Abstract
Haloalkane dehalogenases are microbial enzymes that catalyze cleavage of the carbon-halogen bond by a hydrolytic mechanism. Until recently, these enzymes have been isolated only from bacteria living in contaminated environments. In this report we describe cloning of the dehalogenase gene dhmA from Mycobacterium avium subsp. avium N85 isolated from swine mesenteric lymph nodes. The dhmA gene has a G+C content of 68.21% and codes for a polypeptide that is 301 amino acids long and has a calculated molecular mass of 34.7 kDa. The molecular masses of DhmA determined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and by gel permeation chromatography are 34.0 and 35.4 kDa, respectively. Many residues essential for the dehalogenation reaction are conserved in DhmA; the putative catalytic triad consists of Asp123, His279, and Asp250, and the putative oxyanion hole consists of Glu55 and Trp124. Trp124 should be involved in substrate binding and product (halide) stabilization, while the second halide-stabilizing residue cannot be identified from a comparison of the DhmA sequence with the sequences of three dehalogenases with known tertiary structures. The haloalkane dehalogenase DhmA shows broad substrate specificity and good activity with the priority pollutant 1,2-dichloroethane. DhmA is significantly less stable than other currently known haloalkane dehalogenases. This study confirms that a hydrolytic dehalogenase is present in the facultative pathogen M. avium. The presence of dehalogenase-like genes in the genomes of other mycobacteria, including the obligate pathogens Mycobacterium tuberculosis and Mycobacterium bovis, as well as in other bacterial species, including Mesorhizobium loti, Xylella fastidiosa, Photobacterium profundum, and Caulobacter crescentus, led us to speculate that haloalkane dehalogenases have some other function besides catalysis of hydrolytic dehalogenation of halogenated substances.
Haloalkane dehalogenases catalyze hydrolytic cleavage of carbon-halogen bonds in halogenated aliphatic compounds, leading to the formation of primary alcohols, halide ions, and protons. These enzymes are potentially useful for cleaning up contaminated subsurfaces (32) and for processing by-products of chemical syntheses (33). Haloalkane dehalogenases can serve as a model system for studies of the evolution and distribution of degradation enzymes in the environment since many of these enzymes have already been isolated from different bacterial species originating from geographically distinct areas (26). Haloalkane dehalogenases have primarily been isolated from bacteria colonizing environments contaminated by halogenated substances (12, 16, 22, 27-30, 40). Only recently have the hydrolytic dehalogenation activities of several species of the genus Mycobacterium isolated from clinical material been reported (14). Motivation for the search for haloalkane dehalogenases in clinical samples of mycobacteria came from the identification of dehalogenase-like genes in the genome of Mycobacterium tuberculosis H37Rv resulting from a BLAST search of genetic databases. Now many new dehalogenase-like genes can be identified in the genomes of various bacteria by database searches (Table 1). If biochemical experiments confirm that the translation products of these genes can catalyze hydrolytic dehalogenation, they should become a valuable source of material for protein-engineering studies attempting to develop efficient catalysts for biotechnological applications (9).
TABLE 1.
Haloalkane dehalogenases and putative haloalkane dehalogenasesa
| Organism | Strain | Gene | Protein | Accession no.
|
Reference
|
||
|---|---|---|---|---|---|---|---|
| Gene (EMBL/GenBank/DDBJ) | Protein (SWISS-PROT) | Gene | Protein | ||||
| Sphingomonas paucimobilis | UT26 | linB | LinB | D14594 | P51698 | 23 | 22 |
| Mycobacterium tuberculosis | H37Rv | rv2579 | Rv2579a | Z77724 | Q50642 | 6 | NAb |
| Mycobacterium tuberculosis | CDC1551 | mt2656 | Rv2579a | AE007099 | Q50642 | —c | NA |
| Mycobacterium bovis | MU11 | iso-rv2579 | Iso-Rv2579a | AJ243259 | Q9XB14 | —d | NA |
| Mycobacterium smegmatis | MC2_155 | aal17946 | Aal17946a | AY054120 | AAL17946 | —e | NA |
| Rhodococcus (formerly Corynebacterium) sp. | m15-3 | dhaA | DhaA | NA | Q53042 | 4 | 40 |
| Rhodococcus (formerly Arthrobacter) sp. | HA1 | dhaA | DhaA | NA | Q53042 | 26 | 30 |
| Rhodococcus (formerly Acinetobacter) sp. | GJ70 | dhaA | DhaA | NA | Q53042 | 26 | 12 |
| Rhodococcus sp. | TB2 | dhaA | DhaA | NA | Q53042 | 26 | NA |
| Rhodococcus erythropolis | Y2 | dhaA | DhaA | NA | Q53042 | 26 | 29 |
| Rhodococcus rhodochrous | NCIMB13064 | dhaA | DhaA | AF060871 | Q53042 | 17 | NA |
| Pseudomonas pavonaceae | 170 | dhaA | DhaA | AJ250371 | Q53042 | 28 | 28 |
| Mycobacterium sp. | GP1 | dhaAf | DhaAf | AJ012627 | Q9ZER0 | 27 | NA |
| Mesorhizobium loti | MAFF303099 | mlr5354 | Mlr5354a | AP003006 | Q98C03 | 15 | NA |
| Xylella fastidiosa | 9A5C | xf1965 | Xf1965a | AE004016 | Q9PC20 | 31 | NA |
| Photobacterium profundum | SS9 | aal01057 | Aa101057a | AF409100 | AAL01057 | —f | NA |
| Caulobacter crescentus | CB15 | cc1175 | Cc1175a | AE005795 | Q9A919 | 25 | NA |
| Mycobacterium tuberculosis | H37Rv | rv2296 | Rv2296a | Z77163 | Q50670 | 6 | NA |
| Mycobacterium tuberculosis | CDC1551 | mt2353 | Rv2296a | AE007077 | Q50670 | —c | NA |
| Mycobacterium avium | N85 | dhmA | DhmA | AJ314789 | CAC41377 | This study | This study |
| Mycobacterium avium | 104 | 106 | DhmA | NA | CAC41377 | —c | NA |
| Xanthobacter autotrophicus | GJ10 | dhlA | DhlA | M26950 | P22643 | 13 | 16 |
| Xanthobacter autotrophicus | GJ11 | dhlA | DhlA | NA | P22643 | 35 | NA |
| Ancylobacter aquaticus | AD20 | dhlA | DhlA | NA | P22643 | 35 | NA |
| Ancylobacter aquaticus | AD25 | dhlA | DhlA | NA | P22643 | 35 | NA |
Identified by sequence similarity.
NA, not available.
R. D. Fleischmann et al., unpublished data.
A. Jesenská et al., unpublished data.
E. E. Allen and D. H. Barlett, unpublished data.
Cloning and sequencing of the dhmA haloalkane dehalogenase gene from Mycobacterium avium subsp. avium N85 and its expression and biochemical characterization of DhmA in crude extracts are described in this report. This study confirmed that bacteria isolated from clinical material may express haloalkane dehalogenases.
MATERIALS AND METHODS
Sequence analysis.
Putative haloalkane dehalogenases were identified by iterative searches of nonredundant databases by using PSI-BLAST (1) and the BLOSUM62 substitution matrix. The protein sequences of known (biochemically confirmed) haloalkane dehalogenases served as the query sequences. The protein sequences of haloalkane dehalogenases and putative haloalkane dehalogenases were downloaded from the SWISS-PROT database by using the accession numbers in Table 1. A multiple-sequence alignment was constructed by using CLUSTALX v1.8 (34) and was refined manually. A phylogenetic tree was based on alignment of four motifs corresponding to the epoxidase fingerprint (3) by the neighbor-joining method implemented in CLUSTALX.
Bacterial strains and growth conditions.
M. avium N85 was isolated from swine mesenteric lymph nodes (Svitavy, Czech Republic). M. avium MU1 was isolated from clinical material (Teaching Hospital Bohunice, Brno, Czech Republic). The isolates were identified as M. avium subsp. avium by serotyping (38). The isolates were grown aerobically in liquid Sula's medium and on solid Lowenstein-Jensen medium (18) at 37°C. Escherichia coli GI724 carrying a cloned dhmA gene was cultivated at 37°C in liquid RMG medium (6 g of Na2HPO4 · 12H2O per liter, 0.5 g of NaCl per liter, 1 g of NH4Cl per liter, 0.095 g of MgCl2 per liter, 2 g of Casamino Acids per liter) supplemented with 25 ml of 20% dextrose and 100 μg of ampicillin per ml.
Isolation of total DNA.
Cells were grown in 100 ml of medium at 37°C to the early stationary phase, harvested by centrifugation, and resuspended in TE buffer containing 10 mM Tris and 1 mM EDTA (pH 8.0). Then 50 μg of lysozyme and 60 μl of proteinase K (20 mg/ml) were added, and the culture was incubated at 37°C for 3 h. After addition of 600 μl of 10% sodium dodecyl sulfate (SDS), 2 ml of 5 M NaCl, and 1.6 ml of acetyltrimethylammonium bromide-NaCl, the mixture was incubated at 65°C for 10 min. DNA was extracted twice with equal volumes of phenol, phenol-chloroform (1:1, vol/vol), and chloroform-isoamyl alcohol (24:1, vol/vol). The DNA was precipitated with cold 96% ethanol and washed with 70% ethanol. After centrifugation, the DNA was resuspended in 600 μl of TE buffer.
PCR amplification, cloning, and sequencing.
Oligonucleotides were designed by using the fragment 106 sequence of the unfinished genome of M. avium 104 containing a gene which exhibits very high sequence similarity with currently known genes encoding haloalkane dehalogenases. The primer sequences were as follows: 5′-GCN NNN NTC TAG AGG TCA GAG CAG CGC CTG-3′ (an XbaI restriction site is underlined) and 5′-GCN NNG GTA CCC ATG CAT GTG CTG CGA ACC-3′ (a KpnI restriction site is underlined). DNA samples were amplified in 20-μl PCR mixtures by using a Taq PCR Master Mix kit (QIAGEN, Hilden, Germany), 10 pmol of each primer, and 2 μl of DNA sample. The initial denaturation step consisted of 5 min at 95°C, and this was followed by 35 cycles of denaturation at 95°C for 35 s, annealing at 65°C for 30 s, and extension at 72°C for 90 s and then a final extension step of 72°C for 5 min. The amplification products were separated on a 2% agarose gel, stained with ethidium bromide, and photographed under UV light. The PCR amplification product was purified by using the QIAquick gel extraction kit protocol (QIAGEN) and was cloned between KpnI and XbaI sites behind the PL promoter in the expression vector pAL-781 (Invitrogen, Groningen, The Netherlands) carrying the gene for ampicillin resistance as a selection marker. Transformation of E. coli GI724 cells with a ligation mixture was performed by a heat shock method. Transformants were plated onto RMG medium containing 100 μg of ampicillin per ml. Ampicillin-resistant colonies were screened for the presence of dehalogenating activity for 1,3-dibromopropane by monitoring halide production. For this purpose, the cells were incubated in a microtiter plate with 150 μl of 5 mM 1,3-dibromopropane in 50 mM Tris-sulfate buffer (pH 8.2). The plate was incubated overnight at 30°C, and then 100 ml of 0.25 M NH4(FeSO4)2 in 6 M HNO3 was added, followed by 1 drop of a saturated solution of Hg(SCN)2 in ethanol. A red color indicated the presence of dehalogenase activity. Plasmid DNA was isolated from a colony showing dehalogenase activity, checked by restriction analysis, and used for sequencing. Sequencing reactions were performed with a DNA ABI PRISM 310 genetic analyzer (Perkin-Elmer, Norwalk, Conn.). Strands from both sides were sequenced to ensure accuracy.
Expression and preparation of crude extracts.
Transformed cells of E. coli GI724 were cultured in 10 liters of RMG medium at 37°C. When the culture reached an optical density at 600 nm of 0.6, gene expression was induced with 100 μg of l-tryptophan per ml at 30°C. The cells were harvested 3 h after induction by centrifugation at 15,000 × g for 30 min, washed, and resuspended in 50 mM Tris-sulfate buffer (pH 7.5). The cells were disrupted by sonication with a SONOPLUS GI70 (Bandeline, Berlin, Germany). Intact cells and debris were removed by centrifugation at 40,000 × g for 40 min at 4°C to obtain crude cell extract. The crude extract was stored at −60°C. The same cell disruption and centrifugation procedure was used for preparation of crude extracts of M. avium MU1 cells.
Protein purification.
Crude extract of E. coli GI724 cells was dialyzed for 10 h against a 20-mmol/liter Tris-H2SO4 solution (pH 7.5). The two-step procedure for purification of recombinant DhmA consisted of (i) ion exchange chromatography and (ii) gel permeation chromatography. The dialyzed crude extract was applied to a MONO Q column equilibrated with a 20-mmol/liter Tris-H2SO4 solution (pH 7.5). Elution was carried out by using a linear gradient of Tris-H2SO4 with 1 mol of Na2SO4 per liter. The active fractions were applied to a Superdex 75 column equilibrated with a 50-mmol/liter Tris-H2SO4 solution (pH 7.5) and were eluted with the same buffer. The four-step procedure for purification of DhmA from M. avium MU1 consisted of (i) precipitation by ammonium sulfate, (ii) hydrophobic chromatography, (iii) ion exchange chromatography, and (iv) gel permeation chromatography. The protein concentrations were determined by the Bradford method (5) with bovine serum albumin as a standard.
Activity assays.
Haloalkane dehalogenase activities in crude extracts were determined in triplicate by a microtiter plate colorimetric assay by using the reagents of Iwasaki et al. (11) as described previously by Damborsky et al. (8). A precise activity assay was conducted by using gas chromatography to determine both substrate and product concentrations in the reaction mixture as described by Jesenská et al. (14). Briefly, a 0.2-ml protein preparation which contained between 1.0 and 1.3 mg of protein/ml in 50 mM Tris-H2SO4 buffer (pH 7.5) was incubated with a halogenated substrate at a final concentration of 10 mM. The progress of the reaction was monitored after 15, 30, 45, and 60 min with an HP 6890 gas chromatograph equipped with a flame ionization detector (Hewlett-Packard, Palo Alto, Calif.).
Biochemical characterization.
Characterization of DhmA was conducted in parallel by using crude extracts prepared from E. coli GI724 overexpressing dhmA and M. avium MU1. 1,3-Dibromopropane was used as the substrate whenever appropriate. The pH dependence of the crude extract was investigated by varying the composition of the 50 mM buffer. Sodium acetate was used to cover the pH range from pH 4.5 to 6.0, potassium phosphate was used for pH 6.0 to 8.5, and Tris-H2SO4 was used for pH 8.5 to 9.0. The temperature dependence was determined in 50 mM Tris-H2SO4 buffer (pH 7.5) incubated at 20, 30, 40, 50, and 60°C. Ionic strength was tested in buffers containing 1, 10, 25, 50, 100, and 1,000 mM Tris-H2SO4 (pH 7.5). pH stability was studied by using the buffers used for the pH dependence analysis but with a broader pH range (pH 3.0 to 10.0). Activity was determined under optimal conditions at 0, 24, 48, 72, 96, and 120 h. Temperature stability was tested at −60, 4, and 24°C. The impact of stabilizing additives on the stability of DhmA was tested with 10% glycerol, 1 mM EDTA, and 0.1 mM β-mercaptoethanol. Protein activity was monitored every hour for 6 h.
Nucleotide sequence accession number.
The nucleotide sequence of dhmA has been deposited in the GenBank database under accession number AJ314789.
RESULTS AND DISCUSSION
Identification of the putative haloalkane dehalogenase in M. avium.
The present study was motivated by the recent finding of haloalkane dehalogenase-like genes in M. tuberculosis H37Rv and of dehalogenating activities in 13 different Mycobacterium species (14). M. tuberculosis is pathogenic for humans; thus, for safety reasons M. avium was used for cloning and overexpression of a mycobacterial haloalkane dehalogenase gene and for characterization of a mycobacterial haloalkane dehalogenase. The genome of M. avium 104 has been partially sequenced (http://www.tigr.org/tdb/), providing the data necessary for designing primers complementary to the regions flanking the dehalogenase-like gene of this species. A search of the incomplete genomic database of M. avium 104 (http://www.tigr.org/tdb/) performed with the sequences of genes encoding known and putative haloalkane dehalogenases listed in Table 1 revealed that the translation product of the sequence designated fragment 106 (later designated DhmA) shows 36.7% sequence identity with the haloalkane dehalogenase DhlA of Xanthobacter autotrophicus GJ10 (16), 45.5% sequence identity with the putative haloalkane dehalogenase Cc1175 of Caulobacter crescentus CB15 (25), and 82.4% sequence identity with the putative haloalkane dehalogenase Rv2296 of M. tuberculosis H37Rv (6).
Sequence and phylogenetic analysis of the sequence of DhmA.
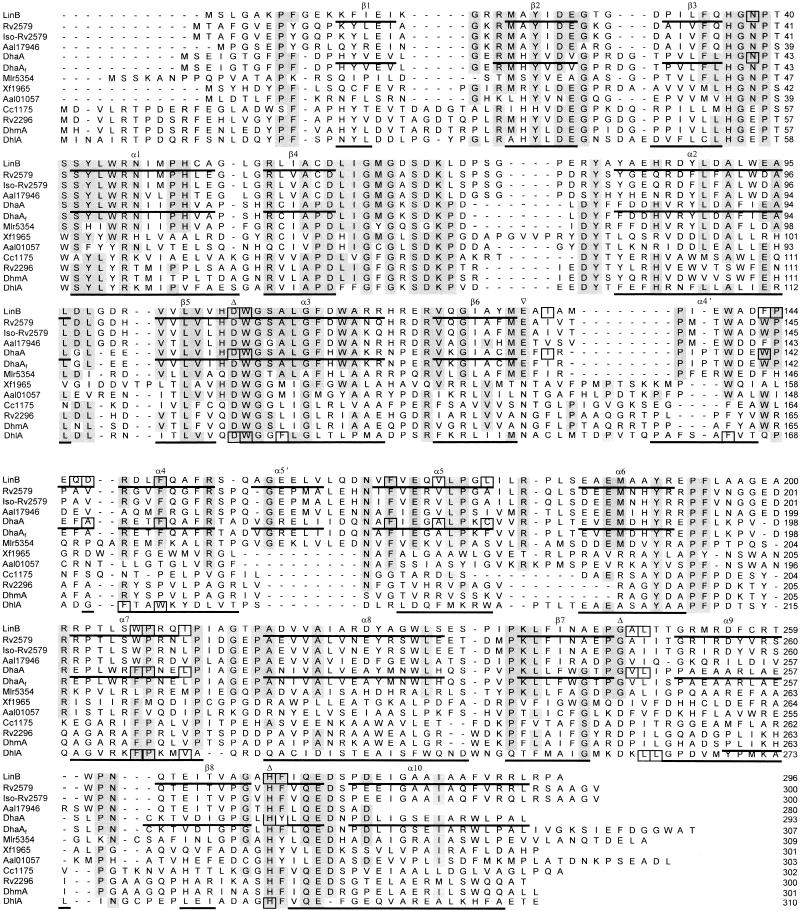
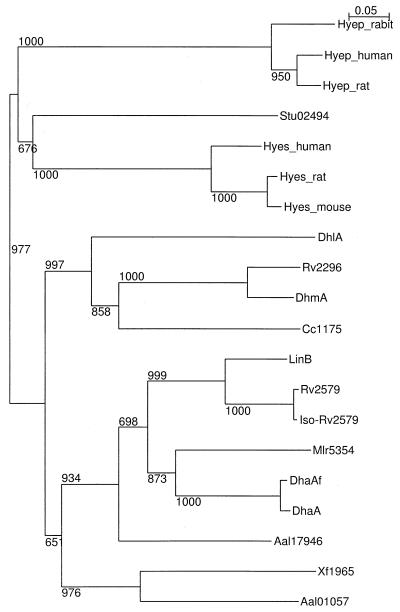
A multiple alignment of the DhmA sequence with the sequences of known and putative haloalkane dehalogenases revealed partially conserved secondary elements and fully conserved catalytic amino acid residues (Fig. 1). The residues essential for catalysis of hydrolytic dehalogenation (7) were identified by comparison of the DhmA sequence with the sequences of haloalkane dehalogenases with known three-dimensional structures (20, 24, 36). We propose that the putative catalytic triad of DhmA consists of Asp123, His279, and Asp250. The putative oxyanion hole consists of Glu55 and Trp124. Trp124 should be also involved in substrate binding and product (halide) stabilization. Site-directed mutagenesis experiments have been initiated to confirm this proposal. The second halide-stabilizing residue is not conserved in DhmA and structurally characterized haloalkane dehalogenases. A phylogenetic tree was constructed to investigate the relationship of the sequence of DhmA with the sequences of haloalkane dehalogenases and epoxidases (Fig. 2), as epoxidases are evolutionarily the proteins that are most closely related to haloalkane dehalogenases (2). The analysis revealed that the sequence encoded by fragment 106 (DhmA) is located on the branch containing the haloalkane dehalogenases, not the branch containing epoxide hydrolases.
FIG. 1.
Multiple alignment of sequences of known and putative haloalkane dehalogenases. The sequence designations are explained in Table 1. Secondary elements deduced from the experimentally determined structures of LinB (20), DhaA (24), and DhlA (36) dehalogenases are indicated under the sequences. Catalytic residues are indicated by triangles above the sequences. Active-site residues are enclosed in boxes. Conserved residues (i.e., identical amino acid residues in more than 50% of the sequences) are shaded.
FIG. 2.
Phylogenetic tree of haloalkane dehalogenases and epoxide hydrolases. The sequence designations for haloalkane dehalogenases are shown in Table 1, and the sequence designations for epoxidases are as follows: Hyes_human, soluble epoxide hydrolase from Homo sapiens; Hyep_human, microsomal epoxide hydrolase from H. sapiens; Hyep_rat, microsomal epoxide hydrolase from rat; Hyes_mouse, soluble epoxide hydrolase from mouse; Hyes_rat, soluble epoxide hydrolase from rat; Hyep_rabit, microsomal epoxide hydrolase from rabbit; and Stu02494, epoxide hydrolase from Solanum tuberosum. The sequences used for fingerprinting of epoxide hydralases were obtained from the Protein Motif Fingerprint Database (3). The numbers at the nodes are the confidence values for the groups derived from 1,000 rounds of the bootstrap procedure.
Cloning and sequencing of dhmA.
The primers designed by using regions flanking the putative dehalogenase gene on fragment 106 of M. avium 104 were used to amplify genes in M. avium N85 whose sequences were similar. M. avium N85 originated from swine mesenteric lymph nodes. The amplification product obtained from M. avium N85 was cloned into E. coli GI724 and designated the dhmA gene. The dhmA gene has a G+C content of 68.21% and codes for a polypeptide that is 301 amino acids long and has a molecular mass of 34.6 kDa. The translated sequence of dhmA was identical to the sequence encoded by the protein-coding region of fragment 106 of M. avium 104.
Overexpression and purification of DhmA.
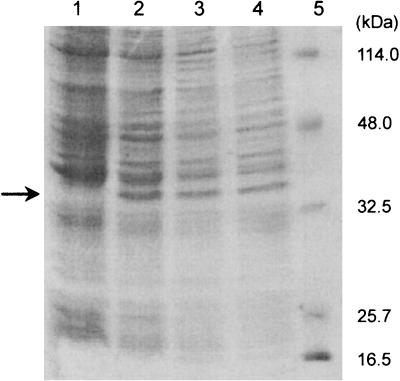
The haloalkane dehalogenase DhmA can be overexpressed in E. coli GI724, as confirmed by SDS-polyacrylamide gel electrophoresis (PAGE) (Fig. 3) and an activity assay with 1,3-dibromopropane (data not shown). The molecular mass of DhmA, as determined by SDS-PAGE, is 34.0 kDa. The recombinant haloalkane dehalogenase was purified to homogeneity from E. coli GI724 by using two chromatographic steps. The molecular mass of pure DhmA determined by gel permeation chromatography was 35.4 kDa. However, the enzyme lost most of its activity during purification. All attempts to obtain a sufficient amount of active haloalkane dehalogenase for biochemical characterization were unsuccessful, irrespective of whether it was purified from E. coli GI724 (dhmA) or M. avium MU1. This was due to the instability of DhmA outside the host cells, as was later confirmed by the stability assay conducted with crude extracts. Preliminary characterization of DhmA was therefore conducted with crude extracts.
FIG. 3.
SDS-PAGE gel after electrophoresis of E. coli GI724 expressing dhmA: total proteins of noninduced E. coli GI724 carrying plasmid pAL781-dhmA (lane 1) and of induced cells withdrawn 2 h (lane 2), 3 h (lane 3), and 4 h (lane 4) after inducer was added. Low-range molecular mass standards (lane 5) were used to estimate the size of the DhmA protein (arrow).
Preliminary biochemical characterization of DhmA.
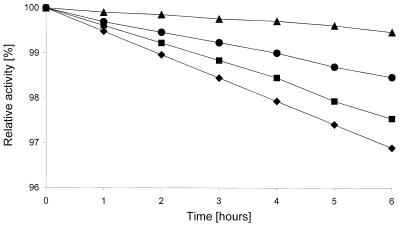
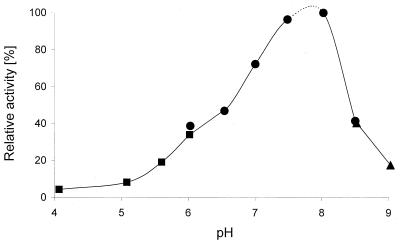
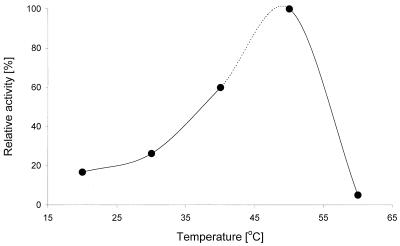
Biochemical characterizations of the haloalkane dehalogenase DhmA conducted in parallel with crude extracts from E. coli GI724 overexpressing dhmA and M. avium MU1 produced very similar results. The values reported below are those obtained with E. coli GI724 (dhmA), unless stated otherwise. The effect of storage temperature on the activity of crude extracts was assessed. The extracts stored at 24°C lost activity after 3 days, the extracts stored at 4°C lost activity after 9 days, and the extracts stored at −60°C lost activity after 3 months. Low stability of DhmA is one of the obvious differences between this protein and previously characterized haloalkane dehalogenases (12, 16, 22, 27-30, 40). The effects of stabilizing additives on the enzyme stored at 4°C were tested. The greatest short-term stabilizing effect was observed with glycerol, while EDTA and 2-mercaptoethanol reduced the enzyme activity (Fig. 4). The effect of pH on the activity of crude extracts was studied by using pH values ranging from 3.06 to 9.94. The dehalogenase exhibited more than 50% of the maximum activity at pH 6.5 to 8 and very low activity at pH values below 5 and above 9.5 (Fig. 5). The highest activity of DhmA was observed at pH 8. The effect of temperature on activity was studied by using temperatures ranging from 20 to 60°C. The dehalogenase activity increased as the temperature increased from 20 to 50°C, but it dropped to zero at 60°C. The highest activity was observed at 50°C (Fig. 6). The effect of ionic strength on activity was tested in Tris-H2SO4 buffer (pH 7.5), and the highest activity was observed in 50 mM buffer (data not shown). The substrate specificity of DhmA was tested with 34 different halogenated compounds selected for testing by a statistical experimental design (21). Halide ions formed by hydrolytic dehalogenation of the substrates by DhmA were detected colorimetrically. The substrate specificity of crude extracts prepared from E. coli GI724 (dhmA) was essentially the same as the substrate specificity of crude extracts prepared from M. avium MU1 (Table 2). The substrate specificity of DhmA is unlike the substrate specificities of LinB, DhaA, and DhlA dehalogenases (8). A biotechnologically interesting observation is the good activity of this protein with the priority pollutant 1,2-dichloroethane (32), which may be related to higher sequence identity between DhmA and DhlA than between DhmA and LinB or DhaA. 1,2-Dichloroethane is efficiently dehalogenated by DhlA but is a poor substrate for all other currently characterized haloalkane dehalogenases.
FIG. 4.
Effects of stabilizing additives on the stability of haloalkane dehalogenase DhmA stored at 4°C. Crude extracts without additives (•), with 10% glycerol (▴), with 1 mM EDTA (▪), and with 0.1 mM mercaptoethanol (♦) were tested. Activities were determined with the substrate 1,3-dibromopropane at 37°C and pH 7.5.
FIG. 5.
Effect of pH on the activity of haloalkane dehalogenase DhmA. Phosphate buffer (•), biphosphate buffer (▴), and acetate buffer (▪) were tested. Activities were determined with the substrate 1,3-dibromopropane at 37°C.
FIG. 6.
Effect of temperature on the activity of haloalkane dehalogenase DhmA. Activities were determined with the substrate 1,3-dibromopropane at pH 7.5.
TABLE 2.
Dehalogenating activities of crude extracts from E. coli GI724 (dhmA) and M. avium MU1
| Substrate | Relative activity (%)
|
|
|---|---|---|
| E. coli GI724 (dhmA) | M. avium MU1 | |
| Monohalogenated alkanes | ||
| 1-Chlorobutane | 100a | 100b |
| 1-Chloropentane | 217 | 197 |
| 2-Chloropropane | 170 | 173 |
| 1-Chloro-2-methylpropane | 3 | 3 |
| Chlorocyclopentane | 58 | 58 |
| Chlorocyclohexane | 16 | 17 |
| 1-Bromoethane | 278 | 282 |
| 1-Bromobutane | 234 | 237 |
| 1-Bromo-2-methylpropane | 59 | 60 |
| Bromocyclohexane | 21 | 21 |
| (1-Bromomethyl)cyclohexane | 10 | 11 |
| 1-Iodopropane | 248 | 251 |
| 1-Iodobutane | 125 | 127 |
| 2-Iodobutane | 171 | 173 |
| Dihalogenated alkanes | ||
| 1,2-Dichloroethane | 200 | 205 |
| 1,2-Dichloropropane | 16 | 17 |
| 1,3-Dichloropropane | 140 | 144 |
| 1,2-Dichlorobutane | 56 | 56 |
| 1-Bromo-2-chloroethane | 77 | 78 |
| 2-Bromo-1-chloropropane | 60 | 62 |
| 1,2-Dibromoethane | 256 | 260 |
| 1,3-Dibromopropane | 326 | 331 |
| 1,2-Dibromopropane | 107 | 108 |
| 1,3-Diiodopropane | 21 | 22 |
| Chlorinated alkenes | ||
| 3-Chloro-2-methylpropene | 78 | 78 |
| 2,3-Dichloropropene | 77 | 78 |
| Trihalogenated alkanes | ||
| 1,2,3-Trichloropropane | 15 | 16 |
| 1,2-Dibromo-3-chloropropane | 39 | 40 |
| 1,2,3-Tribromopropane | 172 | 174 |
| Halogenated nitriles | ||
| Chloroacetonitrile | NAc | NA |
| 4-Chlorobutyronitrile | 84 | 84 |
| 4-Bromobutyronitrile | 38 | 39 |
| Chlorinated ethers | ||
| Bis(2-chloroethyl)ether | 2 | 2 |
| 2-Chloroethylmethylether | NA | NA |
The specific activity of crude extracts of E. coli GI724 (dhmA) was 0.67 mU/mg of protein.
The specific activity of crude extracts of M. avium MU1 was 0.60 mU/mg of protein.
NA, no activity detected.
Function of DhmA in M. avium.
The present study confirmed that the translation product of a dehalogenase-like gene of M. avium has dehalogenase activity. This is the first report of cloning and sequencing of a haloalkane dehalogenase gene and biochemical characterization of a haloalkane dehalogenase from a bacterium that colonizes animal tissues. To the best of our knowledge, all haloalkane dehalogenases described previously originated from bacteria isolated from localities contaminated by halogenated substances (12, 16, 22, 27-30, 40). The presence of haloalkane dehalogenase genes in the genomes of mycobacteria, including the strict pathogens M. tuberculosis and Mycobacterium bovis and the facultative pathogen M. avium, indicated that haloalkane dehalogenases could be involved in protection of mycobacteria against halogenated substances. Humans and other mammals generate halogenated compounds in response to heterogenous microorganisms (10, 37). The active halogen is produced by the myeloperoxidases present in the white blood cells, eosinophils, and neutrophils. Hypochlorous acid and hypobromous acid formed during the inflammation process can further halogenate some biological substrates (19, 39). However, a literature search of the chemical structures of these compounds revealed that they do not resemble usual substrates of haloalkane dehalogenases. Furthermore, identification of dehalogenase-like genes in the genomes of Mycobacterium smegmatis, Mesorhizobium loti, Xylella fastidiosa, Photobacterium profundum, and C. crescentus, which are neither strict pathogens nor colonizers of contaminated environments, provides additional evidence that mycobacterial haloalkane dehalogenases are not involved in dehalogenation reactions which detoxify the halogenated compounds produced by the immune system. We propose that the enzymes encoded by the dehalogenase-like genes are involved in some general biochemical pathway common to many bacterial species.
Acknowledgments
A.J. expresses her sincere thanks to Gerrit J. Poelarends and Dick B. Janssen (Groningen University, Groningen, The Netherlands) for their kind introduction to molecular biology techniques. Kamila Hynkova and Michaela Wimmerova (Masaryk University, Brno, Czech Republic) are gratefully acknowledged for help with the gas chromatography analyses and protein purification, Yuji Nagata (Tohoku University, Sendai, Japan) is acknowledged for critical reading of the manuscript, and Megha Mulchandani is acknowledged for linguistic revision of the manuscript.
This project was supported by the Czech Ministry of Education (grant LN00A016) and by the Czech Ministry of Agriculture (grant MZE-M03-99-01).
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Archelas, A., and R. Furstoss. 1998. Epoxide hydrolases: new tools for the synthesis of fine organic chemicals. Trends Biotechnol. 16:108-116. [DOI] [PubMed] [Google Scholar]
- 3.Attwood, T. K., M. E. Beck, D. R. Flower, P. Scordis, and J. Selley. 1998. The PRINTS protein fingerprint database in its fifth year. Nucleic Acids Res. 26:304-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bosma, T., E. Kruizinga, E. J. de Bruin, G. J. Poelarends, and D. B. Janssen. 1999. Utilization of trihalogenated propanes by Agrobacterium radiobacter AD1 through heterologous expression of the haloalkane dehalogenase from Rhodococcus sp. strain m15-3. Appl. Environ. Microbiol. 65:4575-4581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 6.Cole, S. T., R. Brosch, J. Parkhill, T. Garnier, C. Churcher, D. Harris, S. V. Gordon, K. Eiglmeier, S. Gas, C. E. Barry III, F. Tekaia, K. Badcock, D. Basham, D. Brown, T. Chillingworth, R. Connor, R. Davies, K. Devlin, T. Feltwell, S. Gentles, N. Hamlin, S. Holroyd, T. Hornsby, K. Jagels, A. Krogh, J. McLean, S. Moule, L. Murphy, K. Oliver, J. Osborne, M. A. Quail, M.-A. Rajandream, J. Rogers, S. Rutter, K. Seeger, J. Skelton, R. Squares, S. Squares, J. E. Sulston, K. Taylor, S. Whitehead, and B. G. Barrell. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393:537-544. [DOI] [PubMed] [Google Scholar]
- 7.Damborsky, J., and J. Koca. 1999. Analysis of the reaction mechanism and substrate specificity of haloalkane dehalogenases by sequential and structural comparisons. Protein Eng. 12:989-998. [DOI] [PubMed] [Google Scholar]
- 8.Damborsky, J., E. Rorije, A. Jesenska, Y. Nagata, G. Klopman, and W. J. G. M. Peijnenburg. 2001. Structure-specificity relationships for haloalkane dehalogenases. Environ. Toxicol. Chem. 20:2681-2689. [PubMed] [Google Scholar]
- 9.Gray, K. A., T. H. Richardson, K. Kretz, J. M. Short, F. Bartnek, R. Knowles, L. Kan, P. E. Swanson, and D. E. Robertson. 2001. Rapid evolution of reversible denaturation and elevated melting temperature in a microbial haloalkane dehalogenase. Adv. Synth. Catal. 343:607-617. [Google Scholar]
- 10.Gribble, G. W. 1994. The natural production of chlorinated compounds. Environ. Sci. Technol. 28:311-319. [DOI] [PubMed] [Google Scholar]
- 11.Iwasaki, I., S. Utsumi, and T. Ozawa. 1952. New colorimetric determination of chloride using mercuric thiocyanate and ferric ion. Bull. Chem. Soc. Jpn. 25:226. [Google Scholar]
- 12.Janssen, D. B., J. Gerritse, J. Brackman, C. Kalk, D. Jager, and B. Witholt. 1988. Purification and characterization of a bacterial dehalogenase with activity toward halogenated alkanes, alcohols and ethers. Eur. J. Biochem. 171:67-92. [DOI] [PubMed] [Google Scholar]
- 13.Janssen, D. B., F. Pries, J. Ploeg, B. Kazemier, P. Terpstra, and B. Witholt. 1989. Cloning of 1,2-dichloroethane degradation genes of Xanthobacter autotrophicus GJ10 and expression and sequencing of the dhlA gene. J. Bacteriol. 171:6791-6799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jesenská, A., I. Sedláček, and J. Damborský. 2000. Dehalogenation of haloalkanes by Mycobacterium tuberculosis H37Rv and other mycobacteria. Appl. Environ. Microbiol. 66:219-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kaneko, T., Y. Nakamura, S. Sato, E. Asamizu, T. Kato, S. Sasamoto, A. Watanabe, K. Idesawa, A. Ishikawa, K. Kawashima, T. Kimura, Y. Kishida, C. Kiyokawa, M. Kohara, M. Matsumoto, A. Matsuno, Y. Mochizuki, S. Nakayama, N. Nakazaki, S. Shimpo, M. Sugimoto, C. Takeuchi, M. Yamada, and S. Tabata. 2000. Complete genome structure of the nitrogen-fixing symbiotic bacterium Mesorhizobium loti. DNA Res. 7:331-338. [DOI] [PubMed] [Google Scholar]
- 16.Keuning, S., D. B. Janssen, and B. Witholt. 1985. Purification and characterization of hydrolytic haloalkane dehalogenase from Xanthobacter autotrophicus GJ10. J. Bacteriol. 163:635-639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kulakova, A. N., M. J. Larkin, and L. A. Kulakov. 1997. The plasmid-located haloalkane dehalogenase gene from Rhodococcus rhodochrous NCIMB 13064. Microbiology 143:109-115. [DOI] [PubMed] [Google Scholar]
- 18.MacFaddin, J. F. 1985. Media for isolation-cultivation-identification-maintenance of medical bacteria. Williams & Wilkins, Baltimore, Md.
- 19.Marcinkiewicz, J., A. Grabowska, and B. M. Chain. 1998. Modulation of antigen-specific T-cell activation in vitro by taurine chloramine. Immunology 94:325-330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marek, J., J. Vevodova, I. Kuta-Smatanova, Y. Nagata, L. A. Svensson, J. Newman, M. Takagi, and J. Damborsky. 2000. Crystal structure of the haloalkane dehalogenase from Sphingomonas paucimobilis UT26. Biochemistry 39:14082-14086. [DOI] [PubMed] [Google Scholar]
- 21.Marvanova, S., Y. Nagata, M. Wimmerova, J. Sykorova, K. Hynkova, and J. Damborsky. 2001. Biochemical characterization of broad-specificity enzymes using multivariate experimental design and a colorimetric microplate assay: characterization of the haloalkane dehalogenase mutants. J. Microbiol. Methods 44:149-157. [DOI] [PubMed] [Google Scholar]
- 22.Nagata, Y., K. Miyauchi, J. Damborsky, K. Manova, A. Ansorgova, and M. Takagi. 1997. Purification and characterization of haloalkane dehalogenase of a new substrate class from a γ-hexachlorocyclohexane-degrading bacterium, Sphingomonas paucimobilis UT26. Appl. Environ. Microbiol. 63:3707-3710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nagata, Y., T. Nariya, R. Ohtomo, M. Fukuda, K. Yano, and M. Takagi. 1993. Cloning and sequencing of a dehalogenase gene encoding an enzyme with hydrolase activity involved in the degradation of hexachlorocyclohexane in Pseudomonas paucimobilis. J. Bacteriol. 175:6403-6410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Newman, J., T. S. Peat, R. Richard, L. Kan, P. E. Swanson, J. A. Affholter, I. H. Holmes, J. F. Schindler, C. J. Unkefer, and T. C. Terwilliger. 1999. Haloalkane dehalogenase: structure of a Rhodococcus enzyme. Biochemistry 38:16105-16114. [DOI] [PubMed] [Google Scholar]
- 25.Nierman, W. C., T. V. Feldblyum, M. T. Laub, I. T. Paulsen, K. E. Nelson, J. Eisen, J. F. Heidelberg, M. R. Alley, N. Ohta, J. R. Maddock, I. Potocka, W. C. Nelson, A. Newton, C. Stephens, N. D. Phadke, B. Ely, R. T. DeBoy, R. J. Dodson, A. S. Durkin, M. L. Gwinn, D. H. Haft, J. F. Kolonay, J. Smit, M. B. Craven, H. Khouri, J. Shetty, K. Berry, T. Utterback, K. Tran, A. Wolf, J. Vamathevan, M. Ermolaeva, O. White, S. L. Salzberg, J. C. Venter, L. Shapiro, and C. M. Fraser. 2001. Complete genome sequence of Caulobacter crescentus. Proc. Natl. Acad. Sci. USA 98:4136-4141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Poelarends, G., M. Zandstra, T. Bosma, L. A. Kulakov, M. J. Larkin, J. R. Marchesi, A. J. Weightman, and D. B. Janssen. 2000. Haloalkane-utilizing Rhodococcus strains isolated from geographically distinct locations possess a highly conserved gene cluster encoding haloalkane catabolism. J. Bacteriol. 182:2725-2731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Poelarends, G. J., J. E. T. van Hylckama Vlieg, J. R. Marchesi, L. M. Freitas dos Santos, and D. B. Janssen. 1999. Degradation of 1,2-dibromoethane by Mycobacterium sp. strain GP1. J. Bacteriol. 181:2050-2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Poelarends, G. J., M. Wilkens, M. J. Larkin, J. D. van Elsas, and D. B. Janssen. 1998. Degradation of 1,3-dichloropropene by Pseudomonas cichorii 170. Appl. Environ. Microbiol. 64:2931-2936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sallis, P. J., S. J. Armfield, A. T. Bull, and D. J. Hardman. 1990. Isolation and characterization of a haloalkane halidohydrolase from Rhodococcus erythropolis Y2. J. Gen. Microbiol. 136:115-120. [DOI] [PubMed] [Google Scholar]
- 30.Scholtz, R., T. Leisinger, F. Suter, and A. M. Cook. 1987. Characterization of 1-chlorohexane halidohydrolase, a dehalogenase of wide substrate range from an Arthrobacter sp. J. Bacteriol. 169:5016-5021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Simpson, A. J., and The Xylella fastidiosa Consortium of the Organization for Nucleotide Sequencing and Analysis. 2000. The genome sequence of the plant pathogen Xylella fastidiosa. Nature 406:151-157. [DOI] [PubMed] [Google Scholar]
- 32.Stucki, G., and M. Thuer. 1995. Experiences of a large-scale application of 1,2-dichloroethane degrading microorganisms for groundwater treatment. Environ. Sci. Technol. 29:2339-2345. [DOI] [PubMed] [Google Scholar]
- 33.Swanson, P. E. 1999. Dehalogenases applied to industrial-scale biocatalysis. Curr. Opin. Biotechnol. 10:365-369. [DOI] [PubMed] [Google Scholar]
- 34.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Van den Wijngaard, A. J., K. W. H. J. Kamp, J. Ploeg, F. Pries, B. Kazemier, and D. Janssen. 1992. Degradation of 1,2-dichloroethane by Ancylobacter aquaticus and other facultative methylotrophs. Appl. Environ. Microbiol. 58:976-983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Verschueren, K. H. G., F. Seljee, H. J. Rozeboom, K. H. Kalk, and B. W. Dijkstra. 1993. Crystallographic analysis of the catalytic mechanism of haloalkane dehalogenase. Nature 363:693-698. [DOI] [PubMed] [Google Scholar]
- 37.Winterton, N. 2000. Chlorine: the only green element—towards a wider acceptance of its role in natural cycles. Green Chem. 2:173-225. [Google Scholar]
- 38.Wolinsky, E., and W. B. Schaefer. 1973. Proposed numbering scheme for mycobacterial serotypes by agglutination. Int. J. Syst. Bacteriol. 23:182-183. [Google Scholar]
- 39.Yazdanbakhsh, M., C. M. Eckmann, and D. Roos. 1987. Killing of schistosomula by taurine chloramine and taurine bromamine. Am. J. Trop. Med. Hyg. 37:106-110. [DOI] [PubMed] [Google Scholar]
- 40.Yokota, T., T. Omori, and T. Kodama. 1987. Purification and properties of haloalkane dehalogenase from Corynebacterium sp. strain m15-3. J. Bacteriol. 169:4049-4054. [DOI] [PMC free article] [PubMed] [Google Scholar]