Abstract
Trichoderma reesei strains were constructed for production of elevated amounts of endoglucanase II (EGII) with or without cellobiohydrolase I (CBHI). The endoglucanase activity produced by the EGII transformants correlated with the copy number of the egl2 expression cassette. One copy of the egl2 expression cassette in which the egl2 was under the cbh1 promoter increased production of endoglucanase activity 2.3-fold, and two copies increased production about 3-fold above that of the parent strain. When the enzyme with elevated EGII content was used, an improved stonewashing effect on denim fabric was achieved. A T. reesei strain producing high amounts of EGI and -II activities without CBHI and -II was constructed by replacing the cbh2 locus with the coding region of the egl2 gene in the EGI-overproducing CBHI-negative strain. Production of endoglucanase activity by the EG-transformant strain was increased fourfold above that of the host strain. The filter paper-degrading activity of the endoglucanase-overproducing strain was lowered to below detection, presumably because of the lack of cellobiohydrolases.
The filamentous fungus Trichoderma reesei is known as an efficient producer of cellulases. The cellulolytic system of T. reesei is composed of two cellobiohydrolases (CBHI and CBHII) and at least five endoglucanases (EGI, EGII, EGIII, EGIV, and EGV) (19, 20). Lack of EGII (originally called EGIII [18]) production reduces the endoglucanase activity in the culture supernatant by as much as 55%, whereas lack of EGI reduces it by only 25% (21). Thus, EGII is proposed to account for most of the endoglucanase activity produced by T. reesei (21). T. reesei EGI represents 5 to 10% of the secreted protein (16). The production of EGI has been improved in T. reesei by placing the egl1 gene under the control of the strong promoter of the Trichoderma CBHI (cbh1) gene and by increasing the copy number of the egl1 gene (8).
Cellulases are used widely in the textile industry in treatments of cellulose-containing textile materials during their manufacture and finishing (5). The most well-known application is the use of cellulases in biostoning. Biostoning of fabric means the use of cellulases in place of, or in addition to, the use of pumice stones for the treatment of denim fabric to impart a stonewashed effect. Heikinheimo et al. (7) showed that T. reesei-purified cellulase EGII was the most effective at removing color from denim, producing a good stonewashing effect with the lowest hydrolysis level. Endoglucanases are important also for degradation of β-glucan in feed. Degradation of β-glucan lowers the viscosity of the intestinal contents and this improves the quality of the feed (3).
In this study we have constructed T. reesei strains that produce elevated amounts of endoglucanase activity. The aim of our work was to construct different tailored high endoglucanase activity-producing strains for specific applications. We have improved the production of the EGII enzyme in T. reesei and we have constructed a T. reesei strain that produces high amounts of EGI and -II without any cellobiohydrolases. Cellulase preparations derived from these T. reesei overproduction strains were tested on the biostoning application.
MATERIALS AND METHODS
Microbial strains and plasmids.
Escherichia coli strain XL1-Blue (Stratagene) was used for propagation of plasmids. T. reesei strains VTT-D-79125 (2) and ALKO2698 (8) were used as recipients for transformations (Table 1). T. reesei VTT-D-79125 is a high cellulase activity-producing mutant strain that contains all the identified Trichoderma cellulases, including the main cellulases EGI, EGII, CBHI, and CBHII. ALKO2698 is an EGI-overproducing, cbh1-negative strain in which the cbh1 locus of VTT-D-79125 has been replaced by one copy of an egl1 expression cassette. In the expression cassette, egl1 is under the control of the cbh1 promoter. ALKO2697 (8), used for comparison, is an EGI-overproducing, CBHI-negative strain in which the cbh1 gene of VTT-D-79125 is replaced by two copies of the egl1 expression cassette (Table 1). A cellulase preparation derived from ALKO2656 (8) was used as a control in biostoning experiments. ALKO2656 is a high EGI activity-producing strain which contains three copies of egl1 in the place of cbh1 (Table 1).
TABLE 1.
T. reesei strains used as recipients for transformations and as comparison in cultivations
| Strain | cbh1 | egl1 expression cassette copy no. |
|---|---|---|
| High cellulase-producing mutant strain | ||
| VTT-D-79125 | + | 0 |
| EGI-overproducing strains | ||
| ALKO2698 | − | 1 |
| ALKO2697 | − | 2 |
| ALKO2656 | − | 3 |
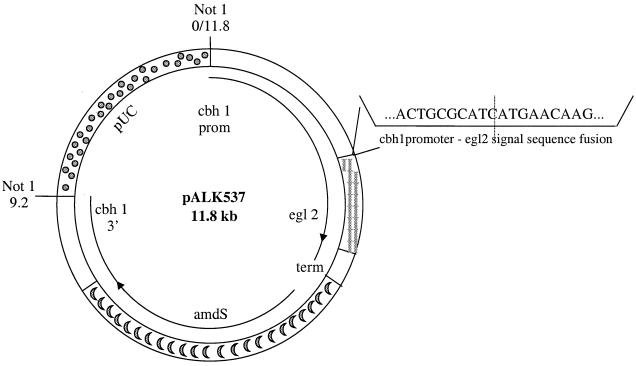
Plasmids were constructed by using pUC19 as a vector backbone, using standard recombinant DNA techniques. The pALK537 and pALK540 plasmids were constructed for EGII overproduction from the strong cbh1 promoter (Fig. 1 and 2). pALK537 and pALK540 can be used to target the expression cassette into the cbh1 and cbh2 loci, respectively, by homologous recombination. The precise fusion between the cbh1 promoter and egl2 cDNA was done with PCR. The SacII site in the cbh1 promoter was included in the 5′ primer, and the HpaI site of the egl2 cDNA (229 nucleotides downstream from the N terminus of the egl2 gene) was included in the 3′ primer. The fusion and the PCR fragment were sequenced to ensure that no mistakes had occurred in the PCR amplification. The plasmid pALK537 contains a 2.2-kb T. reesei cellobiohydrolase (cbh1) promoter and a 0.7-kb AvaII fragment of the cbh1 terminator region (starting 113 bp before the stop codon of cbh1). A 1.4-kb BamHI-EcoRI cbh1 3′ fragment was used together with the promoter to target the expression cassette to the cbh1 locus (21). A 3.1-kb SpeI-XbaI fragment containing the Aspergillus nidulans acetamidase gene from the plasmid p3SR2 (9) was used in the pALK537 plasmid as a marker.
FIG. 1.
Restriction map of the plasmid pALK537. The egl2 cDNA is exactly joined to the cbh1 promoter. A 9.2-kb NotI fragment was isolated from the plasmid for transformation.
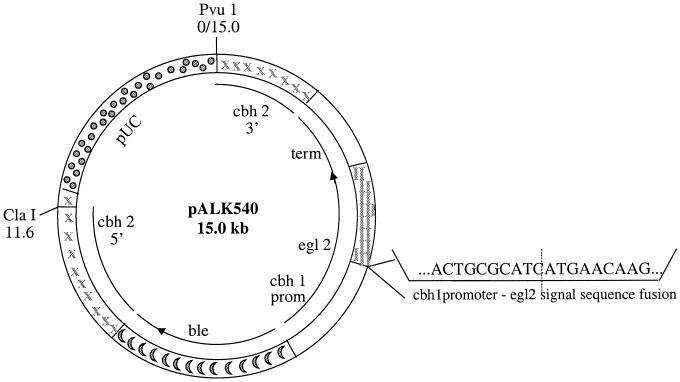
FIG. 2.
Restriction map of the plasmid pALK540. An 11.6-kb ClaI-PvuI fragment was isolated from the plasmid for transformation.
The plasmid pALK540 contains the cbh1 promoter and terminator and egl2 cDNA as in plasmid pALK537. Resistance to phleomycin was used for selecting the transformants. A 3.3-kb XbaI-BglII fragment containing the Streptoalloteichus hindustanus phleomycin gene (ble) from the plasmid pAN8-1 (14) was used in pALK540. In addition, pALK540 contains a 3.4-kb XhoI-PvuII cbh2 5′ fragment (starting 1.4 kb upstream from the cbh2 gene) and a 1.6-kb XbaI-BglII cbh2 3′ fragment (starting 1.1 kb downstream from the cbh2 gene). cbh2 5′ and 3′ fragments were used to target the egl2 expression cassette to the cbh2 locus of ALKO2698.
Inquires concerning the availability of the Trichoderma strains, plasmids, and antisera can be forwarded to Roal Oy, Rajamäki, Finland.
Growth of organisms.
E. coli strains were grown at 37°C overnight in L broth (13) supplemented with 50 μg of ampicillin/ml when needed. Potato dextrose (PD; Difco) agar slants were used for growing the Trichoderma strains. The plates and media for Trichoderma transformations with acetamide selection were essentially as those described by Penttilä et al. (16). MnR medium (per liter, 2.5 g of glucose, 2.5 g of yeast extract, 0.3 g of potassium phthalate, and 15 g of agar) was used in Trichoderma transformations with phleomycin selection. Liquid cultures of T. reesei were started from conidiospores grown on PD agar. A lactose-based complex medium was used for liquid cultivations (21). Cultivations were carried out at 30°C and 250 rpm for 7 days. Mycelia for isolation of the chromosomal DNA from the Trichoderma transformants were grown in shake flasks for 2 days (30°C, 250 rpm) on Trichoderma minimal medium (16) supplemented with 0.2% proteose peptone.
DNA techniques.
DNA manipulations were performed by standard techniques (13). Plasmid DNA from E. coli was isolated by using Qiagen columns (Qiagen GmbH) according to the supplier's instructions. DNA fragments for cloning or transformations were isolated from low-melting-point agarose gels (FMC Bioproducts) by the freeze-thaw phenol method (4). Chromosomal DNA was isolated from T. reesei by using the method of Raeder and Broda (17). For Southern blot analysis the DNA was transferred from agarose gels to nylon membranes by using a VacuGene XL apparatus (Pharmacia). The labeling of the probes with digoxigenin and the hybridization of the filters were performed according to the procedures of Boehringer Mannheim.
The PCRs were performed by using a Techne thermal cycler PHC-2 (Techne Ltd.) in 100-μl volumes. The reaction mixture contained a 0.2 mM concentration of each deoxynucleoside triphosphate (Pharmacia), 20 to 50 pmol of each primer, and 10 ng of plasmid template in 1× buffer supplied by Boehringer. The protocol used was the following: 96°C for 10 min before adding Taq DNA polymerase (2 U; Boehringer) and 100 μl of paraffin oil, denaturation at 95°C for 1 min, annealing at 60°C for 1 min, and extension at 72°C for 1 min for 30 cycles. The PCR fragments were purified by using a Mermaid kit (Bio 101 Inc.) according to the supplier's instructions. The ends of the fragments were filled by using DNA polymerase I Klenow fragment.
Sequencing of the fusion between the cbh1 promoter and egl2 cDNA was carried out by means of pUC/M13 and extension primers using a Taq DyeDeoxy Terminator cycle sequencing kit (Applied Biosystems) and an automated sequencer (model 373A; Applied Biosystems).
Transformation of Trichoderma.
Transformation of T. reesei was carried out by protoplast transformation as described by Penttilä et al. (16) with the modifications described by Karhunen et al. (8). In transformations where phleomycin was used as a selection marker, aliquots of the transformed protoplasts were plated onto the surface of MnR plates osmotically stabilized with 0.44 M sucrose and incubated for 6 h at 30°C prior to the addition of 5 ml of molten MnR (0.6% agar) as an overlay containing 300 μg of phleomycin (Cayla)/ml. The transformants were purified on selective MnR medium supplemented with 50 μg of phleomycin/ml through single spores before transfer to MnR slants containing 50 μg of phleomycin/ml for three generations and after that to PD slants. The acetamidase transformants were purified on selective acetamide-CsCl medium through single spores before transfer to PD slants.
Enzyme activity and protein assays.
The cellulase activities were measured from the culture supernatant as the release of reducing sugars from hydroxyethylcellulose (HEC; Fluka Chemie AG) using 2,4-dinitrosalicylic acid, as described by Bailey and Nevalainen (2) and from filter paper according to the method reported by Mandels et al. (12). Activity against barley β-glucan was measured the same way as activity against HEC, replacing HEC by barley β-glucan (Biocon Biochemicals Ltd.) in the assay. The β-glucosidase activity was measured using 4-nitrophenyl-β-d-glucopyranoside (Merck) as a substrate as described by Bailey and Nevalainen (2). Protein concentrations were determined from the trichloroacetic acid-precipitated T. reesei culture media by the method of Lowry et al. (11), using bovine serum albumin as the standard.
SDS-PAGE and immunological methods.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was performed according to the method of Laemmli (10). For Western blot analysis, purified transformants were grown in 96-well Millititer filtration plates (Millipore Corp.) at 30°C for 7 days. The presence of the CBHII protein was detected by SDS-PAGE followed by Western blotting (23) and immunostaining using monoclonal CII-8 antibody (1) and the ProtoBlot Western blotting AP system (Promega). Dot blot analysis was done with a Minifold Micro-Sample Filtration Manifold (Schleicher & Schull) according to the manufacturer's instructions. Visualization of the CBHI protein was done using the monoclonal mouse antibody CBHI MAb 89 (1) and immunostaining as described above. Quantitation of secreted EGI was carried out by a double-antibody sandwich enzyme-linked immunosorbent assay (ELISA) (6), using the monoclonal anti-EG antibody EI-2 (1) as capture antibody. For quantitation of CBHI and CBHII by ELISA, monoclonal antibodies CI-258 and CII-8, respectively, (1) were used as capture antibodies.
Biostoning.
The color of the desized denim fabric was measured as reflectance values with the Minolta Croma Meter 1000R using the L*a*b* system (illuminant D65) before and after enzyme treatments. In this system, L is the measure of black and white, a is the measure of red and green, and b is the measure of yellow and blue. Cellulase treatments were performed in an LP-2 Launder-Ometer (Atlas). A denim swatch of about 7 g was loaded into a 1.2-liter container that contained 200 ml of 0.05 M citrate buffer (pH 5), and 10 steel balls were added. Cellulase preparations from strains VTT-D-79125, ALKO3529, ALKO3528, and ALKO2656 were used for biostoning. Three and 6 mg of the total protein in cellulase preparations per g of fabric was used in each experiment for 1 and 2 h at 50°C. After cellulase treatment the swatches were soaked for 10 min in 0.01 M NaOH, rinsed with water, and dried.
RESULTS
EGII overproduction. (i) Transformation of T. reesei
For overexpression of the egl2 gene in T. reesei, the powerful promoter of the cbh1 gene of T. reesei was used. The plasmid pALK537 (Fig. 1) was constructed as described in Materials and Methods for expression of the egl2 gene under the control of the cbh1 promoter either in the place of cbh1 or elsewhere in the genome of T. reesei strain VTT-D-79125, depending on homologous or nonhomologous recombination.
For construction of EGII-overproducing CBHI-positive and CBHI-negative strains, the 9.2-kb NotI linear fragment of pALK537 containing the egl2 expression cassette (Fig. 1) was released from the vector backbone and transformed to T. reesei strain VTT-D-79125. The transformation frequency was 20 transformants per μg of DNA.
A total of 119 purified transformants were cultivated in shake flasks on cellulase-inducing medium, and the endoglucanase activity (activity against HEC) was measured from the culture medium of the transformants. The presence of CBHI protein in the culture medium was detected by dot blotting and immunostaining from the 23 best endoglucanase producers. Of these transformants, 61% proved to be CBHI negative, which indicates that in these transformants the expression cassette had replaced the cbh1 gene.
(ii) DNA analysis of transformants.
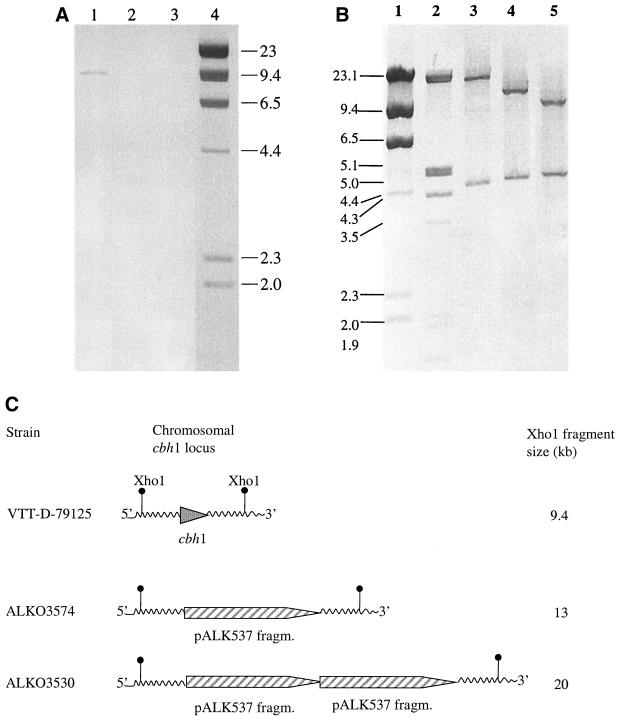
The transformants producing the best endoglucanase activity, ALKO3529 (CBHI positive) and ALKO3530 (CBHI negative), and a transformant strain, ALKO3574, thought to contain one copy of the egl2 expression cassette, were analyzed by Southern blotting to evaluate the copy number of egl2 and the integration of the expression cassettes into the genome.
The ALKO3530 and ALKO3574 strains that did not secrete CBHI according to dot blot analysis were shown by Southern blotting to lack the chromosomal cbh1 gene. The transformants showed no hybridization when the coding region of the cbh1 gene was used as a probe (Fig. 3A). The integration of the expression cassette in these transformants was further studied by Southern blotting using the 9.2-kb NotI fragment of the plasmid pALK537 as a probe. According to the blot analysis (Fig. 3B and C), one copy of the transformed pALK537 fragment replaced the coding region of the cbh1 gene in the ALKO3574 strain, generating a XhoI fragment of about 13 kb. The ALKO3530 strain had two vector fragments (a XhoI fragment of about 20 kb) replacing the cbh1 locus. The 4.7-kb band present in the transformants and VTT-D-79125 is from the wild-type egl2 locus. The 9.4-kb band present in VTT-D-79125 is from the wild-type cbh1 locus. The results were confirmed by hybridizing the chromosomal DNA digested with different restriction enzymes with egl2 and amdS probes (data not shown).
FIG. 3.
Southern analysis of transformants ALKO3530 and ALKO3574 in the which cbh1 locus has been replaced with the 9.2-kb NotI fragment from pALK537. (A) Genomic DNA was digested with XhoI. Hybridization was done with a cbh1 probe. Lane 1, VTT-D-79125; lane 2, ALKO3530; lane 3, ALKO3574; lane 4, molecular weight marker λHindIII. (B) Genomic DNA was digested with XhoI. Hybridization was performed with the 9.2-kb NotI fragment used for the transformations. Lanes 1 and 2, molecular weight markers λHindIII and λEcoRI-HindIII; lane 3, ALKO3530; lane 4, ALKO3574; lane 5, VTT-D-79125. (C) Schematic presentation of the organization of the cbh1 chromosomal locus in the host strain and the transformants, showing the XhoI cleavage site.
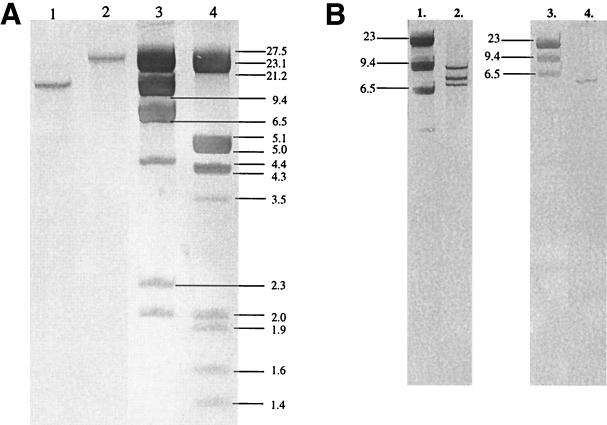
Probing of the XhoI-digested genomic DNA of the CBHI-positive ALKO3529 strain with the cbh1 probe resulted in one band of more than 20 kb while the host strain VTT-79125 gave a 9.4-kb band (Fig. 4A). Thus, the coding region of the cbh1 gene is still present in the ALKO3529 strain. This result suggests that the pALK537 expression cassette has integrated at the cbh1 locus or close to the cbh1 locus, because the band recognized by the cbh1 probe has increased in size. Chromosomal DNA of ALKO3529 was further digested with PvuI and probed with egl2. Since the pALK537 fragment contains one PvuI site (in the amdS gene), a tandem copy of the pALK537 fragment in the genome would generate a band of 9.2 kb and one band of unknown size. One copy of the pALK537 fragment would generate one band of unknown size. In Southern hybridization with egl2, ALKO3529 gave bands of about 7, 8, and 9 kb (Fig. 4B). The 6- to 7-kb band present in ALKO3529 and in VTT-D-79125 is from the wild-type egl2 locus. Thus, it can be concluded that ALKO3529 contains two copies of the transformed vector fragment integrated into the cbh1 locus or close to the cbh1 locus. The results were confirmed by hybridizing the chromosomal DNA digested with different restriction enzymes with amdS and pALK537 fragment probes (data not shown).
FIG. 4.
Southern analysis of the transformant ALKO3529 and the host strain VTT-D-79125. (A) Genomic DNA was digested with XhoI. Hybridization was done with a cbh1 probe. Lane 1, VTT-D-79125; lane 2, ALKO3529; lanes 3 and 4, molecular weight markers λHindIII and λEcoRI-HindIII. (B) Genomic DNA was digested with PvuI. Hybridization was done with an egl2 probe. Lane 1, molecular weight marker λHindIII; lane 2, ALKO3529; lane 3, molecular weight marker λHindIII; lane 4, VTT-D-79125.
(iii) Enzyme production of ALKO3529, ALKO3530 and ALKO3574 transformant strains.
The EGII transformant strains ALKO3529, ALKO3530, and ALKO3574 as well as the parent strain VTT-D-79125 were grown in shake flasks on cellulase-inducing medium. For comparison, the CBHI-negative EGI-overproducing strains ALKO2698, ALKO2697, and ALKO2656 containing one, two, and three egl1 expression cassettes, respectively (8) were also grown in the same cultivations. The results of the measurement of different cellulase activities and ELISA analyses from the culture medium are shown in Table 2.
TABLE 2.
Production of cellulases by the host strain VTT-D-79125, transformants ALKO3529, ALKO3574, and ALKO3530, and T. reesei EGI-overproducing strains ALKO2697, ALKO2698, and ALKO2656a
| Strain | cbh1 Southern analysis result | egl2 cassette copy no. | Secreted protein (mg/ml) | Endoglucanase (HEC) (nkat/ml) | β-glucanase (nkat/ml) | FPU/ml | β-glucosidase (nkat/ml) | CBHI (mg/ml) | CBHII (mg/ml) | EGI (mg/ml) |
|---|---|---|---|---|---|---|---|---|---|---|
| VTT-D-79125 | + | 0 | 9.2 | 1,200 ± 0 | 9,300 ± 500 | 5.3 ± 0.1 | 211 ± 10 | 3.7 | 0.051 | 0.361 |
| ALKO3529 | + | 2 | 10.1 | 3,400 ± 100 | 23,000 ± 2,000 | 6.0 ± 0.7 | 239 ± 5 | 3.4 | 0.069 | 0.587 |
| ALKO3530 | − | 2 | 8.7 | 3,600 ± 100 | 25,000 ± 1,000 | 2.1 ± 0.2 | 320 ± 5 | NAb | 0.085 | 0.590 |
| ALKO3574 | − | 1 | 7.4 | 2,800 ± 50 | 19,400 ± 1,000 | 1.7 ± 0.2 | 281 ± 12 | NA | NA | 0.650 |
| ALKO2697 | − | 0 | 9.2 | 2,600 ± 100 | 20,200 ± 500 | 1.8 ± 0.1 | NA | NA | NA | NA |
| ALKO2698 | − | 0 | 7.6 | 2,300 ± 50 | 15,700 ± 500 | 1.6 ± 0 | 200 ± 5 | NA | NA | NA |
| ALKO2656 | − | 0 | 9.1 | 2,700 ± 500 | 20,200 ± 3,000 | 1.4 ± 0.4 | 231 ± 5 | NA | NA | NA |
Strains were grown for 7 days in cellulase-inducing medium; the results are the average from three flasks. The standard errors are shown.
NA, not analyzed.
The endoglucanase (activity against HEC) and β-glucanase activities produced by the studied EGII transformants correlate to the copy number of the egl2 expression cassette. One copy of the egl2 expression cassette increased the endoglucanase activity 2.3-fold (ALKO3574), and two cassettes (ALKO3529, ALKO3530) increased the activity about 3-fold compared to the VTT-D-79125 strain (Table 2). The β-glucan-hydrolyzing activity was 2.1 times higher in ALKO3574 and 2.5 to 2.7 times higher in ALKO3529 and ALKO3530 than in the VTT-D-79125 host strain. Higher increases in both endoglucanase and β-glucanase activities could be detected in the EGII-overproducing strains compared to EGI-overproducing strains. One additional copy of the egl1 gene expressed under the cbh1 promoter increased the endoglucanase activity by 1.9-fold (ALKO2698), and two copies increased it by 2.2-fold (ALKO2697) (Table 2). The same effect can be seen with β-glucanase: higher β-glucanase activity could be obtained with an additional copy of egl2 (2.1 times) than with an additional copy of egl1 (1.7 times).
The filter paper-hydrolyzing activity (FPU), which is mainly affected by cellobiohydrolases, was decreased 60% in ALKO3530 (two egl2 expression cassettes) and 67% in ALKO3574 (one egl2 cassette), which lack the cbh1 gene. FPU activity of the CBHI-positive ALKO3529 strain was about 10% higher than in the VTT-D-79125 parent strain.
Production of β-glucosidase activity was not significantly changed in transformants ALKO3529 and ALKO3574 compared to the VTT-D-79125 parent strain. In ALKO3530 production of β-glucosidase was 1.5-fold higher than in the host strain.
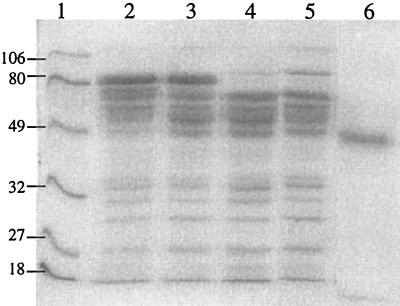
The amount of the secreted EGII protein was roughly evaluated by eye in several SDS-PAGE analyses with a known concentration of purified EGII protein as a standard (Fig. 5). Production of EGII in ALKO3529 and ALKO3530 was about 1.3 mg/ml, in ALKO3574 it was about 0.8 mg/ml, and in VTT-D-79125 it was about 0.4 mg/ml. Thus, one egl2 expression cassette increases the amount of EGII protein by 2-fold and two cassettes increases it up to about 3.2-fold.
FIG. 5.
SDS-PAGE of the samples from the culture supernatants of host strain VTT-D-79125 and the EGII transformants ALKO3529, ALKO3530, and ALKO3574. A total of 20 μg of total secreted protein was loaded in each lane. Lane 1, VTT-D-79125; lane 2, ALKO3529; lane 3, ALKO3530; lane 4, ALKO3574; lane 5, 3 μg of purified EGII protein.
The amounts of CBHI, CBHII, and EGI were analyzed by ELISA from the same culture supernatants from which enzyme activities were analyzed (Table 2). The amount of secreted CBHI was almost the same in the CBHI-positive EGII overproducer ALKO3529 (3.4 mg/ml) as in the VTT-D-79125 parental strain (3.7 mg/ml). Surprisingly, there was an increase in the production of EGI: 1.6-fold by the ALKO3529 and ALKO3530 strains and 1.8-fold by the ALKO3574 strain. The production of CBHII was enhanced in the ALKO3529 and ALKO3530 strains. The lack of cbh1 seemed to increase the amount of CBHII more, resulting in 1.7-fold more in ALKO3530 and 1.3-fold more in ALKO3529.
EGI and -II overproduction without CBHI and -II in T. reesei. (i) Transformation of T. reesei and replacement of cbh2.
The plasmid pALK540 (Fig. 2) was constructed as described in Materials and Methods for replacement of the cbh2 locus of T. reesei ALKO2698 (EGI overproducer, CBHI negative) with the egl2 expression cassette.
For construction of the strain overproducing EGI and EGII without CBHI and CBHII, the 11.6-kb ClaI-PvuI linear fragment of pALK540 containing the egl2 expression cassette was released from the vector backbone and transformed to T. reesei strain ALKO2698 (Fig. 2). The transformation frequency varied from 9 to 42 transformants per μg of DNA. The purified transformants were grown on microtiter plates for detection of the CBHII protein by Western blotting and immunostaining. Twenty-two out of 31 tested transformants were CBHII negative, suggesting that the frequency for targeting of the expression cassette into the cbh2 locus was 71%. The CBHII-negative transformants were grown in shake flask cultivations on cellulase-inducing medium to measure the endoglucanase activity in the culture medium. Strain ALKO3528 produced the highest endoglucanase activity.
(ii) DNA analysis of strain ALKO3528.
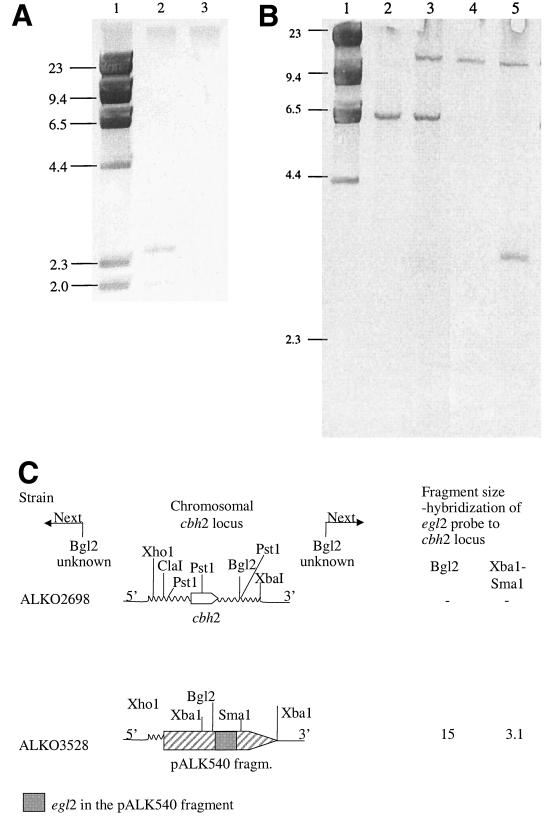
The absence of the chromosomal cbh2 gene from strain ALKO3528 was shown by Southern blot analysis. No hybridization to the chromosomal DNA of ALKO3528 was obtained when probed with the coding region of the cbh2 gene (Fig. 6A). The 2.4- and 2.0-kb bands present in PstI-digested chromosomal DNA of the ALKO2698 host strain are from the wild-type cbh2 locus.
FIG. 6.
Southern analysis of the transformant ALKO3528 and the host strain ALKO2698. (A) Genomic DNA was digested with PstI. Hybridization was done with a cbh2 probe. Lane 1, molecular weight marker λHindIII; lane 2, ALKO2698; lane 3, ALKO3528. (B) Genomic DNA was digested with BglII or XbaI-SmaI. Hybridization was done with an egl2 probe. Lane 1, molecular weight marker λHindIII; lane 2, ALKO2698 digested with BglII; lane 3, ALKO3528 digested with BglII; lane 4, ALKO2698 digested with XbaI-SmaI; lane 5, ALKO3528 digested with XbaI-SmaI. (C) Schematic presentation of the organization of the cbh2 locus in the host strain and the ALKO3528 transformant strain showing different cleavage sites and the fragment sizes from the cbh2 locus when probing with the egl2 probe.
The integration of the expression cassette in ALKO3528 was analyzed further by hybridization of the genomic DNA digested with appropriate restriction enzymes to egl2 and phleomycin probes as well as to the 11.6-kb transformation fragment of the plasmid pALK540. The hybridization patterns with the egl2 probe are shown in Fig. 6B and C. Probing of the BglII-digested genomic DNA of strain ALKO3528 resulted in two bands of 6.6 and about 15 kb. The 6.6-kb band present in BglII-digested chromosomal DNA of the ALKO3528 transformant and ALKO2698 host strain is from the wild-type egl2 locus. The 15-kb band indicates that one copy of a transforming vector had integrated to the genome of strain ALKO3528. Hybridization of the XbaI-SmaI-digested chromosomal DNA of ALKO3528 with the egl2 probe gave bands of 9.2 and 3.1 kb. The 9.2-kb band is from the wild-type egl2 locus and this hybridization was seen also to ALKO2698 DNA. Hybridization of the egl2 gene to the 3.1-kb band is an indication of an intact cbh1 promoter-egl2 fusion. Thus, strain ALKO3528 contains one full-length copy of the egl2 expression cassette in the cbh2 locus.
(iii) Enzyme production of the ALKO3528 transformant strain and the host strains.
EG-overproducing strain ALKO3528 (CBHI and -II negative) and the parent strains ALKO2698 (EGI overproducer) and VTT-D-79125 (parent for ALKO2698) were grown on cellulase-inducing medium for measurement of cellulase activities (Table 3). The endoglucanase activity (measured against HEC) was increased about twofold in strain ALKO3528 above that in the parent strain ALKO2698 and by fourfold above that in VTT-D-79125. The production of β-glucanase activity in ALKO3528 was increased 1.8-fold above that in strain ALKO2698. The filter paper-hydrolyzing activity of strain ALKO3528 was lowered to almost zero because of the lack of the CBHI and CBHII proteins.
TABLE 3.
Production of cellulases by the host strains VTT-D-79125 and ALKO2698 and by the transformant ALKO3528
| Strain | Secreted protein (mg/ml) | Endoglucanase (HEC) (nkat/ml) | β-glucanase (nkat/ml) | FPU/ml | Southern analysis
|
|
|---|---|---|---|---|---|---|
| cbh1 | cbh2 | |||||
| VTT-D-79125 | 9.0 | 1,300 ± 50 | 11,500 ± 1,000 | 4.8 ± 0.8 | + | + |
| ALKO2698 | 7.9 | 2,600 ± 100 | 15,300 ± 1,000 | 1.4 ± 0.1 | − | + |
| ALKO3528 | 8.5 | 5,100 ± 400 | 27,800 ± 2,000 | 0.2 ± 0.1 | − | − |
Use of endoglucanase preparations in biostoning.
Cellulase preparations derived from the endoglucanase-overproducing T. reesei strains ALKO3529 and ALKO3528 were used to impart a stonewashed appearance to denims. The parent strain VTT-D-79125 and the EGI-overproducing strain ALKO2656 were used as controls. Results from the color measurements are shown in Table 4.
TABLE 4.
Color measurements of denim fabrics treated with VTT-D-79125, ALKO2656, ALKO3529, and ALKO3528 cellulase preparationsa
| Preparation and strain | Dosage (mg of total protein/g of fabric) | L, right side of the swatch |
|---|---|---|
| 1 h | ||
| ALKO3529 | 3 | 1.0 |
| 6 | 2.1 | |
| ALKO3528 | 3 | 1.3 |
| 6 | 1.5 | |
| VTT-D-79125 | 3 | 1.5 |
| 6 | 1.5 | |
| ALKO2656 | 3 | 0.1 |
| 2 h | ||
| ALKO3529 | 3 | 3.2 |
| ALKO3528 | 3 | 2.7 |
| VTT-D-79125 | 3 | 1.8 |
| 6 | 2.4 | |
| ALKO2656 | 3 | 2.5 |
The fabrics were treated for 1 and 2 h with doses of 3 and 6 mg of total protein, as explained in Materials and Methods. The results are the average of two parallel treatments. L, lightness unit of the fabric after treatment minus lightness unit of the fabric before the treatment.
Results show that after 1 h of treatment with a 3-mg/g dosage, the stonewashing effects (measured in lightness units) with the EGII-overproducing strain ALKO3529 and the EG-overproducing strain ALKO3528 were almost equal with that of VTT-D-79125. No clear increase in lightness units was obtained with the EGI-overproducing strain ALKO2656. With a 6-mg/g dosage, after 1 h of treatment ALKO3529 showed the highest increase in lightness units, compared to VTT-D-79125 or ALKO3528. After 2 h of treatment with the 3-mg/g dosage, the best stonewashing effect (measured as lightness) was obtained with the ALKO3529 preparation. ALKO3528 was slightly better than ALKO2656. A considerably higher dosage of cellulases from VTT-D-79125 was needed to achieve a comparable stonewashing effect as with the cellulases of the endoglucanase-producing strains.
DISCUSSION
The production of endoglucanase enzymes has been improved in the biotechnically important filamentous fungus T. reesei. By using the strong Trichoderma cbh1 promoter and by adding copy numbers of egl2, high EGII activity-producing strains were obtained. One additional copy of the egl2 gene expressed under the cbh1 promoter in the cbh1 locus increased the endoglucanase activity (activity against HEC) by 2.3-fold, while one additional copy of the egl1 gene expressed under the cbh1 promoter in the cbh1 locus increased the endoglucanase activity by 1.9-fold above that of the parent strain. The same effect could be observed by increasing the copy numbers further: two copies of egl2 increased the endoglucanase activity by 3-fold, while two copies of egl1 increased the endoglucanase activity by 2.2-fold. Thus, it can be concluded that EGII has a major impact on the endoglucanase activity measured as activity against HEC. This is in agreement with the results obtained with cellulase-deletion strains of T. reesei (21). This is also consistent with the specific activities on HEC and on β-glucan: the specific activities of EGII are higher than those of EGI (22). The integration place of the expression cassette had no effect on the endoglucanase activity levels of the two-copy transformants: in ALKO3530 the expression cassettes had replaced the cbh1 locus and in ALKO3529 they had integrated close to the cbh1 locus. EGII also had an effect on the filter paper activity, increasing it by about 10% in the EGII transformant (cbh1 gene present) above that of the parent strain.
By replacing the cbh1 locus with one copy of the egl1 gene under the cbh1 promoter (8) and by replacing the cbh2 locus with one copy of the egl2 gene under the cbh1 promoter, we have been able to construct a T. reesei strain that produces high amounts of pure EGI and -II without any contamination by CBHI or -II. In ALKO3528, the production of endoglucanase activity was increased fourfold above that of the VTT-D-79125 parent strain.
Cellulase preparations derived from the high EGII activity-producing strain ALKO3529 proved to improve the stonewashing effect above that of its parent strain VTT-D-79125 when the same enzyme dosage was used. The same stonewashing effect could be obtained with a considerably lower enzyme dosage when using the EGII cellulase preparation derived from the EGII-overproducing strain than when using the parental strain. Heikinheimo et al. (7) have shown that purified EGII is the most effective of the main cellulases at removing color from denim fabric. Thus, by increasing the relative amount of EGII in the cellulase mixture, an improved stonewashing effect can be obtained. Cellulase enzymes are used in the textile industry for biostoning and also for finishing of cellulosic fibers. Cellulase preparations produced by strains ALKO3529 and ALKO3530 have been tested in cotton finishing (15). The cellulase mixture obtained with the EGII-overproducing strain ALKO3529 proved to reduce pilling with low strength and weight losses on cotton knit fabric. The cellulase preparation of strain ALKO3530 resulted in improved depilling, but at the same time caused relatively high weight loss.
β-Glucanase is an important activity in the degradation of β-glucan in feed. The β-glucanase activity was improved in the EG-overproducing strains. In addition to textile applications, these new preparations can possibly be used for more economical production of β-glucanase for modification of feed.
Acknowledgments
We thank Rolf Bühler for ELISA measurements and Outi Könönen for skillful technical assistance. John Londesborough, Marja Paloheimo, and Jari Vehmaanperä are thanked for valuable comments on the manuscript.
REFERENCES
- 1.Aho, S., V. Olkkonen, T. Jalava, M. Paloheimo, R. Buhler, M.-L. Niku-Paavola, D. Bamford, and M. Korhola. 1991. Monoclonal antibodies against core and cellulose-binding domains of Trichoderma reesei cellobiohydrolases I and II and endoglucanase I. Eur. J. Biochem. 200:643-649. [DOI] [PubMed] [Google Scholar]
- 2.Bailey, M., and H. Nevalainen. 1981. Induction, isolation and testing of stable Trichoderma reesei mutants with improved production of solubilizing cellulase. Enzyme Microb. Technol. 3:153-157. [Google Scholar]
- 3.Bedford, M. 1995. Mechanism of action and potential environmental benefits from the use of feed enzymes. Animal Feed Sci. Technol. 53:145-155. [Google Scholar]
- 4.Benson, S. A. 1984. A rapid procedure for isolation of DNA fragments from agarose gels. Bio/Techniques 2:66-68. [Google Scholar]
- 5.Buchert, J., and L. Heikinheimo. 1998. New cellulase processes for the textile industry. Carbohydr. Europe 22:32-34. [Google Scholar]
- 6.Buhler, R. 1991. Double-antibody sandwich enzyme-linked immunosorbent assay for quantitation of endoglucanase I of Trichoderma reesei. Appl. Environ. Microbiol. 57:3317-3321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Heikinheimo, L., J. Buchert, A. Miettinen-Oinonen, and P. Suominen. 2000. Treating denim fabrics with Trichoderma reesei cellulases. Textile Res. J. 70:969-973. [Google Scholar]
- 8.Karhunen, T., A. Mäntylä, H. Nevalainen, and P. Suominen. 1993. High frequency one-step gene replacement in Trichoderma reesei I. Endoglucanase I overproduction. Mol. Gen. Genet. 241:515-522. [DOI] [PubMed] [Google Scholar]
- 9.Kelly, J. M., and M. J. Hynes. 1985. Transformation of Aspergillus niger by the amdS gene of Aspergillus nidulans. EMBO J. 4:475-479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 11.Lowry, O. H., N. J. Rosebrough, A. L. Farr, and R. J. Randall. 1951. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 193:265. [PubMed] [Google Scholar]
- 12.Mandels, M., R. Andreotti, and C. Roche. 1976. Measurement of saccharifying cellulase, p. 21-33. In E. L. Gaden, M. H. Mandels, E. T. Reese, and L. A. Spano (ed.), Bio/Technology and bioengineering symposium no. 6. John Wiley and Sons, New York, N.Y. [PubMed]
- 13.Maniatis, T., E. Fritsch, and J. Sambrook. 1982. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 14.Mattern, J. E., P. J. Punt, and C. A. M. J. van den Hondel. 1987. A vector of Aspergillus transformation conferring phleomycin resistance. Fungal Genet. Newslett. 35:25. [Google Scholar]
- 15.Miettinen-Oinonen, A., L. Heikinheimo, J. Buchert, J. Morgado, L. Almeida, P. Ojapalo, and A. Cavaco-Paulo. 2001. The role of Trichoderma reesei cellulases in cotton finishing. AATCC Rev. 1:33-35. [Google Scholar]
- 16.Penttilä, M., H. Nevalainen, M. Rättö, E. Salminen, and J. Knowles. 1987. A versatile transformation system for cellulolytic filamentous fungus Trichoderma reesei. Gene 61:155-164. [DOI] [PubMed] [Google Scholar]
- 17.Raeder, U., and P. Broda. 1985. Rapid preparation of DNA from filamentous fungi. Lett. Appl. Microbiol 1:17-20. [Google Scholar]
- 18.Saloheimo, M., P. Lehtovaara, M. Penttilä, T. T. Teeri, J. Ståhlberg, G. Johansson, G. Petterson, M. Claeyssens, P. Tomme, and J. Knowles. 1988. EGIII, a new endoglucanase from Trichoderma reesei: the characterization of both gene and enzyme. Gene 63:11-21. [DOI] [PubMed] [Google Scholar]
- 19.Saloheimo, M., T. Nakari-Setälä, M. Tenkanen, and M. Penttilä. 1997. cDNA cloning of a Trichoderma reesei cellulase and demonstration of endoglucanase activity by expression in yeast. Eur. J. Biochem. 249:584-591. [DOI] [PubMed] [Google Scholar]
- 20.Srisodsuk, M. 1994. Mode of action of Trichoderma reesei cellobiohydrolase I on crystalline cellulose. VTT Publications, Espoo, Finland.
- 21.Suominen, P., A. Mäntylä, T. Karhunen, S. Hakola, and H. Nevalainen. 1993. High frequency one-step gene replacement in Trichoderma reesei. II. Effects of deletions of individual cellulase genes. Mol. Gen. Genet. 241:523-530. [DOI] [PubMed] [Google Scholar]
- 22.Suurnäkki, A., M. Tenkanen, M. Siika-aho, M.-L. Niku-Paavola, L. Viikari, and J. Buchert. 2000. Trichoderma reesei cellulases and their core domains in the hydrolysis and modification of chemical pulp. Cellulose 7:189-209. [Google Scholar]
- 23.Towbin, H., T. Staehelin, and J. Gordon. 1979. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc. Natl. Acad. Sci. USA 76:4350-4354. [DOI] [PMC free article] [PubMed] [Google Scholar]