlap DNAse1-protected footprinted sequences of the transcription factors mentioned above (underlined MCSs in Kr730, Fig. 2). For example, the 5′- and 3′-most Kr730 MCSs identified by the EvoP are overlapping Bicoid (Bcd)/Hb and Knirps (Kni)/Bcd DNA-binding sites, respectively (35, 36). Analysis of the MCSs positioned within the CD2/AD1 and AD2/NS2 also identify multiple potential DNA-binding sites for homeodomain, Hb, Bcd, and POU domain transcription factors. For example, the 5′-most MCS in the AD2/NS2 region contains a consensus Hb docking site (TTTTATG), and the third 5′-most MCS in this region contains a canonical POU domain DNA-binding Octamer motif (ATTTGCAT) (Hb and POU binding sites, underlined in Fig. 2). Interestingly, CNS cis-regulatory elements map to this region (34), and studies have shown that Kr neuroblast expression is preceded by expression of Hb, which is a known repressor of Kr, and temporally followed by the Octamer-binding POU domain transcription factors Pdm-1 and Pdm-2 (ref. 33 and see ref. 37 for review). evoprinter analysis of other characterized Drosophila genes has identified MCSs within all examined enhancer regions (10 genes and 20 enhancers; data not shown).
Within the Kr transcribed sequence, EvoP identifies multiple clusters of MCSs, many of which encode essential amino acids as identified by their wobble signatures (identified by two or more 2-bp MCSs separated by single nonconserved nucleotides; Fig. 2). The region that encodes the five consecutive Zn-fingers spanning 146 aa (highlighted in yellow in Fig. 2) is especially prominent. Excluding four nonwobble methionine (ATG) codons in the Zn-finger domain, the genomic EvoP reveals that only 39 of the remaining 142 codons are invariant in all species for all three codon positions. Of these 39 codons, 28 of the encoded amino acids have restricted wobble, allowing for only two different nucleotide substitutions in the third codon position. Although the genomic EvoP found conserved amino acids within the Zn-finger region and in the immediate flanking domains, two additional conserved protein domains were missed either partially or completely [the N-terminal transrepressor1-transactivator1 region (38, 39) and the C-terminal C64 repressor domain (40, 41), respectively (Fig. 2)]. However, when D. virilis and D. pseudoobscura are excluded from the EvoP, both the N- and C-terminal encoding domains are revealed by invariant amino acid wobble signatures (data not shown).
In the 3′ untranslated sequence of the Kr transcribed region, the EvoP identified a single MCS (Fig. 2). A genomewide search for 3′ UTR microRNA binding sites (12) identified a potential miR-34 mRNA binding-site that overlaps the first seven nucleotides of this MCS (data not shown). Interestingly, this MCS also contains a bHLH E-box consensus DNA-binding site (CAATTG) and when D. yakuba and D. ananassae are excluded from the EvoP, the MCS includes two additional 5′ nucleotides and now contains a second E-box (CAGCTG) (both E-boxes are underlined, Fig. 2). Additional MCSs were detected 3′ to the transcribed region (Fig. 2). Although a recent study (42) did not detect any posttranslational regulation of Kr mRNA in the embryo, and analysis of the 3′ downstream intragenic region did not identify any additional embryonic cis-regulatory elements (34), the possibility remains that some or all of these MCSs may have a role in controlling larval or adult Kr expression.
evoprinter Uncovers a Small Peptide Gene Not Identified in the Current FlyBase Annotation of the Drosophila Genome. The EvoP has the potential to discover small protein-encoding genes that had been previously unannotated by conventional gene-prediction methods. For example, EvoP exploration of the 12.9-kb intragenic region between the Drosophila beta amyloid protein precursor-like gene (43) and the ventral nervous system defective (vnd) gene (44) has identified a cluster of MCSs that were invariant in the D. melanogaster, D. simulans, D. yakuba, D. ananassae, D. pseudoobscura, D. virilis, and Drosophila mojavensis species. Positioned 8.5 kb upstream of the vnd transcribed sequence, portions of the MCS cluster possessed all of the hallmarks of an ORF that encodes short runs of conserved amino acids (multiple 2-bp MCSs separated by single nonconserved nucleotides) (Fig. 3). Further analysis of this region revealed that most, but not all, of the MCSs are part of an ORF that codes for a 40-aa peptide in all species. As indicated by the genomic EvoP, a protein EvoP of the predicted amino acid sequence (shown in Fig. 3) reveals that all but four of the residues are invariant in the seven species. The genomic EvoP also revealed that the translation stop codon in one or more of the species had diverged. Subsequent analysis of the different test species blat readouts revealed that both D. virilis and D. mojavensis use TGA as their termination codon, whereas the others use TAA as the stop codon (data not shown).
Fig. 3.
evoprinter identifies a small peptide gene not annotated in the Berkeley Drosophila Genome Project (BDGP) database. EvoP analysis of the intragenic 12.9-kb region between the Drosophila Appl and vnd genes uncovered a small peptide gene conserved in D. melanogaster, D. simulans, D. yakuba, D. ananassae, D. pseudoobscura, D. virilis, and D. mojavensis species. Shown is 1.5-kb of the D. melanogaster reference species (nucleotides -9,154 to -7,670 5′ to the vnd transcribed region). MCSs shared by all species are identified by uppercase, black nucleotides. evodif analysis of individual species revealed that one 5′ upstream MCS was not conserved in D. mojavensis but is present in all other species (lowercase red nucleotides). Underlined sequence in this MCS represents a consensus Hb DNA-binding motif. A protein EvoP of the encoded 40-aa peptide is also shown. Aligned with the codons, invariant amino acids residues are shown as uppercase black letters, and residues that are different in at least one of the six species tested above are shown as lowercase gray letters.
Although the conserved ORF was not identified in the recent FlyBase genome annotation release 3.1, a GenBank blast homology search using the predicted protein sequence revealed that the Heidelberg Prediction, Heidelberg Collection (HDC) had identified the ORF as the HDC16822 gene (45). The presence of HDC16822 was initially predicted by using a lower-stringency, ab initio gene-prediction algorithm (fgenesh; ref. 46) and then confirmed by whole-transcriptome microarray analysis (41). The genomic EvoP analysis also revealed the presence of additional upstream MCSs that may harbor HDC16822 cis-regulatory elements (Fig. 3). For example, core homeodomain DNA-binding motifs (ATTA) exist in four of these MCSs. Interestingly, evodif analysis of the D. mojavensis species revealed that an additional upstream 5′ MCS is conserved in all species except for D. mojavensis (red lowercase nucleotides in Fig. 3). The fact that this sequence contains a consensus DNA-binding motif for Hb (TTTTATG) suggests that Hb may have a role in the regulation of HDC16822 expression in at least six of the seven species.
Summary. We have developed a simple, yet effective, comparative genomics tool for identifying MCSs shared among related DNAs. Generated from multiple pairwise blat alignments of a reference DNA to different test genomes, the EvoP presents an ordered, uninterrupted representation of the evolutionarily resilient sequences within the reference DNA. By superimposing the different species evolutionary histories, the combined mutagenic force reveals DNA sequences that are essential for gene expression and function. Also, the evodif algorithm reveals the degree of molecular divergence between species by identifying individual species differences to the EvoP. When compared with other multispecies-alignment tools, the two principal advantages of evoprinter are its speed (derived from the speed of a blat alignment) and the fact that only a single curated genomic sequence is required to initiate the analysis of orthologous DNAs from multiple species. Based on the success of the evoprinter identification of MCSs within known vertebrate and Drosophila cis-regulatory elements, we believe that this tool could be of great use to understanding gene regulation in all animals.
Acknowledgments
We thank L. Elnitski, J. Kassis, S. Landis, M. Muenke, H. Nash, and A. Raldow for helpful discussions; L. Elnitski and H. Nash for critically reading the manuscript; and J. Brody for help with the evoprinter web site construction and editorial assistance. This work was supported by the National Institutes of Health National Institute of Neurological Disorders and Stroke and National Institute of Medical Health Intramural Research Program.
Abbreviations: MSC, multispecies-conserved sequence; evodif, evodifference; bHLH, basic helix-loop-helix; Kr, Krüppel; Hb, Hunchback.
References
- 1.Tagle, D. A., Koop, B. F., Goodman, M., Slightom, J. L., Hess, D. L. & Jones, R. T. (1988) J. Mol. Biol. 203, 439-455. [DOI] [PubMed] [Google Scholar]
- 2.Wasserman, W. W., Palumbo, M., Thompson, W., Fickett, J. W. & Lawrence, C. E. (2000) Nat. Genet. 26, 225-228. [DOI] [PubMed] [Google Scholar]
- 3.Yuh, C. H., Brown, C. T., Livi, C. B., Rowen, L., Clarke, P. J. & Davidson, E. H. (2002) Dev. Biol. 246, 148-161. [DOI] [PubMed] [Google Scholar]
- 4.Grad, Y. H., Roth. F. P., Halfon, M. S. & Church, G. M. (2004) Bioinformatics 20, 2738-2750. [DOI] [PubMed] [Google Scholar]
- 5.Berezikov, E., Guryev, V., Plasterk, R. H. & Cuppen, E. (2004) Genome Res. 14, 170-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blanchette, M., Kent, W. J., Riemer, C., Elnitski, L., Smit, A. F., Roskin, K. M., Baertsch, R., Rosenbloom, K., Clawson, H., Green, E. D., et al. (2004) Genome Res. 14, 708-715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blanchette, M. & Tompa, M. (2003) Nucleic Acids Res. 31, 3840-3842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sinha, S., Blanchette, M. & Tompa, M. (2004) BMC Bioinformatics 5, 170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Altschul, S. F., Madden, T. L., Schaffer, A., Zhang, J., Zhang, Z., Miller, W. & Lipman, D. J. (1997) Nucleic Acids Res. 25, 3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kent, W. J. (2002) Genome Res. 12, 656-664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cartharius, K., Frech, K., Grote, K., Klocke, B., Haltmeier, M., Klingenhoff, A., Frisch, M., Bayerlein, M. & Werner, T. (2005) Bioinformatics 21, 2933-2942. [DOI] [PubMed] [Google Scholar]
- 12.Enright, A. J., John, B., Gaul, U., Tuschl, T., Sander, C. & Marks, D. S. (2003) Genome Biol. 5, R1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.John, B., Enright, A. J., Aravin, A., Tuschl, T., Sander, C. & Marks, D. S. (2004) PLoS Biol. 2, e363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Casarosa, S., Fode, C. & Guillemot, F. (1999) Development (Cambridge, U.K.) 126, 525-534. [DOI] [PubMed] [Google Scholar]
- 15.Torii, M., Matsuzaki, F., Osumi, N., Kaibuchi, K., Nakamura, S., Casarosa, S., Guillemot, F. & Nakafuku M. (1999) Development (Cambridge, U.K.) 126, 443-445. [DOI] [PubMed] [Google Scholar]
- 16.Tuttle, R., Nakagawa, Y., Johnson, J. E. & O'Leary, D. D. (1999) Development (Cambridge, U.K.) 126, 1903-1916. [DOI] [PubMed] [Google Scholar]
- 17.Lo, L. C., Johnson, J. E., Wuenschell, C. W., Saito, T. & Anderson, D. J. (1991) Genes Dev. 5, 1524-1537. [DOI] [PubMed] [Google Scholar]
- 18.Guillemot, F. & Joyner, A. L. (1993) Mech. Dev. 42, 171-185. [DOI] [PubMed] [Google Scholar]
- 19.Borges, M., Linnoila, R. I., van de Velde, H. J., Chen, H., Nelkin, B. D., Mabry, M., Baylin, S. B. & Ball, D. W. (1997) Nature 386, 852-855. [DOI] [PubMed] [Google Scholar]
- 20.Verma-Kurvari, S., Savage, T., Gowan, K. & Johnson, J. E. (1996) Dev. Biol. 180, 605-617. [DOI] [PubMed] [Google Scholar]
- 21.Verma-Kurvari, S., Savage, T., Smith, D. & Johnson, J. E. (1998) Dev. Biol. 197, 106-116. [DOI] [PubMed] [Google Scholar]
- 22.Kurokawa, D., Takasaki, N., Kiyonari, H., Nakayama, R., Kimura-Yoshida, C., Matsuo, I. & Aizawa, S. (2004) Development (Cambridge, U.K.) 131, 3307-3317. [DOI] [PubMed] [Google Scholar]
- 23.Ball, D. W., Azzoli, C. G., Baylin, S. B., Chi, D., Dou, S., Donis-Keller, H., Cumaraswamy, A., Borges, M. & Nelkin, B. D. (1993) Proc. Natl. Acad. Sci. USA 90, 5648-5652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen, H., Biel, M. A., Borges, M. W., Thiagalingam, A., Nelkin, B. D., Baylin, S. B. & Ball, D. W. (1997) Cell Growth Differ. 8, 677-686. [PubMed] [Google Scholar]
- 25.Breslin, M. B., Zhu, M., Notkins, A. L. & Lan, M. S. (2002) Nucleic Acids Res. 30, 1038-1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu, F., Pouponnot, C. & Massague, J. (1997) Genes Dev. 11, 3157-3167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Breslin, M. B., Zhu, M. & Lan, M. S. (2003) J. Biol. Chem. 278, 38991-38997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wieschaus, E., Nusslein-Volhard, C. & Kluding, H. (1984) Dev. Biol. 104, 172-186. [DOI] [PubMed] [Google Scholar]
- 29.Rivera-Pomar, R. & Jackle, H. (1996) Trends Genet. 12, 478-483. [DOI] [PubMed] [Google Scholar]
- 30.Gaul, U. & Weigel, D. (1990) Mech. Dev. 33, 57-67. [DOI] [PubMed] [Google Scholar]
- 31.Ruiz-Gomez, M., Romani, S., Hartmann, C., Jackle, H. & Bate, M. (1997) Development (Cambridge, U.K.) 124, 3407-3414. [DOI] [PubMed] [Google Scholar]
- 32.Romani, S., Jimenez, F., Hoch, M., Patel, N. H., Taubert, H. & Jackle, H. (1966) Mech. Dev. 60, 95-107. [DOI] [PubMed] [Google Scholar]
- 33.Isshiki, T., Pearson, B., Holbrook, S. & Doe, C. Q. (2001) Cell 106, 511-521. [DOI] [PubMed] [Google Scholar]
- 34.Hoch, M., Schroder, C., Seifert, E. & Jackle, H. (1990) EMBO J. 9, 2587-2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hoch, M., Seifert, E. & Jackle, H. (1991) EMBO J. 10, 2267-2278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hoch, M., Gerwin, N., Taubert, H. & Jackle, H. (1992) Science 256, 94-97. [DOI] [PubMed] [Google Scholar]
- 37.Brody, T. & Odenwald, W. F. (2002) Development (Cambridge, U.K.) 129, 3763-3770. [DOI] [PubMed] [Google Scholar]
- 38.Licht, J. D., Grossel, M., Figge, J. & Hansen, U. M. (1990) Nature 346, 76-79. [DOI] [PubMed] [Google Scholar]
- 39.Licht, J. D., Hanna-Rose, W., Reddy, J. C., English, M. A., Ro, M., Grossel, M., Shaknovich, R. & Hansen, U. (1994) Mol. Cell. Biol. 14, 4057-4066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sauer, F. & Jackle, H. (1993) Nature 364, 454-457. [DOI] [PubMed] [Google Scholar]
- 41.Hanna-Rose, W., Licht, J. D. & Hansen, U. (1997) Mol. Cell. Biol. 17, 4820-4829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Grosskortenhaus, R., Pearson, B. J., Marusich, A. & Doe, C. Q. (2005) Dev. Cell 8, 193-202. [DOI] [PubMed] [Google Scholar]
- 43.Rosen, D. R., Martin-Morris, L., Luo, L. Q. & White, K. (1989) Proc. Natl. Acad. Sci. USA 86, 2478-2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim, Y. & Nirenberg, M. (1989) Proc. Natl. Acad. Sci. USA 86, 7716-7720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hild, M., Beckmann, B., Haas, S. A., Koch, B., Solovyev, V., Busold, C., Fellenberg, K., Boutros, M., Vingron, M., Sauer, F., et al. (2003) Genome Biol. 5, R3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Salamov, A. A. & Solovyev V. V. (2000) Genome Res. 10, 516-522. [DOI] [PMC free article] [PubMed] [Google Scholar]