Abstract
Transcriptional activity is often used as a surrogate for gene expression in environmental microbial communities. We developed a real-time PCR assay in which the ABI-Prism (PE Applied Biosystems) detection system is used for quantification of large-subunit ribulose-1,5-bisphosphate caboxylase/oxygenase (rbcL) mRNA in diatoms and pelagophytes both in cultures and from natural phytoplankton communities. Plasmid DNA containing rbcL inserts, as well as in vitro transcribed mRNA of the plasmids, was used to generate standard curves with a dynamic range of more than 6 orders of magnitude with high accuracy and precision (R2 = 0.998). Expression levels in a cultured diatom (Phaeodactylum tricornutum) were quantified through one light-dark cycle by using traditional 35S-labeled oligonucleotide hybridization and real-time PCR. The mRNA levels detected by the two techniques were similar and correlated well (R2 = 0.95; slope = 1.2). The quantities obtained by hybridization were slightly, yet significantly, larger (t = 5.29; P = 0.0011) than the quantities obtained by real-time PCR. This was most likely because partially degraded transcripts were not detected by real-time PCR. rbcL mRNA detection by real-time PCR was 3 orders of magnitude more sensitive than rbcL mRNA detection by hybridization. Diatom and pelagophyte rbcL mRNAs were also quantified in a profile from an oligotrophic site in the Gulf of Mexico. We detected the smallest amount of diatom rbcL expression in the surface water and maximum expression at a depth that coincided with the depth of the subsurface chlorophyll maximum. These results indicate that real-time PCR may be utilized for quantification of microbial gene expression in the environment.
One of the continuing challenges in microbial ecology is to estimate microbial activity. Even small water or soil samples usually contain large numbers of diverse microbial species, yet it is difficult to determine the individual contributions of the species to particular processes by using bulk assay techniques. While tremendous advances have been made since quantitative methods for analysis of nucleic acids via various hybridization techniques (27, 28, 34) became available, applications have generally been limited by the sensitivity of the procedures. Such methods for quantification of gene expression have traditionally involved the use of radiolabeled probes for detection of a particular mRNA cross-linked to charged filters. These protocols are time-consuming and costly and involve generation of radioactive waste. PCR technology has greatly increased the sensitivity of methods for gene detection, but it is inherently nonquantitative. Quantitative PCR combines the sensitivity of PCR with real-time measurement of amplification and thus allows quantification of the original target concentration.
A PCR-based quantitative assay, first described by Holland et al. (13) and referred to as a real-time PCR, has recently emerged (6, 11). While this technique offers all the advantages of conventional PCR, such as high sensitivity and specificity, it also allows quantification of PCR product formation during the exponential phase of the reaction. PCR product formation is monitored by determining the increase in fluorescence either due to binding of the amplicon to a fluorescent DNA stain, such as SYBR green (12), or due to the release of a fluorescent moiety from an oligonucleotide probe (i.e., a TaqMan probe) specific for the amplicon. TaqMan probes are short oligonucleotides which are labeled with a fluorescent chromophore and a quencher at the 5′ and 3′ ends, respectively. During template elongation the probe is cleaved by the 5′→3′ exonuclease activity of Taq DNA polymerase, which releases the 5′-linked dye from the 3′-linked quencher, resulting in an increase in fluorescence with product formation. Even though real-time PCR was originally developed for clinical applications, it has recently been applied to microbial ecology. For example, it has been used for detection of small-subunit rRNA (31) and for determination of nirS gene copy abundance (7) in natural microbial communities. Both of these applications of real-time PCR technology are directed at quantifying DNA gene copy abundance in environmental microbial communities. The work described here was directed at detection of mRNA as a surrogate for gene expression and functional activity of phytoplankton in the marine environment.
Concerns about global warming have led to interest in processes that might influence the potential removal and/or sequestration of carbon dioxide from the oceans and the atmosphere. By far, the most important global carbon sink is photosynthetic carbon dioxide fixation, almost half of which is performed by oceanic phytoplankton (3). Phytoplankton primary production is thought to lead to burial and sequestration of significant amounts of organic carbon in marine sediments and consequently to the permanent removal of carbon from the immediate carbon cycle (14, 16). Indeed, it is thought that most of the carbon sequestered by the oceans is buried in sediments along continental margins (10), where high rates of nutrient input, as a result of continental runoff, lead to correspondingly high rates of new production in the water column. Since coastal phytoplankton communities are dominated by the activity of eukaryotic algae, such as diatoms and phytoflagellates (17, 37), measuring the expression of carbon fixation genes in diatoms may be a useful indicator of new production. We developed a method for detection of rbcL (the gene encoding the large subunit of ribulose-1,5-bisphospate carboxylase/oxygenase [RubisCO]) transcript abundance in diatoms and pelagophytes by real-time PCR.
RubisCO is the key enzyme of the Calvin-Benson-Basham cycle, catalyzing the first step, in which CO2 is reductively assimilated into organic carbon. Several forms of RubisCO exist in nature. Virtually all oceanic picophytoplankton contain a form I RubisCO, which is a 550-kDa holoenzyme composed of eight large and eight small subunits (15). The genes encoding the small and large subunits are designated rbcS and rbcL, respectively (in proteobacteria they are designated cbbS and cbbL, respectively). Phylogenetic analysis revealed that there are four distinct evolutionary lineages within the form I clade of rbcL sequences (20, 32). Form IA is found in most marine picocyanobacteria in the genus Prochlorococcus, as well as some Synechococcus species, while it appears that all green algae and other cyanobacteria contain a form IB rbcL. Chromophytic (nongreen) algae (prymnesiophytes, pelagophytes, diatoms, eustigmatophytes, etc.) all contain form ID rbcL, whose sequences are closely related to the proteobacterial form IC sequences. Understanding the factors that influence the transcription of rbcL in these important phytoplankton groups may provide useful information about the factors that control oceanic primary productivity and phytoplankton biocoenosis structure.
We have previously described PCR primers (23) that amplify a wide range of rbcL genes from chromophytic algae. Combining these primers with a diatom- and pelagophyte-specific TaqMan probe provided a means to estimate rbcL gene expression by these organisms by real-time PCR. The results were compared to results obtained with more traditional mRNA detection assays in which hybridization and probing were used. Our results indicate that real-time PCR shows great promise for estimating gene expression by transcript abundance as a measure of the functional activities of specific groups of phytoplankton both in the marine environment and in culture.
MATERIALS AND METHODS
Algal cultures and diel experiments.
Phaeodactylum tricornutum CCMP 630 was obtained from the Provasoli-Guillard Center for Culture of Marine Phytoplankton (West Boothbay Harbor, Maine) and was grown in F/2 media (8). Cultures were maintained at 25°C and illuminated with cool white fluorescent tubes (25 to 40 microeinsteins/m2) during cycles consisting of 12 h of light and 12 h of darkness. For diel experiments 2-liter cultures were grown in 4-liter Erlenmeyer flasks. Each of the cultures was stirred with a magnetic stir bar and constantly aerated with sterile filtered air. Growth of the cultures was monitored spectrophotometrically at 480 nm. Once a culture had reached sufficient cell density (late log phase), it was sampled every 2 h to determine whole-cell carbon fixation, DAPI (4′,6′-diamidino-2-phenylindole) cell counts, enzyme activity, and rbcL gene expression.
Primers and probe design.
Degenerate PCR primers for amplification of rbcL from form ID-containing phytoplankton have been described elsewhere (20; B. Wawrik, J. H. Paul, L. Campbell, D. Griffin, L. Houchin, A. Fuentes-Ortega, and F. Mueller-Karger, unpublished data). These primers amplify a 554-bp fragment of rbcL, which includes the functional site of the enzyme (forward primer, 5′-GATGATGARAAYATTAACTC-3′; reverse primer, 5′-ATTTGDCCACAGTGDATACCA-3′). We have found that these primers amplify sequences from a great diversity of chromophytic algae in environmental samples, including diatoms, pelagophytes, prymnesiophtes, and eustigmatophytes (20; Wawrik et al., unpublished). The TaqMan probe was designed by obtaining all available diatom rbcL sequences (as of July 2000) in the GenBank database (www.ncbi.nlm.nih.gov). A conserved site 15 bp downstream of the 5′ form ID rbcL primer was located. One degeneracy was introduced by inserting inosine (I) at an ambiguous position (probe sequence, 5′-TGCGTTGGAGAGAICGTTTCTTA-3′). All primer and probe sequences given here are described with International Union of Pure and Applied Chemistry degeneracies. The TaqMan probe was 5′ labeled with the fluorescent dye 6-carboxyfluorescein and 3′ labeled with the quencher 6-carboxytetramethyl-rhodamine. Oligonucleotides and probes were obtained from Operon Technologies (Alameda, Calif.).
PCR.
For PCR 5 μl of sample was directly added to 45 μl of a reverse transcription (RT)-PCR mixture prepared from 2× RT-PCR TaqMan master mixture (PE Applied Biosystems, Foster City, Calif.) containing each primer at a concentration of 1 μM, 2 mM MgCl, and 100 μM probe. The cycle parameters were as follows: 3 min at 95°C and 40 cycles of 1 min at 95°C, 1 min at 52°C, and 1 min 30 s at 72°C. Cycling was preceded by incubation for 10 min at 95°C to activate AmpliTaq Gold. For RT all of these steps were preceded by 30 min of incubation at 48°C. Amplification mixtures were analyzed by using the ABI Prism detection system.
RNA sampling and extraction.
For RNA extraction triplicate 20-ml samples of culture were removed and filtered onto 25-mm-diameter, 0.45-μm-pore-size HV polyvinylidene difluoride filters (Millipore Durapore). The filters were then placed into 2.0-ml screw-cap tubes containing 0.1-mm-diameter muffled glass beads (Biospec Products), 750 μl of RLT lysis buffer (Qiagen, Valencia, Calif.), and 7.5 μl of β-mercaptoethanol. The tubes were frozen in liquid nitrogen and stored at −80°C until extraction. The glass beads were baked overnight at 450°C in a muffle furnace. Cells were disrupted by bead beating (19), and 500 μl of lysate was extracted by using RNeasy spun columns (Qiagen) as recommended by the manufacturer. For real-time PCR analysis of cultures, DNA was removed by 15 min of DNase digestion on the RNeasy columns with RNase-free DNase from Qiagen. This removed the small amounts of DNA contamination usually present in RNeasy extracts. To double-check that extracted RNA was not contaminated with DNA, a digestion control was included. A crude lysate was split equally, and individual aliquots were not digested or were digested with DNase, RNase, or RNase plus DNase. Extracts were then amplified by RT-PCR to show the purity of the extracted RNA.
Gulf of Mexico samples were obtained on a cruise during July 2001 of the RV Walton-Smith (25°18.781′N, 84°13.213′W) on a calm day during the early morning hours. Between 200 and 800 ml of seawater was filtered, and filters were stored as indicated above for culture samples. Cells were disrupted by bead beating (19), and 500-μl portions of lysate were extracted by using RNeasy spun columns (Qiagen) as recommended by the manufacturer.
Quantitative hybridization.
mRNAs from cultures and the environment were extracted by using RNeasy columns as indicated above. However, samples were not digested with DNase on the columns but were divided into three aliquots. In each case the first aliquot remained undigested on ice, the second aliquot was digested with DNase, and the third aliquot was digested with RNase. Extracts were then dot blotted onto Zeta-Probe charged nylon filters (Bio-Rad). The filters were dried under a heat lamp, and the RNA was cross-linked to the filters with a UV cross-linker (Fisher Scientific) by using the optimal cross-link setting on the equipment. Each filter was then hybridized with a probe derived from Cylindrotheca sp. strain N1 (23). A 35S-labeled riboprobe was generated by in vitro transcription of linearized plasmid DNA with 35S-UTP. Probing occurred at 55°C as previously described (19). Dotted RNA was quantified with a Bio-Rad model GS363 molecular imager. Standard curves were generated by in vitro transcription of the same rbcL clone employed to make the probe. Standards were dotted in duplicate by using amounts ranging from 500 to 0.5 pg/dot.
TaqMan probe specificity.
To screen for the specificity of our TaqMan probe, a representative subset of rbcL-containing clones obtained from the Gulf of Mexico (Wawrik et al., unpublished) were screened by colony amplification. These clones are known to be amplifiable with the form ID rbcL primers that have been described. Amplification was detected with the ABI Prism detection system as described above. To verify amplification and correct amplicon size, aliquots from real-time PCR were electrophoresed on a 1% agarose gel stained with ethidium bromide.
Standard curves for real-time PCR.
Standard curves for rbcL DNA were generated by using plasmid DNA (PCR2.1; Invitrogen, Carlsbad, Calif.) containing a diatom rbcL insert cloned from a natural Gulf of Mexico population (Wawrik et al., unpublished). Highly purified plasmid was obtained by extracting 500 ml of an Escherichia coli culture (clone P99FH13) with a Plasmid Maxi kit (Qiagen). DNA was quantified by using Hoechst 33258 (22). To produce RNA standards, plasmid DNA was linearized with BamHI, and in vitro transcriptions were performed by using T7 RNA polymerase for 2.5 h at 37°C. To eliminate DNA contamination, RNA was digested for 15 min with RQ1 DNase (Promega). RNA was purified by using an RNeasy column (Qiagen) as recommended by the manufacturer and was quantified by using a Ribogreen RNA quantification kit (Molecular Probes, Eugene, Oreg.) and the 16S rRNA standard included with the kit. Serial dilutions of DNA and RNA were prepared by using sterile RNase-free water. Five microliters of diluted nucleic acid was then added to 45 μl of RT-PCR master mixture, and PCR was performed as described above.
Competitive PCR.
To determine the effect of chromophytic rbcL DNA from algae other than diatoms and pelagophytes on the quantification of diatom DNA by real-time PCR, an experiment was performed with a range of concentrations of plasmid DNA containing an rbcL sequence not detected by our probe. We were concerned that coamplification of such nontarget nucleic acid in environmental samples might interfere with the quantification of diatom and pelagophyte rbcL target sequences. One picogram of diatom rbcL-containing plasmid DNA (clone P99FH13) was added to 0.4, 4, 40, and 400 pg of nontarget plasmid DNA in triplicate. The nontarget DNA clones chosen (P99BH7 and P99FH16) should be amplified by PCR primers but should not be detected by the probe. To a third series of tubes we added 0.4, 4, 40, and 400 pg of a 1:1 mixture of nontarget plasmid DNA from the two clones. Amplification was performed as described above.
Whole-cell carbon fixation.
Photosynthetic carbon fixation was measured by a method described by Strickland and Parson (29), as modified by Carpenter and Lively (2). Ten-milliliter portions of culture were incubated in acid-leached liquid scintillation vials. Light vials were incubated under an average light flux of 80 microeinsteins m−2 s−1 for 30 min, while dark vials were shielded from light by using black electrical tape. Radioactive 14C-labeled bicarbonate (Amersham Corp., Arlington Heights, Ill.) was added to both types of vials to a final concentration of 0.5 μCi/ml (18.5 kBq/ml). Before incubation and after 30 min, duplicate 2-ml samples of culture were removed and filtered onto 25-mm-diameter, 0.22-μm-pore-size GS filters (Millipore Corp.). The filters were then placed in 0.5 ml of 0.5 N hydrochloric acid to drive off residual bicarbonate, and radioactive counts were obtained by liquid scintillation counting.
RubisCO enzyme activity.
Duplicate 20-ml samples of culture were placed in sterile Oakridge tubes and centrifuged for 10 min at 10,000 × g to collect cells. The harvested cells were washed in 50 mM Tris-HCl (pH 8.0) containing 1 mM EDTA (pH 8.0), 10 mM MgCl2, 50 mM NaHCO3, and 1 mM dithiothreitol. The cells were then centrifuged as described above, and the cell pellets were stored in liquid nitrogen at −80°C until they were assayed. For enzyme assays cells were thawed, resuspended in the buffer described above (33), and then assayed for RubisCO enzyme activity by using a radiometric assay (5).
Cell counting.
Cell density was monitored by autofluorescent cell counting. One milliliter of culture was diluted in fresh medium and preserved with 3.7% formalin. The cells were then filtered onto black polycarbonate, 25-mm-diameter, 0.2-μm-pore-size Poretics filters (Osmonics, Minnetonka, Minn.). The filters were placed on glass microscope slides, and cells were counted by using oil immersion (magnification, ×1,000) and blue excitation. To check for bacterial contamination of algal cultures, samples were DAPI stained and slides were inspected by epifluorescence microscopy (24).
Flow cytometry.
Twenty microliters of 10% paraformaldehyde was added to 1 ml of seawater. Samples were incubated at room temperature for 10 min and frozen in liquid nitrogen until they were analyzed in the lab. For processing, samples were shipped on dry ice to the Flow Cytometry Lab of Texas A&M University. A FACSCalibur flow cytometer (Becton Dickinson, San Jose, Calif.) equipped with a 488-nm, 15-mW argon laser was used to quantify Prochlorococcus, Synechococcus, and picoeukaryotic algal populations. The fluorescence parameters measured for each event were forward and right angle light scatter for green (530 ± 30 nm), orange (585± 30 nm), and red (650 ± 30 nm) excitation wavelengths. For signal normalization of fluorescence signals, purple-yellow calibration beads (diameter, 2.2 μm; Spherotech Inc., Libertyville, Ill.) were added to each sample. Data collection was performed by using CellQuest software (version 3; Becton Dickinson), and data were analyzed by using CYTOWIN software (35; http://www.sb-oscoff.fr/Phyto/cyto.html#cytowin). Event rates were recorded for each sample. Amounts were corrected for the volume analyzed and the enumeration efficiency factor. Event rates and counts obtained with a series of known concentrations of calibration beads were used to determine efficiency factors.
RESULTS
TaqMan probe specificity.
The specificity of our probe was confirmed by amplification of a representative sample of rbcL-containing clones obtained from an ambient population in the Gulf of Mexico. Table 1 shows a list of the clones screened, their GenBank accession numbers, and their phylogenetic affinities (Wawrik et al., unpublished). Seven diatom and two pelagophyte sequences were positively identified by our probe. Prymnesiophyte and eustigmatophyte sequences were also amplified with our primer set, as detected by agarose gel electrophoresis (data not shown), yet amplification was not detected by the ABI Prism detection system.
TABLE 1.
Specificity of real-time PCR for diatom and pelagophyte DNA
| Clone | Phylogenetic association | GenBank accession no. | Amplification | Detection by real-time PCR |
|---|---|---|---|---|
| Positive controls | ||||
| + | + | |||
| P994AH8 | Diatom | AF381735 | + | + |
| P994CH1 | Pelagophyte | AF381685 | + | + |
| P994CH2 | Diatom | AF381686 | + | + |
| P994CH3 | Diatom | AF381677 | + | + |
| P994CH22 | Diatom | AF381676 | + | + |
| P994FH13 | Diatom | AF381681 | + | + |
| P994HH14 | Diatom | AF381684 | + | + |
| P994HH7 | Pelagophyte | AF381691 | + | + |
| P994AH8 | Diatom | AF381735 | + | + |
| Negative controls | ||||
| P994AH1 | Prymnesiophyte | AF381673 | + | − |
| P994AH5 | Prymnesiophyte | AF381733 | + | − |
| P994AH7 | Prymnesiophyte | AF381734 | + | − |
| P994AH12 | Prymnesiophyte | AF381673 | + | − |
| P994AH28 | Prymnesiophyte | AF381732 | + | − |
| P994CH9 | Prymnesiophyte | AF381741 | + | − |
| P994CH25 | Eustigmatophyte | AF381740 | + | − |
| P994HH1 | Prymnesiophyte | AF381752 | + | − |
| P994FY9 | Prochlorococcus | AF381660 | − | − |
Standard curves.
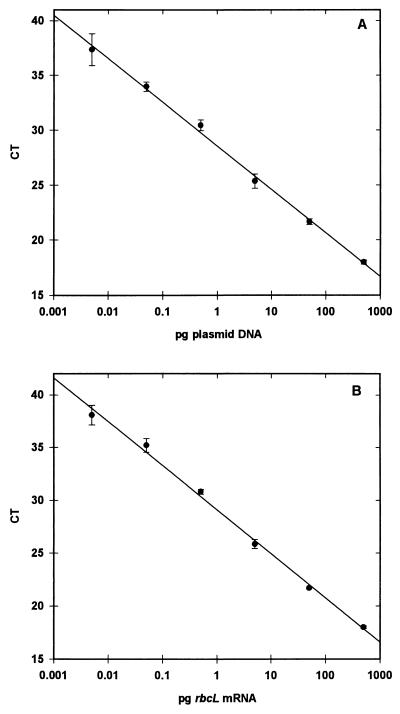
Standard curves were generated with good precision through a dynamic range of 6 orders of magnitude by using either plasmid DNA (Fig. 1A) or in vitro transcribed RNA (Fig. 1B) with our diatom TaqMan probe and the ABI Prism 7700 detection system. Coefficients of determination (R2) for the standard curves were more than 0.99 for both types of standards. Larger quantities of DNA or RNA led to nonlinear amplification due to overloading of the reaction mixture with target. Quantities smaller than the low end of our standards (<1,000 targets for plasmid DNA) were detected, but reliable quantification was not possible.
FIG. 1.
Standard curves generated from plasmid DNA containing a diatom rbcL insert (A) and in vitro transcribed mRNA of a diatom rbcL clone obtained from an ambient phytoplankton population in the Gulf of Mexico (B). The target nucleic acid concentration is plotted against the CT value. All values are averages of triplicate measurements. The error bars indicate 1 standard deviation.
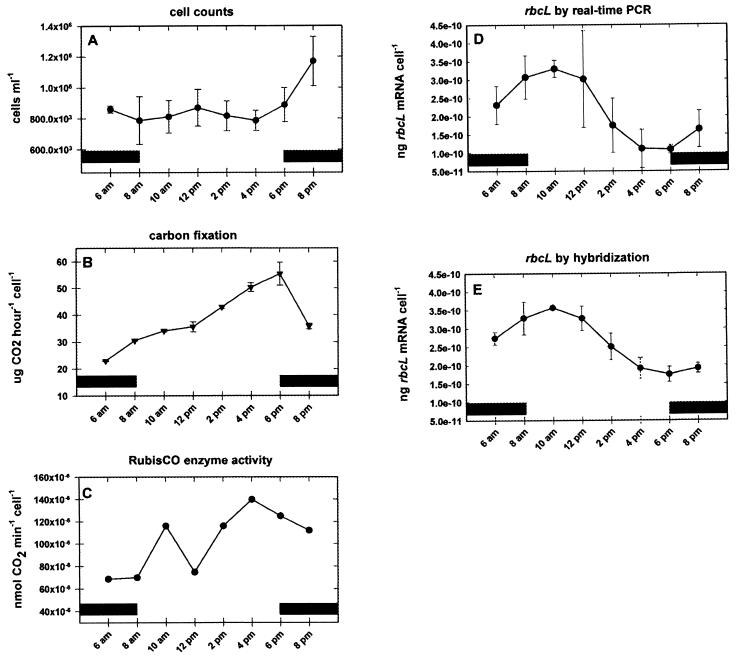
Culture experiments.
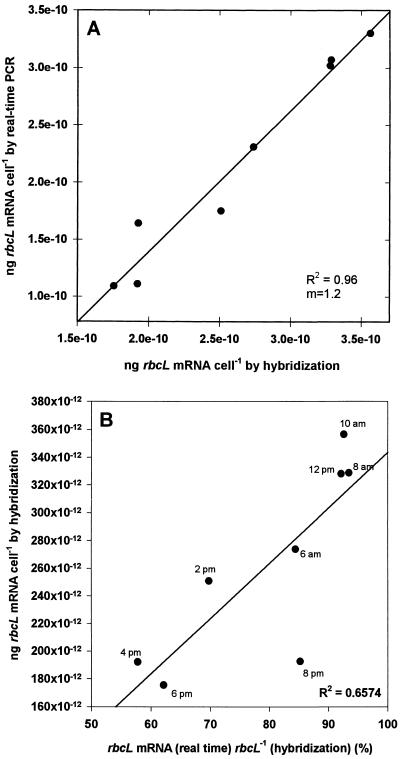
In order to quantify and compare rbcL mRNA by using either real-time PCR or hybridization with radiolabeled riboprobes, a culture of P. tricornutum was sampled throughout one light-dark cycle. Cell counts changed little throughout most of the sampling period but increased towards the end of the experiment (Fig. 2A). Cellular carbon fixation rose steadily throughout the illumination period, peaking in the late afternoon and decreasing thereafter (Fig. 2B). Cellular RubisCO enzyme activity also peaked during the late afternoon (Fig. 2C). During these time periods, rbcL mRNA levels were quantified by real-time PCR (Fig. 2D) and hybridization with 35S-labeled riboprobes (Fig. 2E). Clear diel regulation of transcript abundance was indicated by both methods; transcript abundance reached a maximum in the early morning hours and decreased to a low around 4 p.m. The quantities measured by the two methods were, in general, within similar ranges. However, a paired t test analysis indicated that the quantities obtained by hybridization were significantly greater than those obtained by real-time PCR at the 99% confidence interval (t = 5.293; P = 0.00113). The quantities measured by hybridization ranged from 1.75 × 10−10 to 3.56 × 10−10 ng of rbcL mRNA/cell, while the quantities obtained by real-time PCR varied between 1.09 × 10−10 and 3.30 × 10−10 ng of rbcL mRNA/cell. Paired t test analysis of the standard deviations for the two methods revealed no significant difference in precision at the 99% confidence interval (t = 2.923; P = 0.0223). Regression analysis revealed a good correlation between the two types of measurements (Fig. 3A) (R2 = 0.96; slope = 1.2). The increased slope was due to the relatively larger decrease in the amount of transcript detected by real-time PCR than by hybridization as transcript levels decreased and reached their minimum values during the afternoon hours. Figure 3B shows the relationship of the discrepancy between the two methods as a function of the total amount of rbcL mRNA detected by hybridization. During the peak expression in the early morning hours, real-time PCR accounted for more than 90% of the rbcL mRNA detected by hybridization. In the afternoon, however, when mRNA levels decreased, the quantities obtained by real-time PCR decreased more and accounted for only 58% in the 4 p.m. sample. The discrepancy between the two methods declined again in early evening hours, and real-time PCR accounted for ca. 85% of the rbcL mRNA in the 8 p.m. sample.
FIG. 2.
Growth parameters and rbcL mRNA contents of a P. tricornutum CCMP630 culture during a 16-h sampling period. (A) Epifluorescence cell counts for a culture containing 106 cells ml−1; (B) amount of CO2 fixed per 106 cells; (C) RubisCO enzyme activity; (D) rbcL mRNA expression as measured by using real-time PCR; (E) amount of rbcL mRNA per cell as measured by dot blotting and hybridization with a 35S-labeled probe. The error bars indicate 1 standard deviation calculated from triplicate samples. The solid bars indicate dark periods.
FIG. 3.
(A) Relationship between rbcL mRNA quantification determined by hybridization and rbcL mRNA quantification determined by real-time PCR. (B) Percentage of rbcL mRNA detected by real-time PCR. The quantities of rbcL mRNA detected by hybridization equal to 100% are plotted versus the quantities obtained by hybridization.
Environmental samples.
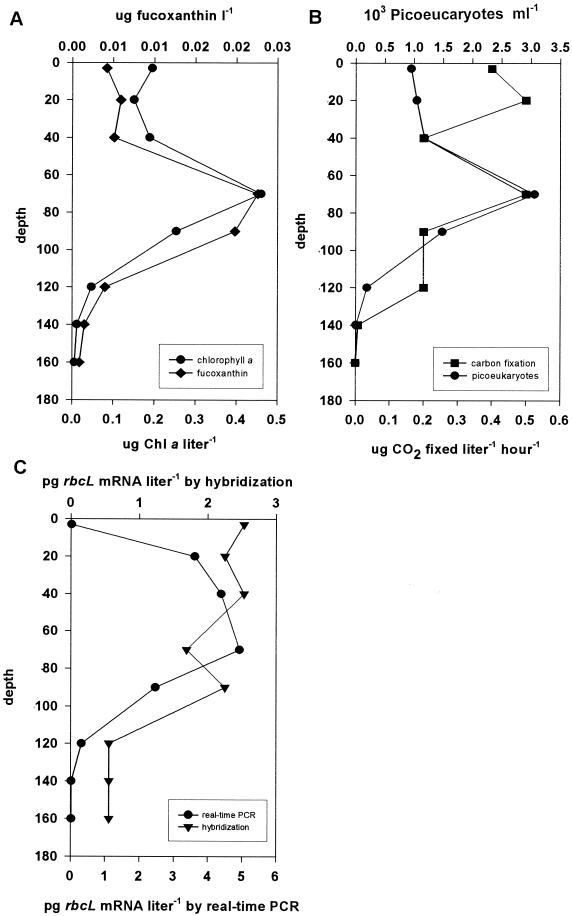
Figure 4 shows the biological parameters measured at station 8 in the Gulf of Mexico. Pigment profiles are shown in Fig. 4A. The levels of chlorophyll a and fucoxanthin (a diatom pigment) both peaked at a depth of ca. 80 m, indicating the position of the subsurface chlorophyll maximum (SCM). Flow cytometry (Fig. 4B) showed that there was a good correlation between the concentration of chlorophyll a and the abundance of picoeukaryotes (R2 = 0.983). The picoeukaryotic concentration was about 103 cells/ml at the surface, and it was more than threefold greater at the SCM around a depth of 80 m. At depths below the SCM, the cell concentration rapidly declined. Primary production at station 8 exhibited a bimodal distribution in the water column, peaking at a depth of 20 m and at the SCM. Figure 4C shows the amounts of rbcL mRNA detected by real-time PCR and hybridization. The mRNA expression measured by real-time PCR was low in the surface water, increased rapidly at middepth, and reached the maximum value at the SCM, below which it rapidly declined, like the abundance of picoeukaryotes. The quantities determined by hybridization were greatest at the surface, remained similar to a depth below the SCM, and then declined.
FIG. 4.
Biological parameters measured at station 8, an oligotrophic station in the Gulf of Mexico. (A) Levels of the pigments chlorophyll a (Chl a) and fucoxanthin (indicative of diatoms); (B) flow cytometry and primary productivity results; (C) levels of rbcL mRNA measured by real-time PCR and hybridization.
Competitive PCR experiments.
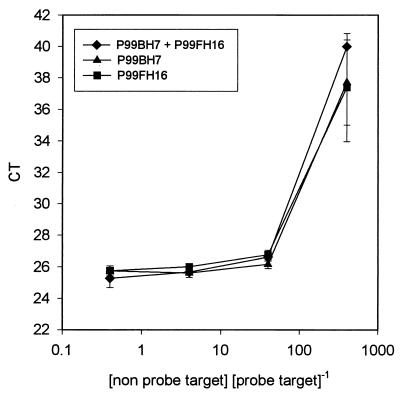
Our degenerate primer set was capable of amplifying not only the rbcL gene sequences of diatoms and pelagophytes but also sequences from a large array of other chromophytic algae (Wawrik et al., unpublished). We were concerned that the presence of competitive PCR targets in environmental samples, which might not be detected by our TaqMan probe, could influence the detection of diatom and pelagophyte rbcL mRNA. Figure 5 shows the behavior of the threshold cycle (CT value) as a function of a competitive PCR target. When the diatom rbcL DNA accounted for at least 2.5% of the total rbcL DNA in the reaction mixture, detection was possible and the CT value changed little with competitor concentration. At lower relative concentrations detection rapidly degraded and the CT value became erratic or was equal to the total number of PCR cycles (the ABI Prism detection system gives a CT value equal to the total number of cycles if no target is detected).
FIG. 5.
Change in CT value in real-time PCR as a function of competitive PCR target not detected by our TaqMan probe. The x-axis values are the ratio of the amount of total PCR target DNA to the amount of DNA recognized by the diatom probe. The error bars indicate 1 standard deviation calculated from triplicate samples.
DISCUSSION
A real-time PCR method was developed to detect diatom and pelagophyte rbcL gene expression with great precision and dynamic range. Using our diatom TaqMan probe, we were able to distinguish diatom and pelagophyte rbcL sequences form nontarget DNA. In culture experiments the precision of real-time PCR proved to be similar to the precision of hybridization analysis, yet real-time PCR provided a dynamic range that was 3 orders of magnitude greater. Quantities obtained by real-time PCR were slightly, yet significantly, lower than the quantities obtained by hybridization, a difference that can most likely be attributed to a potential bias of hybridization experiments towards partial or degraded target sequences. Alternatively, there may have been partial internal homology of the Cylindrotheca-derived riboporbe. Such homology can lead to the internal probe binding to itself before binding to target on the filter and thus lead to an increased signal.
One of the most important benchmarks of an environmental detection method is specificity (i.e., the ability to distinguish between the target sequence and other closely related nontarget organisms present in a sample). We screened 20 rbcL clones obtained from an ambient phytoplankton community in the Gulf on Mexico, which revealed the fidelity of this technique. It is important to note that our primer set amplifies rbcL from a much broader range of phytoplankton than our TaqMan probe is designed to detect. This approach allows the development of multiple probes for different phytoplankton groups, while only a limited number of primer sets is used.
Equally important are sensitivity and dynamic range. Real-time PCR detection of rbcL target nucleic acid has proven to be extremely sensitive; in addition, a large dynamic range with a quantitative detection limit of approximately 1,000 gene copies has been observed. The assay is linear through at least 6 orders of magnitude covering the entire range of traditional probing assays while being almost 3 orders of magnitude more sensitive. Similar amplification and detection characteristics have been reported by Gruntzig et al. (7) for amplification of nirS in Pseudomonas stutzeri and by Becker et al. (1) for amplification of 16S ribosomal DNA first internal transcribed spacer from Synechococcus sp. These results suggest that the lower detection limit of real-time PCR-based assays is approximately 102 targets. This value seems high compared to the theoretical limit of just one copy per reaction tube but was not viewed as a problem because of the abundance of rbcL mRNA in natural samples and our ability to increase sensitivity by a variety of concentration protocols (e.g., vacuum or tangential flow filtration). Real-time PCR detection of a particular gene sequence is therefore more likely to be limited by adequate nucleic acid extraction protocols and primer and probe design and not by the sensitivity of the real-time PCR.
rbcL mRNA expression is known to exhibit strong diurnal regulation both in picoplankton cultures (21) and in samples taken from the environment (23, 39). For marine Synechococcus species, the rbcL mRNA levels increased before sunrise, reaching a maximum at daybreak (39). In the form IB-containing marine cyanobacterium Synechococcus sp. strain PCC7002, the cellular rbcL mRNA content was highest during the mid-afternoon, preceding the maximum for cellular CO2 fixation (21). Ambient form IB-containing phytoplankton populations near Cape Hatteras have been observed to exhibit peak rbcL expression during the early afternoon hours (23), while chromophytic, form ID-containing picoplankton displayed the highest rbcL mRNA levels during the late afternoon and early night hours (23), similar to the patterns observed for the prymnesiophyte Pavlova gyrans (21). P. tricornutum rbcL expression increased just before the light period, resulting in the appearance of peak cellular mRNA concentrations during the early morning (Fig. 2D and E). These observations indicate that there are interesting differences in the time of appearance of the rbcL transcript in different phytoplankton groups. The early morning to sunrise hours appear to be utilized by form IA-containing cyanobacteria for peak rbcL mRNA synthesis. Shortly thereafter, diatoms (or at least Phaeodactylum) reach their expression maximum. Throughout the early afternoon hours form IB-containing phytoplankton (mainly green algae) most actively transcribe rbcL, while prymnesiophytes are transcriptionally most active late in the day.
In Phaeodactylum rbcL transcription is not well correlated with carbon fixation and RubisCO enzyme activity (Fig. 2). Net cellular carbon fixation peaked during the late afternoon together with RubisCO enzyme activity. This suggests that some type of posttranscriptional regulatory mechanism modulates the production of functional RubisCO. Such decoupling of rbcL mRNA expression from enzyme activity has also been observed in the chromophytic alga P. gyrans, in which peak RubisCO enzyme activity actually precedes mRNA expression (21). Possible regulatory mechanisms which could explain these observations are known to exist in higher plants (25) and may involve regulation of the carbamylation step, as well as binding of sugar phosphates to RubisCO (9). The significance of the decoupling, however, remains poorly understood. Perhaps decoupling is common in eukaryotes, while regulation in cyanobacteria seems to be at the level of transcription (21, 36).
We observed a good correlation between standard hybridization results and real-time PCR results (R2 = 0.95). The quantities obtained by these two methods were also of similar magnitudes, but they were significantly different. Real-time PCR consistently produced lower estimates, which ranged from 58 to 94% of the estimates obtained by hybridization. The discrepancy between the two methods was correlated (R2 = 0.65; slope ≈ 0.2 × 10−10 ng/cell) with the amount of rbcL transcript detected by hybridization (Fig. 3B). A strict correlation between the methods should have yielded a regression line with a slope of approximately zero in Fig. 3B. A possible explanation for the deviation from direct correlation of the two detection methods is the formation of partial rbcL transcripts as the result of normal cellular degradation of mRNA. The quantities obtained by hybridization measurements are thus necessarily higher, because no distinction is made between partial and complete mRNA transcripts. PCR, on the other hand, requires a continuous, uninterrupted nucleic acid molecule (at least for the sequence between the primer sites) for proper amplification of the target sequence. It should be noted that this requirement gives a distinct advantage to real-time PCR methods over hybridization techniques, since partial targets are, of course, not translated into functional enzymes. The standard deviations of the measurements obtained by the two methods were not significantly different, indicating that the two methods exhibit similar precision. This demonstrates that real-time PCR measurements represent a real alternative to conventional hybridization measurements and should be accepted with a similar degree of confidence.
It was also important to compare the rbcL mRNA levels obtained by using the two methods with a natural phytoplankton community. Such communities are dominated by picoplankton (diameters, 0.2 to 2 μm) in offshore, nutrient-depleted environments. Picoeukaryotes (including diatoms and pelagophytes) are found throughout the photic zone and numerically dominate the phytoplankton community at the SCM. The diversity of picoeukaryotes at any particular site in the ocean may be quite large; the population may simultaneously include green algae, coccolithophorids, and a great diversity of haptophytes, such as prymnsiophytes, pelagophytes, eustigmatophytes, and diatoms (18, 20; Wawrik et al., unpublished). Little is known, however, about the numerical contributions of these groups to oceanic picoplankton communities, nor is there information concerning the relative contributions of these different organisms to primary productivity. When several biological parameters were measured at an oligotrophic station in the Gulf of Mexico (Fig. 4), we observed a strong chlorophyll maximum at a depth of 80 m coinciding with a maximum level of fucoxanthin (a pigment indicative of diatoms). The levels of both pigments correlated well with the amounts of picoeukaryotes throughout the water column. This is not necessarily expected for a natural population, since there is usually considerable photoadaptation of cellular pigment content with depth. Primary productivity was bimodally distributed, peaking at the SCM and at a depth of 20 m. While the peak primary productivity at the SCM was most likely caused by the activity of picoeukaryotes, near the surface the productivity was likely due to Prochlorococcus, which is abundantly found in this region of the water column (data not shown). Real-time PCR demonstrated that the highest level of diatom and pelagophyte rbcL mRNA expression was at the SCM. The levels of expression above the SCM were slightly higher than the levels expected from a direct correlation between picoeukaryotic abundance or chlorophyll a level and rbcL expression. This suggests that diatoms and pelagophytes might contribute more actively to primary productivity than other picoeukaryotes in the portion of the water column dominated by Prochlorococcus. The levels of expression in the surface water were very low, suggesting that diatom and pelagophyte success in the surface layer might have been limited. We have, however, in the past had difficulty amplifying rbcL mRNA from Gulf of Mexico surface water, which led us to believe that the surface rbcL expression value reported here might be underestimated. This interpretation is supported by the observation that the highest hybridization levels were in fact observed in the surface sample. The rbcL hybridization levels were consistently high throughout most of the water column and declined below the SCM. Direct correlation between the two methods was poor (R2 = 0.34); however, if the questionable surface water value was removed from the data set, the two methods correlated reasonably well (R2 = 0.74), and the data were not significantly different as determined by paired t test analysis (P = 0.21; t = 1.42). It is possible that DNA damage in the surface water, due perhaps to high levels of UV radiation or enhanced nuclease levels, could lead to the formation of poor, partial, or erroneous transcripts. Such damaged transcripts would be included in the hybridization signal yet would not be amplifiable. Alternatively, the discrepancy might be explained by the presence of other chromophytic algae, which cross-hybridized with the diatom-derived riboprobe but were not detected by our diatom real-time PCR probe.
PCR-based techniques used in the study of environmental communities are subject to many biases. PCRs are often inhibited by contaminants, and clone libraries often include artifacts due to chimera formation in PCR mixtures (38). Related sequences are not always amplified with similar efficiencies, and not all target molecules are equally accessible to PCR primers, perhaps because primers do not bind with equal efficiencies or because extension efficiencies are not always equal for all templates (30). For example, Suzuki and Giovannoni (30) have shown that the proportion of products formed in PCR may be biased towards a 1:1 ratio regardless of the initial ratio of templates. The G+C content of the template DNA has also been suggested to influence PCR amplification (4, 26).
Figure 5 demonstrates another type of PCR bias in mixed-template experiments. It appears that amplification of minor components in mixed assemblages of targets may be completely repressed by the presence of a dominant template. This has great implications for interpretation of clone libraries since sequences present only at low levels may not be recovered. Also, the inability to quantitatively detect minor targets in real-time PCR applications may limit the capacity to use nonspecific primers for amplification.
Regardless of the shortcomings of PCR, real-time PCR is an extremely sensitive, reliable, and accurate technique for estimation of microbial activity in the environment and has great potential for use in studying the diversity and contributions of genes encoding important biogeochemical processes.
Acknowledgments
This work was funded by the Biotechnological Investigations-Ocean Margins Program, by Biological and Environmental Research, by the U.S. Department of Energy (grants DE-FG02-97ER62452 and DE-FG02-97ER62454), and by the National Science Foundation.
Flow cytometry was performed by Lisa Campbell and her coworkers at the Flow Cytometry Lab of Texas A&M University.
REFERENCES
- 1.Becker, S., P. Boger, R. Oehlmann, and A. Ernst. 2000. PCR bias in ecological analysis: a case study for quantitative Taq nuclease assays in analyses of microbial communities. Appl. Environ. Microbiol. 66:4945-4953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carpenter, E. J., and J. S. Lively. 1980. Review of estimates of algal growth using 14C-tracer techniques. Plenum Press, New York, N.Y.
- 3.Chisholm, S. W. 2000. Stirring times in the Southern Ocean. Nature 407:685-689. [DOI] [PubMed] [Google Scholar]
- 4.Dutton, C. M., C. Paynton, and S. Sommer. 1993. General method for amplifying regions of very high G + C content. Nucleic Acids Res. 21:2953-2954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gibson, J. L., D. L. Falcone, and F. R. Tabita. 1991. Nucleotide sequence, transcriptional analysis, and expression of genes encoded within the form I CO2 fixation operon of Rhodobacter sphaeroides. J. Biol. Chem. 266:14646-14653. [PubMed] [Google Scholar]
- 6.Gibson, U. E., C. A. Heid, and P. M. Williams. 1996. A novel method for real time quantitative RT-PCR. Genome Res. 6:995-1001. [DOI] [PubMed] [Google Scholar]
- 7.Gruntzig, V., S. C. Nold, J. Zhou, and J. M. Tiedje. 2001. Pseudomonas stutzeri nitrite reductase gene abundance in environmental samples measured by real-time PCR. Appl. Environ. Microbiol. 67:760-768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guillard, R. R. L. 1975. Culture of phytoplankton for feeding marine invertebrates, p. 26-60. In Culture of marine invertebrate animals. Plenum Press, New York, N.Y.
- 9.Hartman, F. C., and M. R. Harpel. 1994. Structure, function, regulation, and assembly of d-ribulose-1,5-bisphosphate carboxylase/oxygenase. Annu. Rev. Biochem. 63:197-234. [DOI] [PubMed] [Google Scholar]
- 10.Hedges, J. I., and R. G. Keil. 1995. Sedimentary organic matter preservation: an assessment and speculative analysis. Mar. Chem. 49:81-115. [Google Scholar]
- 11.Heid, C. A., J. Stevens, K. J. Livak, and P. M. Williams. 1996. Real time quantitative PCR. Genome Res. 6:986-994. [DOI] [PubMed] [Google Scholar]
- 12.Higuchi, R., G. Dollinger, P. S. Walsh, and R. Griffith. 1992. Simultaneous amplification and detection of specific DNA sequences. Bio/Technology 10:413. [DOI] [PubMed] [Google Scholar]
- 13.Holland, P. M., R. D. Abramson, R. Watson, and D. H. Gelfand. 1991. Detection of specific polymerase chain reaction product by utilizing the 5′-3′ exonuclease activity of Thermus aquaticus DNA polymerase. Proc. Natl. Acad. Sci. USA 88:7276-7280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Karl, D. M., D. V. Hebel, K. Bjoerkman, and R. M. Letelier. 1998. The role of dissolved organic matter release in the productivity of the oligotrophic North Pacific Ocean. Limnol. Oceanogr. 43:1270-1286. [Google Scholar]
- 15.Knight, S., I. Anderson, and C. I. Branden. 1990. Crystallographic analysis of ribulose 1,5 bisphosphate carboxylase from spinach at 2.4 C resolution. J. Mol. Biol. 215:113-160. [DOI] [PubMed] [Google Scholar]
- 16.Legendre, L., and J. Michaud. 1998. Flux of biogenic carbon in oceans: size-dependent regulation by pelagic food webs. Mar. Ecol. Prog. Ser. 164:1-12. [Google Scholar]
- 17.Marshall, H., and K. K. Nesius. 1996. Phytoplankton composition in relation to primary production in Chesapeake Bay. Mar. Ecol. Prog. Ser. 78:11-22. [Google Scholar]
- 18.Moon-van der Staay, S. Y., R. De Wachter, and D. Vaulot. 2001. Oceanic 18S rDNA sequences from picoplankton reveal unsuspected eukaryotic diversity. Nature 409:607-610. [DOI] [PubMed] [Google Scholar]
- 19.Paul, J. H. 2001. Marine microbiology, vol. 30. Dowden, Hutchinson & Ross, New York, N.Y.
- 20.Paul, J. H., A. Alfreider, and B. Wawrik. 2000. Micro- and macrodiversity in rbcL sequences in ambient phytoplankton populations from southeastern Gulf of Mexico. Mar. Ecol. Prog. Ser. 198:9-18. [Google Scholar]
- 21.Paul, J. H., J. B. Kang, and F. R. Tabita. 2000. Diel patterns of regulation of rbcL transcription in a cyanobacterium and a prymnesiophyte. Mar. Biotechnol. 2:429-436. [DOI] [PubMed] [Google Scholar]
- 22.Paul, J. H., and B. Myers. 1982. Use of Hoechst dyes 33258 and 33342 for the enumeration of attached and pelagic bacteria. Appl. Environ. Microbiol. 43:1393-1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Paul, J. H., S. L. Pichard, J. B. Kang, G. M. F. Watson, and F. R. Tabita. 1999. Evidence for a clade-specific temporal and spatial separation in ribulose bisphosphate carboxylase gene expression in phytoplankton populations off Cape Hatteras and Bermuda. Limnol. Oceanogr. 44:12-23. [Google Scholar]
- 24.Porter, K. G., and Y. S. Feig. 1980. The use of DAPI for identifying and counting aquatic microflora. Limnol. Oceanog. 25:943-948. [Google Scholar]
- 25.Portis, A. R. 1992. Regulation of ribulose 1,5-bisphospate carboxylase/oxygenase activity. Annu. Rev. Plant Physiol. 43:415-437. [Google Scholar]
- 26.Reysenbach, A. L., L. J. Giver, G. S. Wickham, and N. R. Pace. 1992. Differential amplification of rRNA genes by polymerase chain reaction. Appl. Environ. Microbiol. 58:3417-3418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sharp, P. A., A. J. Berk, and S. M. Berget. 1980. Transcription maps of adenovirus. Methods Enzymol. 65:750-768. [DOI] [PubMed] [Google Scholar]
- 28.Southern, E. M. 1975. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J. Mol. Biol. 98:503-517. [DOI] [PubMed] [Google Scholar]
- 29.Strickland, J. D. H., and T. R. Parson. 1968. Inorganic carbon assimilation by marine biota. Fish. Res. Board Can. Bull. 167:311. [Google Scholar]
- 30.Suzuki, M. T., and S. J. Giovannoni. 1996. Bias caused by template annealing in the amplification of mixtures of 16S rRNA genes in PCR. Appl. Environ. Microbiol. 62:625-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Suzuki, M. T., L. T. Taylor, and E. F. DeLong. 2000. Quantitative analysis of small-subunit rRNA genes in mixed microbial populations via 5′-nuclease assays. Appl. Environ. Microbiol. 66:4605-4614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tabita, F. R. 1999. Microbial ribulose 1,5-bisphosphate carboxylase/oxygenase: a different perspective. Photosynth. Res. 60:1-28. [Google Scholar]
- 33.Tabita, F. R., P. Caruso, and W. Whitman. 1978. Facile assay of enzymes unique to the Calvin cycle in intact cells, with special reference to ribulose 1,5-bisphosphate carboxylase. Anal. Biochem. 84:462-472. [DOI] [PubMed] [Google Scholar]
- 34.Thomas, P. S. 1980. Hybridization of denatured RNA and small DNA fragments transferred to nitrocellulose. Proc. Natl. Acad. Sci. USA 77:5201-5205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vaulot, D., C. Courties, and F. Partensky. 1989. A simple method to preserve oceanic phytoplankton for flow cytometric analyses. Cytometry 10:629-635. [DOI] [PubMed] [Google Scholar]
- 36.Watson, G. M., and F. R. Tabita. 1997. Microbial ribulose 1,5-bisphosphate carboxylase/oxygenase: a molecule for phylogenetic and enzymological investigation. FEMS Microbiol. Lett. 146:13-22. [DOI] [PubMed] [Google Scholar]
- 37.Webber, D. F., and J. C. Roff. 1996. Influence of Kingston Harbor on the phytoplankton community of the near-shore Hellshire Coast, southeast Jamaica. Bull. Mar. Sci. 59:245-258. [Google Scholar]
- 38.Winzingerode, F. V., U. B. Gobel, and E. Stackebrandt. 1997. Determination of microbial diversity in environmental samples: pitfalls of PCR-based rRNA analysis. FEMS Microbiol. Rev. 21:213-229. [DOI] [PubMed] [Google Scholar]
- 39.Wyman, M. 1999. Diel rhythms in ribulose-1,5-bisphosphate carboxylase/oxygenase and glutamine synthetase gene expression in a natural population of marine picoplanktonic cyanobacteria (Synechococcus spp.). Appl. Environ. Microbiol. 65:3651-3659. [DOI] [PMC free article] [PubMed] [Google Scholar]