Abstract
Virus replication in higher vertebrates is restrained by IFNs that cause cells to transcribe genes encoding antiviral proteins, such as 2′-5′ oligoadenylate synthetases. 2′-5′ oligoadenylate synthetase is stimulated by dsRNA to produce 5′-phosphorylated, 2′-5′-linked oligoadenylates (2-5A), whose function is to activate RNase L. Although RNase L is required for a complete IFN antiviral response and mutations in the RNase L gene (RNASEL or HPC1) increase prostate cancer rates, it is unknown how 2-5A affects these biological endpoints through its receptor, RNase L. Presently, we show that 2-5A activation of RNase L produces a remarkable stimulation of transcription (≥20-fold) for genes that suppress virus replication and prostate cancer. Unexpectedly, exposure of DU145 prostate cancer cells to physiologic levels of 2-5A (0.1 μM) induced approximately twice as many RNA species as it down-regulated. Among the 2-5A-induced genes are several IFN-stimulated genes, including IFN-inducible transcript 1/P56, IFN-inducible transcript 2/P54, IL-8, and IFN-stimulated gene 15. 2-5A also potently elevated RNA for macrophage inhibitory cytokine-1/nonsteroidal antiinflammatory drug-activated gene-1, a TGF-β superfamily member implicated as an apoptotic suppressor of prostate cancer. Transcriptional signaling to the macrophage inhibitory cytokine-1/nonsteroidal antiinflammatory drug-activated gene-1 promoter by 2-5A was deficient in HeLa cells expressing a nuclease-dead mutant of RNase L and was dependent on the mitogen-activated protein kinases c-Jun N-terminal kinase and extracellular signal-regulated kinase, both of which were activated in response to 2-5A treatments. Because 2-5A and RNase L participate in defenses against viral infections and prostate cancer, our findings have implications for basic cellular mechanisms that control major pathogenic processes.
Keywords: prostate, macrophage inhibotory cytokine 1, HPC1, RNASEL
IFN mechanism of action and RNA biology converge on a uniquely regulated RNA cleavage pathway known as the 5′-phosphorylated, 2′-5′-linked oligoadenylate (2-5A) system (1). Exposure of human cells to IFN induces transcription of three 2′-5′ oligoadenylate synthetase genes (OAS1-OAS3), resulting in eight to 10 OAS isoforms owing to alternative mRNA splicing (2). Viral infections produce dsRNA, a pathogen-associated molecular pattern that stimulates OAS to produce 2-5A [px5′A(2′p5′A)n, where x = 1-3 and n ≥ 2] from ATP. 2-5A functions through its receptor, the 2-5A-dependent ribonuclease (RNase L), a ubiquitous 83-kDa protein that dimerizes into its catalytically active form upon binding 2-5A (3, 4). The 2-5A system has been shown to promote survival from some viral infections in an animal model consisting of wild-type and RNase L-/- mice infected with the picornaviruses, encephalomyocarditis virus, and Coxsackievirus B4 (5, 6). Recently, RNase L has also been implicated as a possible suppressor of prostate cancer, the second leading cause of cancer-related deaths in men in the United States. Positional cloning determined that the human RNase L gene (RNASEL) maps to HPC1 (the hereditary prostate cancer 1 gene) on chromosome 1q25 (7, 8). For example, in a large sibling-controlled study, individuals that were homozygous for the missense variant of RNase L, R462Q, which reduces catalytic activity, had a 2-fold increased incidence of prostate cancer (9, 10). In addition, RNase L was shown to contribute to apoptosis of prostate cancer cells in response to 2-5A, TNF-related apoptosis-inducing ligand, or topoisomerase I inhibitors (11). Mutations in RNASEL/HPC1 are associated with an increased risk of prostate cancer in some, but not all, populations, possibly owing to complex genetics and environmental factors, such as infections (12).
Current understanding of the effects of RNase L activation by 2-5A on RNA metabolism is limited to examination of a small number of natural and synthetic RNA substrates. 2-5A activation of RNase L results in cleavage of single-stranded RNA species 3′ of UU and UA dinucleotides (13-15). Some viral infections induce 2-5A synthesis that activates RNase L, causing both viral RNAs and rRNA in intact ribosomes to be cleaved (16, 17). However, a systematic investigation of the impact of 2-5A activation of RNase L on cellular RNA profiles has not been reported. Therefore, to better understand the molecular pathways that underlie both the antiviral and tumor suppressor functions of the 2-5A system, we have performed such an analysis using a custom DNA microarray strategy. Remarkably, for every RNA species that declined in response to 2-5A activation of RNase L, two RNA species were increased in their amounts. One of these genes, macrophage inhibitory cytokine 1/nonsteroidal antiinflammatory drug-activated gene 1 (MIC-1/NAG-1; also known as PTFG-β, GDF15, PLAB, and PDF) (18, 19), is shown to be transcriptionally activated by 2-5A in a stress-response pathway requiring RNase L and the mitogen-activated protein (MAP) kinases, c-Jun N-terminal kinase (JNK) and extracellular signal-regulated kinase (ERK). Interestingly, mutations or variants in MIC-1/NAG-1 and RNase L are associated with prostate cancer risk (20).
Materials and Methods
Reagents, Antibodies, and Plasmids. Monoclonal antibody to human RNase L and anti-human-NAG-1 antibody were as described in refs. 4 and 19. Antibody to β-actin was from Sigma. All other antibodies were from Cell Signaling Technology, Beverly, MA. Inhibitors to JNK (SP600125), p38(SB203580), and ERK1/2 (PD98059) were from Calbiochem. 2-5A was prepared enzymatically from ATP with recombinant 2-5A synthetase (21). Individual 2-5A oligomers were purified as described in ref. 11. p35′A(2′p5′A)2 was dephosphorylated with calf alkaline phosphatase (New England Biolabs) and purified with a Dionex HPLC column. The luciferase constructs containing the MIC-1/NAG-1 promoter were as described in ref. 19, except pNAG-492/+1 was constructed by PCR amplification of the -492/+1 region using primers with NheI and HindIII sites and cloned into pcDNA3.1 digested with NheI and HindIII and sequenced. pcDNA3.1 expressing RNase L, R462Q, R667A, and H672A mutations were as described in refs. 9 and 22.
Cell Culture, Transfections, and Monitoring RNase L-Mediated rRNA Cleavages in Intact Cells. DU145 cells were grown in RPMI medium 1640 with 10% FBS (Invitrogen). HeLa M cells were grown in DMEM with 10% FBS. Transfection of 2-5A or plasmids was done with Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol. The cell-based RNase L assay using RNA chips (Agilent Technologies, Palo Alto, CA) was performed as described in ref. 9.
Western Blots. Protein (80 μg) in cell extracts were separated in 10% SDS/polyacrlyamide gels, transferred to nitrocellulose membranes (Schleicher & Schuell, Keene, NH), and incubated with various primary antibodies. Secondary antibodies were either goat anti-mouse antibody or goat anti-rabbit antibody tagged with horseradish peroxidase (Cell Signaling Technology). Immunoreactive bands were detected by enhanced chemiluminescence (ECL, Amersham Biosciences) and exposed to x-ray film (Eastman Kodak).
Luciferase Assays. DU145 cells were plated at 105 cells per well. After 16 h, plasmid mixtures containing 1.0 μg of MIC-1/NAG-1 promoter linked to firefly luciferase cDNA and 0.1 μg of pRL-null (Renilla luciferase vector) (Promega) were cotransfected with Lipofectamine 2000 for 6 h, then media was replaced with media plus serum and incubated for a further 16-18 h. Subsequently, p35′A(2′p5′A)2 or A(2′p5′A)2 were transfected for 4 h and media was replaced. Inhibitors were added to some experiments 30 min before 2-5A transfection. Cells were harvested in luciferase lysis buffer at 16-18 h after 2-5A transfection, and luciferase activity was determined and normalized to Renilla luciferase activity by using a dual luciferase assay kit (Promega).
RNA Isolation and Microarray Analysis. Array construction, RNA labeling, and acquisition and normalization of data were as described in ref. 23. The array comprised 950 genes containing adenylate/uridylate-rich elements (24), 855 IFN-stimulated genes (ISGs) (25), 288 dsRNA responsive genes (26), and 85 housekeeping genes. Cells were transfected with p35′A(2′p5′A)2 or A(2′p5′A)2 at the indicated concentrations (see Fig. 1) and for various times. Total RNA was isolated with TRIzol reagent (Invitrogen) according to the manufacturer's instructions. Data presented in Fig. 1 and in Tables 1-3, which are published as supporting information on the PNAS web site, were generated with amplified target RNA in T7 polymerase-based linear amplification reactions (23). Data were acquired with a GenePix 4000B laser scanner and genepix pro 5.1 software as described in ref. 24. Raw data were imported into genespring 7.0 software (Agilent Technologies) and normalized based on the distribution of all values with locally weighted linear regression (LOWESS) before further analysis.
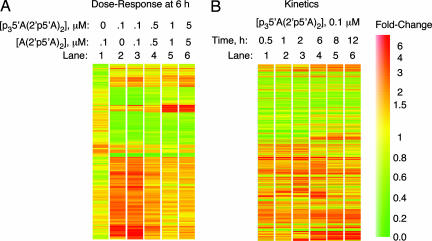
Fig. 1.
2-5A induces gene expression changes in DU145 prostate carcinoma cells in a dose- and time-dependent manner. (A) A gene tree representing ≥2-fold changes in RNA levels in response to increasing dosage of 2-5A, measured at 6 h. (B) RNA level expression changes of ≥2-fold in response to 0.1 μM active 2-5A measured at different time points. A heat bar indicating the colors associated with the fold change in expression levels is shown to the right.
Results
RNA Profiles of a Prostate Cancer Cell Line Treated with 2-5A Reveal Induced and Repressed Species of RNA. To determine the effect of 2-5A on RNA profiles, DU145 prostate cancer cells were transfected with the trimer species of 2-5A, p35′A(2′p5′A)2. As a comparison, cells were treated with dephosphorylated 2-5A, A(2′p5′A)2, which fails to activate RNase L. RNA was analyzed on a custom innate immune response cDNA microarray representing 2,178 genes (Materials and Methods). Changes in RNA profiles were determined either as a function of 2-5A concentration or time. RNA species that were induced or repressed by ≥2-fold are illustrated (Fig. 1). Transfection of inactive 2-5A, A(2′p5′A)2, at 0.1 μM for 6 h had only minimal effects on gene expression when compared with untreated cells (Fig. 1 A, lane 1). In contrast, whereas 0.1 μM p35′A(2′p5′A)2 induced 140 RNA species by ≥2-fold, there were only 63 RNA species that declined in amounts by at least a factor of two (Fig. 1 A, lane 2, and Tables 1-3). Nearly identical results were obtained when cells treated with 0.1 μM p35′A(2′p5′A)2 were compared with untreated cells or with cells treated with 0.1 μM A(2′p5′A)2 (Fig. 1 A, lanes 2 and 3). Increasing concentrations of p35′A(2′p5′A)2, from 0.1 to 5 μM produced decreased levels of some RNA species but further increased amounts of other RNA molecules (Fig. 1 A, lanes 3-6). A kinetic analysis of cells treated for 30 min to 12 h with 0.1 μMp35′A(2′p5′A)2 was performed, and the results were compared with those from untreated cells (Fig. 1B). Most RNA species that changed in amounts by a factor of two or greater were apparent by 30 min of 2-5A treatment; some decreased at first, then increased in amounts, whereas others increased up to 2 h and then declined. At 8 h, there were 99 RNA species up-regulated and 56 species down-regulated. Interestingly, several ISGs were induced by 2-5A treatment, including IFN-inducible transcript (IFIT)2/P54, IFIT1/P56, IL-8, and ISG15/IFI-15K. P54 was the most highly up-regulated gene, peaking at a >20-fold increase by 6 h of 2-5A treatment. Only six RNA species were down-regulated by 5- to 10-fold in response to 0.1 μM p35′A(2′p5′A)2 (Tables 1-3). These findings suggest that the biological activities of 2-5A are due to a combination of gene induction and repression.
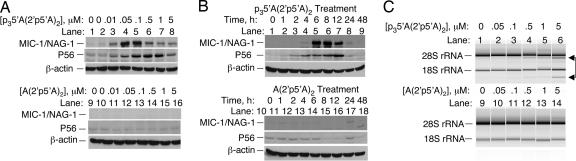
2-5A Induction of MIC-1/NAG-1 and P56 Proteins in DU145 Cells. Array experiments on 2-5A-transfected DU145 cells using a modified cDNA labeling method (without probe amplification) detected 2-5A induction of MIC-1/NAG-1, a member of the TGF-β superfamily. MIC-1/NAG-1 mRNA was induced 5.7- and 11.5-fold at 2 and 4 h, respectively, after 2-5A treatment (data not shown). In addition, 2-5A potently induced MIC-1/NAG-1 mRNA levels in DU145 cells as determined by RT-PCR and by Northern blotting (data not shown). To establish MIC-1/NAG-1 and P56 induction at the protein level, DU145 cells were transfected for 6 h with different amounts of p35′A(2′p5′A)2 or A(2′p5′A)2. MIC-1/NAG-1 protein, the 35-kDa pro-form, was clearly detected after transfecting for 6 h with 10 nM p35′A(2′p5′A)2 but was highly induced with 50 or 100 nM p35′A(2′p5′A)2 (Fig. 2A). Higher concentrations of p35′A(2′p5′A)2 (0.5 to 5 μM), produced less MIC-1/NAG-1 induction, perhaps because of the RNA-degrading activity of RNase L. P56 was induced with p35′A(2′p5′A)2 between 0.01 to 0.05 μM, with maximal induction at 0.5 μM (Fig. 2 A). MIC-1/NAG-1 was first observed at 4 h of treatment with 0.1 μM p35′A(2′p5′A)2 and reached maximum levels between 6 and 8 h, whereas P56 was induced by 2-4 h and levels peaked at 12 h (Fig. 2B). A(2′p5′A)2 failed to induce MIC-1/NAG-1 or P56 protein, suggesting that RNase L activation is necessary (Fig. 2 A Lower and B Lower). To verify activation of RNase L in the intact cells, specific rRNA cleavage products were observed in cells transfected with p35′A(2′p5′A)2 but were not seen with A(2′p5′A)2 (Fig. 2C) (17).
Fig. 2.
Induction of MIC-1/NAG-1 and P56 proteins by 2-5A as determined in Western blots. (A) Dose-dependent induction of MIC-1/NAG-1 and P56 by p35′A(2′p5′A)2 (lanes 1-8) compared with A(2′p5′A)2 (lanes 9-16) (see Materials and Methods). (B) Kinetics of MIC-1/NAG-1 and P56 induction by 0.1 μM p35′A(2′p5′A)2 compared with 0.1 μM A(2′p5′A)2 for the indicated times. (C) Cleavage of 28S and 18S rRNA in DU145 cells transfected with the indicated concentrations of p35′A(2′p5′A)2 (lanes 1-6) or A(2′p5′A)2 (lanes 9-14) for 4 h before isolating RNA. RNase L-mediated cleavage products of rRNA are indicated to the right (arrows).
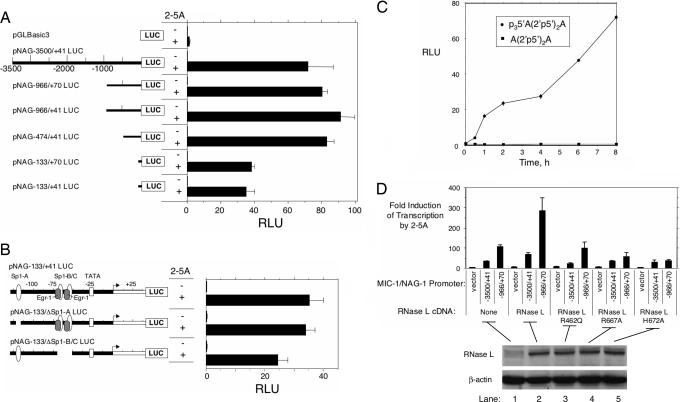
Induction of the MIC-1/NAG-1 Promoter by 2-5A Activation of RNase L. To determine whether MIC-1/NAG-1 induction was transcriptional, constructs from the MIC-1(NAG-1) promoter fused to luciferase cDNA were transiently transfected into DU145 cells (19). Cells were transfected with MIC-1/NAG-1 promoter constructs, followed by 100 nM p35′A(2′p5′A)2 (Fig. 3A). Constructs with 3,500, 966, and 474 nt of the MIC-1/NAG-1 promoter were remarkably induced by 2-5A. A minimal MIC-1/NAG-1 promoter (133 nt) was induced to ≈50% of the extent of the longer constructs. To determine whether Sp1 and Egr1 binding sites in the 133-nt promoter contributed to 2-5A induction, these sites were deleted (Fig. 3B). Although removal of the Sp1-A site was without effect, removal of the Sp1-B, Sp1-C, and Egr-1 sites reduced 2-5A-induced transcription by approximately one-third. However, a very substantial 2-5A induction remained, indicating the presence of additional 2-5A/RNase L responsive sites in the minimal pNAG-133/ΔSp1-B/C promoter segment. The kinetics of induction for the 474-nt MIC-1/NAG-1 promoter demonstrated luciferase expression by 1 h of 2-5A treatment, with maximum expression by 8 h (Fig. 3C).
Fig. 3.
Transcriptional activation of MIC-1/NAG-1 promoter constructs in response to 2-5A requires RNase L. (A) DU145 cells were transfected with the constructs indicated on the left. After 16 h, cells were transfected with p35′A(2′p5′A)2 or p35′A(2′p5′A)2 (see Materials and Methods). (B) Effect of deleting Sp1 and Egr-1 sites in the minimal MIC-1/NAG-1 promoter on 2-5A induction determined as described for A. (C) Kinetics of induction of the -474/+41 promoter by p35′A(2′p5′A)2 and lack of induction by A(2′p5′A)2 was performed as described for A. (D) Catalytically active RNase L is required for 2-5A induction of the MIC-1/NAG-1 promoter. HeLa M cells were transiently transfected with constructs of wild-type or mutant human RNase L cDNA along with MIC-1/NAG-1 promoter constructs and subsequently transfected with 0.1 μMp35′A(2′p5′A)2 (see Materials and Methods). Expression of RNase L was determined on Western blots. RLU, relative luciferase units represent firefly luciferase activity normalized for Renilla luciferase activity.
To further establish that the signal generated by 2-5A is mediated through RNase L, experiments were done in a human HeLa cell line, clone M, that contains low levels of RNase L (Fig. 3D). In the HeLa cells, the expression of the 966-nt MIC-1/NAG-1 promoter segment exceeded that of the 3,500-nt promoter by 2- to 3-fold. Expression of RNase L cDNA produced a 3-fold stimulation of the 2-5A-induced transcription. However, the RNase L variant R462Q did not affect 2-5A induction of transcription (9, 10). In contrast, 2-5A induction of the MIC-1/NAG-1-966/+70 promoter was substantially reduced (2- to 3-fold) by the nuclease-dead RNase L mutants, R667A or H672A (22). These data indicate that the signal relayed by 2-5A through RNase L requires amino acid residues necessary for the catalytic function of RNase L.
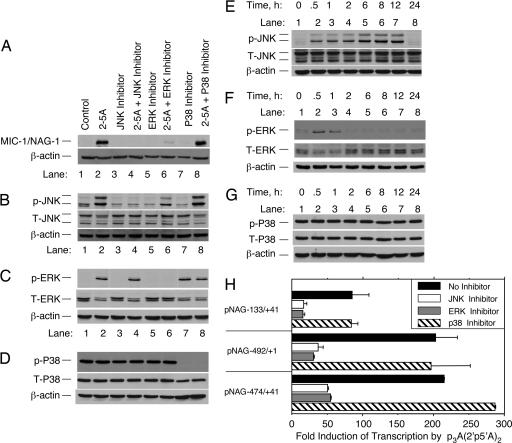
2-5A Signaling Is a Stress-Response Pathway That Requires MAP Kinases JNK and ERK. The question of how 2-5A activation of RNase L leads to a transcriptional response was further investigated. Prior studies showed that 2-5A treatments of cells led to activation of the MAP kinase JNK (11, 27). To determine whether JNK and other MAP kinases participate in the signal transduction pathway initiated by 2-5A, DU145 cells were treated with 2-5A in the presence or absence of different kinase inhibitors. The JNK inhibitor SP600125 completely prevented induction of MIC-1/NAG-1 protein and phosphorylation of JNK in response to 2-5A treatments (Fig. 4 A and B). Similarly, the ERK inhibitor PD98059 greatly reduced MIC-1/NAG-1 induction and completely prevented ERK phosphorylation by 2-5A (Fig. 4 A and C). In contrast, inhibition with SB203580 of p38, which is constituitively active in these cells, had no effect on MIC-1/NAG-1 induction by 2-5A (Fig. 4 A and D). JNK activation by 2-5A occurred within 30 min and was sustained for at least 12 h after treatment with 2-5A (Fig. 4E). In contrast, ERK phosphorylation peaked at 30 min and then rapidly declined (Fig. 4F). P38 levels and its phosphorylation state were unaffected by 2-5A (Fig. 4G). The JNK and ERK inhibitors, but not the p38 inhibitor, reduced by 4- to 5-fold the induction by 2-5A of the 474- or 133-nt MIC-1/NAG-1 promoters (Fig. 4H). In addition, a promoter construct (pNAG -492/+1), which lacks all of the MIC-1/NAG-1 5′ UTR, was highly induced by 2-5A, and it was also repressed by the JNK or ERK inhibitors (Fig. 4H). These findings demonstrate that JNK and ERK contribute to the 2-5A-mediated induction of the MIC-1/NAG-1 promoter.
Fig. 4.
JNK and ERK contribute to induction of the MIC-1/NAG-1 promoter in response to 2-5A. DU145 cells were treated for 30 min with either JNK inhibitor SP600125 (25 μM), ERK1/2 inhibitor PD98059 (25 μM), or p38 inhibitor SB203580 (20 μM) before transfection with 0.1 μMp35′A(2′p5′A)2 for 4 h (A-D)orinfor the indicated times (E-G). (A-G) MIC-1/NAG-1 protein (A), phosphorylation of JNK1/2(B and E), phosphorylation ERK1/2(C and F), and phosphorylation of p38 in Western blots (D and G) were probed with the indicated antibodies. T, total protein; p, phosphorylated protein. (H) Inhibition of 2-5A induction of the -133/+41, -492/+1, and -474/+41 MIC-1/NAG-1 promoter luciferase constructs by JNK and ERK inhibitors but not by the p38 inhibitor.
Discussion
2-5A Mediates a Potent Transcriptional Response Through RNase L and MAP Kinases JNK and ERK. Our findings reveal a signaling pathway in which activation of RNase L by 2-5A induces a range of different genes, including several ISGs and MIC-1/NAG-1. 2-5A activation of RNase L stimulates JNK and ERK, leading to transcriptional induction of the MIC-1/NAG-1 gene. Ribonuclease activity is strongly implicated in the signaling pathway because two separate nuclease-dead missense mutations in RNase L (R667A and H672A) failed to activate and, instead, inhibited 2-5A induction of the MIC-1/NAG-1 -966/+70 promoter (22) (Fig. 3D). In contrast, expression of the R462Q variant of RNase L, which reduces ribonuclease activity by 2- to 3-fold, had no effect (9, 10). These findings suggest that, although ribonuclease activity is required for signaling, it may not be sufficient. For instance, our results do not rule out the possibility of 2-5A-regulated protein-protein interaction in the signaling pathway. Although the specific proximal RNA substrate(s) of RNase L required for the transcriptional response are unknown, is worth noting that JNK is activated in response to different treatments that damage rRNA (e.g., ricin A, α-sarcin, UV light, and RNase L) (27-29). Because RNase L cleaves 28S and 18S rRNA at specific sites in intact ribosomes (17), the ribosome may be a sentinel for different RNA damage-response pathways. Alternatively, it is possible that RNase L cleaves an inhibitory RNA, such as a microRNA, leading to translation of a transcriptional activator or a repressor mRNA allowing transcription of different genes.
Transcription factors NF-κB, p53, Sp1, and Egr-1 have been implicated in MIC-1/NAG-1 induction (30). The -133 nucleotide minimal 2-5A-responsive MIC-1/NAG-1 promoter construct has binding sites for Sp1 and Erg-1 but not for NF-κB and p53. The 2-5A pathway is independent of p53 because DU145 cells are mutant for p53 and because 2-5A also induced MIC-1/NAG-1 in the p53 mutant cell line, HCT-116 p53-/- (31) (data not shown). However, NF-κB could be responsible for the enhanced 2-5A induction observed with longer MIC-1/NAG-1 promoter constructs, such as the -474 nucleotide promoter through the κB site at nucleotides -277 to -286, compared with the -131 nucleotide construct. The Sp1-A site (nucleotides -114 to -121) was dispensable, whereas the Sp1-B, Sp1-C, and Egr-1 sites (nucleotides -51 to -70) contributed to, but were not essential for, the 2-5A transcriptional response. Optimal induction of MIC-1/NAG-1 by 2-5A required JNK and ERK. In contrast, 12-O-tetradecanoylphorbol-13-acetate stimulation of MIC-1/NAG-1 occurs through NF-κB and is independent of these MAP kinases (30). On the other hand, MIC-1/NAG-1 induction by troglitazone through Egr-1 was ERK-dependent (32). These findings indicate that the MIC-1/NAG-1 gene contains multiple promoter elements that can respond to different types of signals. Although the MIC-1/NAG-1 is the focus of this study, 2-5A induction of other genes, such as IFIT1/P56, IFIT2/P54, IL8, and ISG15, likely involves additional or alternative intermediate signaling molecules and promoter elements.
Implications for MIC-1/NAG-1 Induction by RNase L in Prostate Cancer. MIC-1/NAG-1 inhibits prostate cancer proliferation and adhesion, induces apoptosis, and, in macrophages, inhibits lipopolysaccharide-induced TNF-α production (18, 33, 34). However, MIC-1/NAG-1 expression is also up-regulated during prostate cancer progression and has been suggested as a potential biomarker (35). The RNase L gene (RNASEL) maps to the HPC1 gene, and a variant of MIC-1/NAG-1 (H6D) is associated with prostate cancer risk (20). Furthermore, other genes involved in host defense to infections and/or inflammation are implicated as prostate cancer susceptibility genes, including TLR4 (Toll-like receptor 4 gene) and MSR1 (macrophage scavenger receptor 1 gene) (36, 37). Proliferative inflammatory atrophy is observed adjacent to prostate intra-epithelial neoplasia and may be initiated by infections (38). 2-5A produced in response to viral infections of prostate could induce the antiinflammatory, proapoptotic MIC-1/NAG-1 gene and antiviral ISGs, such as P56 and ISG15 (39, 40). Our findings suggest a connection between RNase L and MIC-1/NAG-1. We show here that when RNase L is mutated, induction of the MIC-1/NAG-1 promoter is deficient (Fig. 3D). Perhaps, when RNase L is mutated in prostate, MIC-1/NAG-1 levels would be reduced, thereby allowing inflammation and initiation of prostatic carcinogenesis (38).
Biological Roles of 2-5A and RNase L Are Due to Induction and Repression of RNA Species. 2-5A, the only known nucleic acid with more than one 2′-5′ internucleotide linkage, is produced from an IFN-induced enzyme (OAS) when stimulated by the pathogen-associated molecular pattern dsRNA. Although tetramer (data not shown) and trimer 2-5A are active in inducing MIC-1/NAG-1 transcription, it will be interesting to further study the effect of 2-5A oligomer size on the transcriptional response. In addition, RNase L-/- cells reconstituted with different mutant forms of RNase L will be useful for determining the precise molecular mechanism for the transcriptional effects of 2-5A (5). The range of 2-5A concentrations (10-100 nM) required for the induction of MIC-1/NAG-1 and other RNAs is the same as that observed in encephalomyocarditis virus-infected cells (41). Therefore, physiological concentrations of 2-5A cause RNase L to stimulate a potent transcriptional response. Interestingly, transcription of MIC-1/NAG-1 (referred to as PLAB and prostate differentiation factor in ref. 42) was induced 3.5-fold in the human hepatoma cell line Huh7 in response to infection for 2 h with severe acute respiratory syndrome-coronavirus (42). It is possible that the induction of MIC-1/NAG-1 in that study was due to viral stimulation of the 2-5A/RNase L pathway. It is intriguing to think that perhaps the dominant function of the RNase L is to induce gene expression rather than to destroy RNA species. The consequence of activation of RNase L by 2-5A is induction of genes, such as MIC-1/NAG-1, P56, IL-8, and ISG15, that have a profound effect on viruses, inflammation, and tumorigenesis.
Supplementary Material
Acknowledgments
We thank Babal Kant Jha and Kevin (Dalei) Ni for preparation of the 2-5A compounds and Ganes Sen (Cleveland Clinic Foundation) for the antibody to P56. This work was supported by National Institutes of Health Grants R01 CA44059 (to R.H.S.) and NIH P01 CA62220 (to B.R.G.W.).
Abbreviations: 2-5A, 2′-5′-linked oligoadenylate; OAS, 2′-5′ oligoadenylate synthetase; MIC-1, macrophage inhibitory cytokine 1; NAG-1, nonsteroidal antiinflammatory drug-activated gene 1; MAP, mitogen-activated protein; JNK, c-Jun N-terminal kinase; ERK, extracellular signal-regulated kinase; ISG, IFN-stimulated gene; IFIT, IFN-inducible transcript.
References
- 1.Kerr, I. M. & Brown, R. E. (1978) Proc. Natl. Acad. Sci. USA 75, 256-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mashimo, T., Glaser, P., Lucas, M., Simon-Chazottes, D., Ceccaldi, P. E., Montagutelli, X., Despres, P. & Guenet, J. L. (2003) Genomics 82, 537-552. [DOI] [PubMed] [Google Scholar]
- 3.Zhou, A., Hassel, B. A. & Silverman, R. H. (1993) Cell 72, 753-765. [DOI] [PubMed] [Google Scholar]
- 4.Dong, B. & Silverman, R. H. (1995) J. Biol. Chem. 270, 4133-4137. [DOI] [PubMed] [Google Scholar]
- 5.Zhou, A., Paranjape, J., Brown, T. L., Nie, H., Naik, S., Dong, B., Chang, A., Trapp, B., Fairchild, R., Colmenares, C. & Silverman, R. H. (1997) EMBO J. 16, 6355-6363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Flodstrom-Tullberg, M., Hultcrantz, M., Stotland, A., Maday, A., Tsai, D., Fine, C., Williams, B., Silverman, R. & Sarvetnick, N. (2005) J. Immunol. 174, 1171-1177. [DOI] [PubMed] [Google Scholar]
- 7.Carpten, J., Nupponen, N., Isaacs, S., Sood, R., Robbins, C., Xu, J., Faruque, M., Moses, T., Ewing, C., Gillanders, E., et al. (2002) Nat. Genet. 30, 181-184. [DOI] [PubMed] [Google Scholar]
- 8.Silverman, R. H. (2003) Biochemistry 42, 1805-1812. [DOI] [PubMed] [Google Scholar]
- 9.Xiang, Y., Wang, Z., Murakami, J., Plummer, S., Klein, E. A., Carpten, J. D., Trent, J. M., Isaacs, W. B., Casey, G. & Silverman, R. H. (2003) Cancer Res. 63, 6795-6801. [PubMed] [Google Scholar]
- 10.Casey, G., Neville, P. J., Plummer, S. J., Xiang, Y., Krumroy, L. M., Klein, E. A., Catalona, W. J., Nupponen, N., Carpten, J. D., Trent, J. M., et al. (2002) Nat. Genet. 32, 581-583. [DOI] [PubMed] [Google Scholar]
- 11.Malathi, K., Paranjape, J. M., Ganapathi, R. & Silverman, R. H. (2004) Cancer Res. 64, 9144-9151. [DOI] [PubMed] [Google Scholar]
- 12.Maier, C., Haeusler, J., Herkommer, K., Vesovic, Z., Hoegel, J., Vogel, W. & Paiss, T. (2005) Br. J. Cancer 92, 1159-1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wreschner, D. H., McCauley, J. W., Skehel, J. J. & Kerr, I. M. (1981) Nature 289, 414-417. [DOI] [PubMed] [Google Scholar]
- 14.Floyd-Smith, G., Slattery, E. & Lengyel, P. (1981) Science 212, 1030-1032. [DOI] [PubMed] [Google Scholar]
- 15.Han, J. Q., Wroblewski, G., Xu, Z., Silverman, R. H. & Barton, D. J. (2004) J. Interferon Cytokine Res. 24, 664-676. [DOI] [PubMed] [Google Scholar]
- 16.Li, X. L., Blackford, J. A. & Hassel, B. A. (1998) J. Virol. 72, 2752-2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wreschner, D. H., James, T. C., Silverman, R. H. & Kerr, I. M. (1981) Nucleic Acids Res. 9, 1571-1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bootcov, M. R., Bauskin, A. R., Valenzuela, S. M., Moore, A. G., Bansal, M., He, X. Y., Zhang, H. P., Donnellan, M., Mahler, S., Pryor, K., et al. (1997) Proc. Natl. Acad. Sci. USA 94, 11514-11519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Baek, S. J., Horowitz, J. M. & Eling, T. E. (2001) J. Biol. Chem. 276, 33384-33392. [DOI] [PubMed] [Google Scholar]
- 20.Lindmark, F., Zheng, S. L., Wiklund, F., Bensen, J., Balter, K. A., Chang, B., Hedelin, M., Clark, J., Stattin, P., Meyers, D. A., et al. (2004) J. Natl. Cancer Inst. 96, 1248-1254. [DOI] [PubMed] [Google Scholar]
- 21.Hartmann, R., Justesen, J., Sarkar, S. N., Sen, G. C. & Yee, V. C. (2003) Mol. Cell 12, 1173-1185. [DOI] [PubMed] [Google Scholar]
- 22.Dong, B., Niwa, M., Walter, P. & Silverman, R. H. (2001) RNA 7, 361-373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dong, B., Zhou, Q., Zhao, J., Zhou, A., Harty, R. N., Bose, S., Banerjee, A., Slee, R., Guenther, J., Williams, B. R., et al. (2004) J. Virol. 78, 8983-8993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Frevel, M. A., Bakheet, T., Silva, A. M., Hissong, J. G., Khabar, K. S. & Williams, B. R. (2003) Mol. Cell. Biol. 23, 425-436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.de Veer, M. J., Holko, M., Frevel, M., Walker, E., Der, S., Paranjape, J. M., Silverman, R. H. & Williams, B. R. (2001) J. Leukoc. Biol. 69, 912-920. [PubMed] [Google Scholar]
- 26.Elco, C. P., Guenther, J. M., Williams, B. R. & Sen, G. C. (2005) J. Virol. 79, 3920-3929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li, G., Xiang, Y., Sabapathy, K. & Silverman, R. H. (2004) J. Biol. Chem. 279, 1123-1131. [DOI] [PubMed] [Google Scholar]
- 28.Iordanov, M. S., Pribnow, D., Magun, J. L., Dinh, T. H., Pearson, J. A. & Magun, B. E. (1998) J. Biol. Chem. 273, 15794-15803. [DOI] [PubMed] [Google Scholar]
- 29.Iordanov, M. S., Pribnow, D., Magun, J. L., Dinh, T. H., Pearson, J. A., Chen, S. L. & Magun, B. E. (1997) Mol. Cell. Biol. 17, 3373-3381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shim, M. & Eling, T. E. (2005) J. Biol. Chem. 280, 18636-18642. [DOI] [PubMed] [Google Scholar]
- 31.Isaacs, W. B., Carter, B. S. & Ewing, C. M. (1991) Cancer Res. 51, 4716-4720. [PubMed] [Google Scholar]
- 32.Baek, S. J., Wilson, L. C., Hsi, L. C. & Eling, T. E. (2003) J. Biol. Chem. 278, 5845-5853. [DOI] [PubMed] [Google Scholar]
- 33.Tan, M., Wang, Y., Guan, K. & Sun, Y. (2000) Proc. Natl. Acad. Sci. USA 97, 109-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu, T., Bauskin, A. R., Zaunders, J., Brown, D. A., Pankhurst, S., Russell, P. J. & Breit, S. N. (2003) Cancer Res. 63, 5034-5040. [PubMed] [Google Scholar]
- 35.Nakamura, T., Scorilas, A., Stephan, C., Yousef, G. M., Kristiansen, G., Jung, K. & Diamandis, E. P. (2003) Br. J. Cancer 88, 1101-1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zheng, S. L., Augustsson-Balter, K., Chang, B., Hedelin, M., Li, L., Adami, H. O., Bensen, J., Li, G., Johnasson, J. E., Turner, A. R., et al. (2004) Cancer Res. 64, 2918-2922. [DOI] [PubMed] [Google Scholar]
- 37.Xu, J., Zheng, S. L., Komiya, A., Mychaleckyj, J. C., Isaacs, S. D., Hu, J. J., Sterling, D., Lange, E. M., Hawkins, G. A., Turner, A., et al. (2002) Nat. Genet. 32, 321-325. [DOI] [PubMed] [Google Scholar]
- 38.Nelson, W. G., De Marzo, A. M. & Isaacs, W. B. (2003) N. Engl. J. Med. 349, 366-381. [DOI] [PubMed] [Google Scholar]
- 39.Das, D., Shah, R. B. & Imperiale, M. J. (2004) Oncogene 23, 7031-7046. [DOI] [PubMed] [Google Scholar]
- 40.Zambrano, A., Kalantari, M., Simoneau, A., Jensen, J. L. & Villarreal, L. P. (2002) Prostate 53, 263-276. [DOI] [PubMed] [Google Scholar]
- 41.Knight, M., Cayley, P. J., Silverman, R. H., Wreschner, D. H., Gilbert, C. S., Brown, R. E. & Kerr, I. M. (1980) Nature 288, 189-192. [DOI] [PubMed] [Google Scholar]
- 42.Tang, B. S., Chan, K. H., Cheng, V. C., Woo, P. C., Lau, S. K., Lam, C. C., Chan, T. L., Wu, A. K., Hung, I. F., Leung, S. Y. & Yuen, K. Y. (2005) J. Virol. 79, 6180-6193. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.