Abstract
A long-standing model postulates that X-chromosome dosage compensation in Drosophila occurs by twofold up-regulation of the single male X, but previous data cannot exclude an alternative model, in which male autosomes are down-regulated to balance gene expression. To distinguish between the two models, we used RNA interference to deplete Male-Specific Lethal (MSL) complexes from male-like tissue culture cells. We found that expression of many genes from the X chromosome decreased, while expression from the autosomes was largely unchanged. We conclude that the primary role of the MSL complex is to up-regulate the male X chromosome.
Keywords: X-chromosome dosage compensation, MSL complex
X-chromosome dosage compensation was first postulated by H.J. Muller in 1932 to explain the difference in phenotype between male and female Drosophila carrying a single copy of a partially defective X-linked white gene (Muller 1932). Since that time, it has become clear that dosage compensation is an essential, sex-specific process that has evolved to equalize X-linked gene expression in male and female (or hermaphrodite) flies, nematodes, and mammals. Specific modes of regulation differ among these model systems. However, in each organism, dosage compensation is controlled by genes with sex-specific lethal phenotypes that encode proteins and RNAs localized along the length of the regulated X chromosome (for review, see Cline and Meyer 1996; Meller and Kuroda 2002; Nusinow and Panning 2005). In flies, the MSL (Male-Specific Lethal) complex is postulated to up-regulate X-linked gene expression in males to equal that in females, and is composed of at least five proteins (collectively called the MSL proteins) and two noncoding roX (RNA on X) RNAs. MSL1 and MSL2 are thought to form the essential core of the complex, and MSL3 (a chromodomain protein), MOF (a MYST histone acetyltransferase), MLE (a helicase), and either roX1 or roX2 noncoding RNAs are all required for normal targeting of the male X chromosome.
The model for dosage compensation in Drosophila by hypertranscription of the male X predates the discovery of the MSL complex and was based on incorporation of 3H-uridine into nascent RNA over the polytene X and autosomes in male and female nuclei (Mukherjee and Beermann 1965). Consistent with the hypertranscription model, the male polytene X exhibits a diffuse morphology generally associated with increased gene expression that is lost in dosage compensation mutants (Belote and Lucchesi 1980a). However, analyses of selected genes in msl males have revealed a lack of consensus regarding how gene expression changes in these mutants (Breen and Lucchesi 1986; Hiebert and Birchler 1994; Bhadra et al. 1999; Chiang and Kurnit 2003; Pal Bhadra et al. 2005). Notably, the conclusion of Birchler and colleagues is that individual autosomal genes increase more often than X-linked genes decrease expression in msl mutants (Hiebert and Birchler 1994; Bhadra et al. 1999; Pal Bhadra et al. 2005). One explanation might be that msl mutants exhibit both direct and indirect effects from failure to establish compensation of a large segment (∼16%) of their genome over many days of development (Chiang and Kurnit 2003). Alternatively, the action of the MSL complex may be more complicated than proposed based on its X chromosomal localization. The alternative, inverse dosage model is that all chromosomes tend to be up-regulated in male nuclei, so that autosomal gene expression in males must be down-regulated to provide balanced gene expression (Birchler et al. 2003). In this model, the primary role of the MSL complex is to sequester positive factors away from the autosomes.
The hypertranscription and inverse models make very different predictions regarding the effect on X and autosomal gene expression when the MSL complex is removed from male cells. However, previous studies have faced significant technical challenges. First, the expression changes to be measured are quite small (twofold or less). Second, analysis of msl mutant males is complicated by the fact that they are developmentally delayed and dying. Third, a limited set of individual genes was monitored, so global trends could not be thoroughly assessed. Here, we have directed our efforts to eliminating, as much as possible, indirect or cumulative effects of defective dosage compensation by utilizing RNA interference (RNAi) in male tissue culture cells to uncouple loss of MSL function from perturbations in development. Furthermore, we assay global gene expression changes on microarrays, in which overall trends of even small changes in X and autosomal gene expression, if they exist, should be readily apparent.
Results and Discussion
msl2 RNAi reduces msl2 RNA and protein expression, removing MSL complexes and histone H4K16ac modification from the X
We chose MSL2 as our target for RNAi because it is the key limiting factor in the MSL complex. MSL2 protein is expressed in males but is normally repressed in females by the female-specific factor Sex Lethal (SXL) (Bashaw and Baker 1995; Kelley et al. 1995; Zhou et al. 1995). Since females lack MSL2, they do not form MSL complexes. However, when MSL2 is ectopically expressed in females, full complexes form on both X chromosomes, leading to delayed development and substantial lethality (Kelley et al. 1995). Therefore, the absence of MSL2 is normally sufficient to completely block dosage compensation by the MSL complex.
We performed our assays in SL2 tissue culture cells, which display potent RNAi activity (Clemens et al. 2000; Kiger et al. 2003). SXL is absent in SL2 cells (Ryner and Baker 1991), while all of the known MSL protein components are expressed, form complexes, and are localized to the X chromosome, strongly suggesting a male identity (Copps et al. 1998; A. Alekseyenko and M.I. Kuroda, unpubl.).
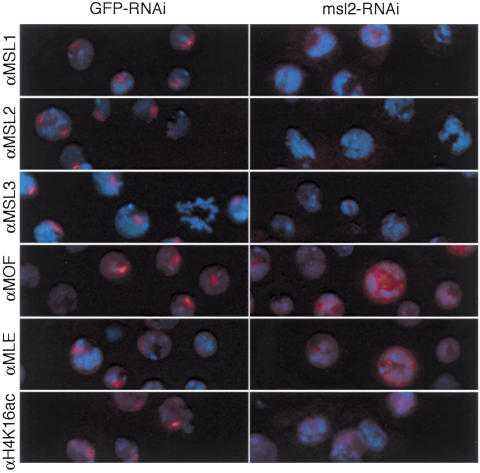
RNAi for msl2 was previously demonstrated in SL2 cells by Buscaino et al. (2003). Using a similar approach, we first assayed the effect of msl2 RNAi, or RNAi for an irrelevant gene (GFP) on the localization of MSL complexes to a nuclear subdomain presumed to be the X chromosome. After 4 d we found that RNAi against msl2 largely eliminated the localization of MSL complexes (Fig. 1). More than 90% of treated cells lacked MSL2 staining on the X chromosome and showed diminished overall signal within 4 d of double-stranded RNA (dsRNA) treatment. Treatment with msl2 dsRNA also resulted in a similar loss of MSL1 and MSL3 from the X chromosome. Immunostaining varied from cell to cell, but in general MLE and MOF appeared to be released from the X and redistributed throughout the nucleoplasm.
Figure 1.
Delocalization of MSL complexes from the X chromosome following RNAi for msl2 in Drosophila SL2 cells. SL2 cells were treated with GFP-dsRNA or msl2-dsRNA, grown 4 d, fixed, and immunostained with affinity purified rabbit anti-MSL1, anti-MSL2, anti-MSL3, anti-MOF, anti-MLE, or anti-histone H4K16ac followed by anti-rabbit secondary antibodies labeled with Texas Red. DNA was counterstained with DAPI (blue).
One of the known consequences of MSL action on the X chromosome is the site-specific acetylation of histone H4 at Lys 16 by the MOF acetyltransferase (Turner et al. 1992; Bone et al. 1994; Hilfiker et al. 1997; Akhtar and Becker 2000; Smith et al. 2000). Therefore, we immunostained SL2 cells after RNAi treatment with antibodies that specifically recognize histone H4K16ac (Fig. 1). In control cells, H4K16ac is concentrated on the X chromosome and also is variably detected throughout the nucleus, presumably on all chromosomes at a lower level (Turner et al. 1992). After msl2 RNAi treatment, tight foci of H4K16ac disappeared. H4K16ac staining was no longer detectable in ∼25% of cells, while ∼75% retained weak staining throughout the nucleus. In both control and experimental samples, a fraction of nuclei (∼8%–13%) were very brightly staining, with no evident subnuclear localization (data not shown). The significance of these brightly staining nuclei is not known, but since they were present in both control and experimental samples, they have not been considered further in this analysis.
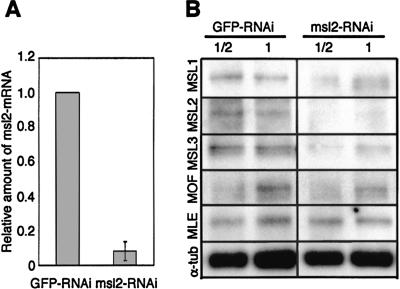
To confirm that the RNAi effects correlated with a loss of msl2 mRNA, we performed quantitative real-time RT–PCR analysis (Fig. 2A). Four days after RNAi treatment, we found that msl2 RNA levels were reduced ∼92% in the experimental cells compared with control cells. To determine the consequence of decreased msl2 RNA levels on protein levels of MSL2 and the other MSL proteins, we performed Western analyses (Fig. 2B). We found that lack of MSL2 protein resulted in lower levels of MSL1 and MSL3, similar to what is seen in wild-type females or msl2 mutants (Palmer et al. 1994; Gorman et al. 1995). In contrast, MOF and MLE levels appeared unchanged in the absence of MSL2. Therefore, our RNAi conditions appear to mimic the female state, in which MSL2 is absent, MSL1 and MSL3 are present only in low levels, and MOF and MLE have similar abundance to males but are delocalized.
Figure 2.
Changes in MSL protein abundance following RNAi for msl2. (A) Real-time RT–PCR quantification of msl2 mRNA. Total cellular RNA was isolated from parallel cultures of cells 4 d after treatment with GFP-dsRNA or msl2-dsRNA. For each sample, transcript levels were normalized to the internal control mRNA pka. The standard deviation was calculated based on three independent experiments. (B) Western analysis of MSL proteins. Crude lysates were prepared from cells 4 d after treatment with GFP-dsRNA or msl2-dsRNA and separated by SDS-PAGE. Western blots were incubated with anti-MSL1, anti-MSL2, anti-MSL3, anti-MOF, anti-MLE, or α-tubulin. Protein extracts were diluted twofold as indicated. Detection was as described in Materials and Methods.
msl2 mutant cells appear to have a severe growth disadvantage during Drosophila development, based on the failure to recover homozygous msl2 mutant clones in most adult tissues of males following induction of mitotic recombination in heterozygous individuals (Belote and Lucchesi 1980b). msl2 mutant males do survive to the larval stages, but they are severely delayed in development. Therefore, we asked whether depletion of MSL2 from cells in culture would lead to changes in their doubling time. We found that cell doubling was indistinguishable comparing mock-treated cells with cells treated with msl2 dsRNA through the 4-d course of this experiment (data not shown). We propose that the lack of a requirement to execute sensitive developmental pathways allows cells without dosage compensation to remain healthy, at least within this limited time frame, and thus has allowed us to separate the direct from the majority of indirect effects caused by depletion of MSL2.
Differential effects of MSL2 RNAi treatment on X-linked genes and autosomal genes
Total RNA was extracted after msl2 or GFP RNAi treatment of cells, and fluorescently labeled cDNA was produced and hybridized in parallel to Affymetrix Drosophila Genome 2.0 microarrays. Experiments were performed three times, with RNAi and GFP control in each case, for a total of six arrays.
A key step in microarray analysis is to identify the genes that have a sufficiently consistent signal to be measured accurately. Genes with undetectable or low expression are removed from the analysis. The 18,000 genes on the array were filtered using Affymetrix Present/Absent calls, resulting in 7923 genes counted as present. A second filter was employed to ensure the consistency of fold ratios in the three experiments (see Statistical Analysis for details). After the second filter, ∼5400 genes with reliable fold ratios were divided into those on the X chromosome (897) and those on the autosomes (4484), and the distribution of the ratios were plotted for each case in the log 2 scale. The two distributions were estimated using the median ratio among the triplicates by a smoothing technique. In Figure 3A, the log fold ratios are centered roughly on zero and are symmetric for the autosomes (black line, peak at x = 0.08 or 20.08 = 1.06-fold), and they are clearly shifted to the left for the X chromosome (red line, peak at x = -0.36 or 2-0.36 = 0.78-fold). This indicates that genes on the X chromosome are down-regulated compared with autosomal genes. The microarray data were processed in many different ways but the results remained the same (see Supplemental Material).
Figure 3.
Microarray gene expression analysis after RNAi for msl2. (A) Distribution of fold change ratios from the X chromosome (red line) and the autosomes (black line). The log fold ratios are centered roughly on zero and are symmetric (black line) for the autosomes, and they are clearly shifted to the left for the X chromosome (red line). In either the t-test or Kolmogorov-Smirnov test, the p-value is <10-15. (B) Changes in the number of genes reliably detected on microarrays after RNAi removal of MSL2. The black, dark-gray, and light-gray bars indicate the percentage of change in transcripts present for the whole genome, the X chromosome, and the autosomes, respectively. In each case, the decrease is either mostly or entirely from the X chromosome.
To determine whether such a large difference in the distributions could have arisen by chance, we calculated the p-value for the null hypothesis that the underlying distributions are the same. Using either the t-test or Kolmogorov-Smirnov test, the p-value is <10-15. This is the probability of observing the difference in distribution as shown in Figure 3A if they were in fact the same.
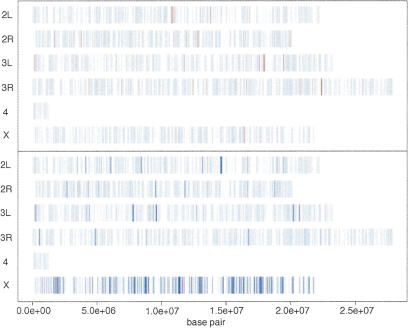
Overall, using a threshold change of 1.4-fold, we found that many X chromosomal genes (∼29%) exhibited a decrease in transcript levels (261 out of 897), while only a small number of autosomal transcripts (∼1.2%) decreased (56 out of 4484). Both X and autosomes had some transcripts that increased slightly under these conditions (∼0.2% and 1.6%, respectively). Another way to examine the changes in expression is to locate the genes with differential expression by their chromosomal location. In Figure 4, we marked the genes up-regulated by 1.4-fold or more in red; we marked those that are down-regulated by 1.4-fold or more in blue. The strongly down-regulated genes are concentrated on the X chromosome. We used the 1.4 threshold for this figure but other threshold values result in a similar picture (see Supplemental Material).
Figure 4.
Chromosomal location of genes affected by RNAi for msl2. Genes up-regulated >1.4-fold are indicated in red, and genes down-regulated >1.4-fold are indicated in dark blue. The strongly down-regulated genes are concentrated on the X chromosome.
We compared our data for individual genes with the results of Straub et. al. (2005), who performed a similar msl2 RNAi analysis in SL2 cells but assayed specific MSL targets for expression changes by real-time RT–PCR analysis. Each individual X-linked gene, CG14804, mRpL16, and Arm, showed a 1.3- to 1.5-fold decrease in our experiments, in agreement with their results. In addition, the autosomal RpII140 gene showed no change in either study. We further validated the Affymetrix values for five X and one autosomal gene by quantitative real-time RT–PCR (see Supplementary Fig. H). Our complete data set for individual genes is available at http://chip.tch.harvard.edu/~ppark/KurodaLab.
Validation that X-linked gene expression decreases by normalization-independent analysis
When microarrays are processed, there is a natural fluctuation in the overall signal intensity among different arrays, due to variations both in the samples and in the experimental procedures. Even if the samples are the same, the labeling and scanning steps can introduce enough variability so that some type of normalization must be performed. There are a number of different methods to adjust for variability among arrays, but a common method implemented in most software as the default is to set the average of the signal intensities across all genes to be the same, after outlier data points are eliminated. An assumption required in this step, however, is that most genes are not differentially expressed. While this assumption holds in the vast majority of expression profiling studies, it would be violated if the autosomal genes are up-regulated and the X-linked genes remain unchanged in the current experiment. Therefore, to unambiguously determine whether X-chromosome gene expression decreased or autosomal gene expression increased after msl2 RNAi, we sought a method that did not require normalization.
We based our normalization-independent analysis on the raw intensities of signals within individual arrays. In Affymetrix arrays, a “probe” for a gene is in fact a set of probes, consisting of 14 distinct “probe pairs” for the Drosophila array, and an expression value is in fact a summary measure based on these multiple probes (http://www.affymetrix.com). Each probe pair consists of a 25-mer probe (PM for “perfect match”) and the same probe with a base pair flip in the middle oligonucleotide to control for cross-hybridization (MM for “mismatch”). By examining the pattern across these multiple probes for a single transcript, it becomes possible to estimate the probability that the transcript is reliably detected. This is the basis for the Present/Marginal/Absent calls and their associated p-values in the Affymetrix software for data processing (see Supplemental Material). Consistently higher PM probe values compared with their counterpart MM probe values leads to a low p-value, which means that the transcript is likely to be present. Therefore, we examined these p-values to determine whether the number of transcripts detected reliably increased or decreased in each RNAi experiment. If autosomal gene expression increased globally after msl2 RNAi, we would expect the primary effect to be an absolute increase in the number of autosomal transcripts detected, as marginal genes would now be counted as present. Conversely, if X gene expression decreased globally after msl2 RNAi, we would expect that the primary effect would be an absolute decrease in the number of X chromosomal transcripts detected.
In Figure 3B, the first bar (black) for each experiment represents the change in the number of genes thought to be present in the RNAi sample compared with the GFP sample, at a threshold p-value of 0.01. The second bar (dark gray) and the third bar (light gray) in Figure 3B represent the changes for the X chromosomal genes and the autosomal genes, respectively. For the first experiment, for example, there is an ∼0.8% decrease in the number of transcripts reliably detected overall, but this number is ∼5% for the genes on the X chromosome, accounting for nearly all of the overall decrease. The changes in the autosomes are negligible. The second experiment actually showed a small increase in the number of detected genes in the autosomes, but the dominant feature is that fewer X-chromosome genes are detected reliably in the RNAi sample compared with the GFP control. The p-value threshold used in Figure 3B is 0.01 but the pattern persists for lower thresholds, even though the absolute magnitude of the percentages increases (see Supplemental Material). Therefore, results shown in Figure 3B strongly support our initial conclusion that X transcripts specifically decrease in the absence of MSL complexes. Using independent methodologies, Straub et al. (2005) have reached the identical conclusion.
Are most genes on the X-chromosome regulated by the MSL complex?
In each replicate of our experiment, we detected changes in mRNA levels from many but not all X-linked genes, dependent on the threshold value that we set. For example, ∼19% of the expressed genes on the X changed expression <1.1-fold, suggesting that not all X-linked genes are controlled by the MSL complex. However, the partial dosage compensation seen here may also be an inevitable result of our methodologies combined with the difficulties in measuring small changes in gene expression. For example, RNAi often results in partial loss-of-function phenotypes, and in our experiments, msl2 RNA was reduced but not completely eliminated (on average to 8% of wild type). Therefore, the technology implemented does not currently allow us to define the precise number of genes affected by the MSL complex.
We previously proposed that a significant subset of X-linked genes, carrying at least three SXL-binding sites in their 3′ UTRs, might be regulated by a distinct dosage compensation pathway (Kelley et al. 1995). This SXL-dependent, MSL-independent pathway was proposed to operate by down-regulation of target transcript stability or translation in females, rather than by up-regulation in males. We found that nine of the previously identified X-linked genes with SXL-binding sites were expressed in SL2 cells. Surprisingly, four of the nine showed a 1.4-fold or greater reduction in expression after RNAi for msl2, suggesting that they are normally up-regulated by the MSL complex. This was substantiated by analysis of a more comprehensive list of genes with SXL-binding sites generated from the annotated Drosophila genomic sequence (C. Stuckenholz and M.I. Kuroda, unpubl.). Therefore, if MSL-independent regulation of this subset of genes does occur, it may be at a specific time or place during development not represented by SL2 cells grown in culture.
In conclusion, we have tested current models for the mechanism of dosage compensation in Drosophila by performing the first global analysis of gene expression after removal of the MSL complex in male cells. Our results demonstrate that X-linked genes are widely affected as a group when MSL2 levels are reduced, consistent with the striking localization of the MSL complex to sites along the length of the male polytene X chromosome (Kuroda et al. 1991; Palmer et al. 1993; Bashaw and Baker 1995; Gorman et al. 1995; Kelley et al. 1995; Zhou et al. 1995; Gu et al. 1998) and the morphological changes to that chromosome when MSL proteins are depleted or overexpressed (Belote and Lucchesi 1980a; Oh et al. 2003). Our results clearly support a role for the MSL complex in up-regulation of X-linked genes in Drosophila males. The specific decrease in X-linked gene expression that we see following msl2 RNAi is incompatible with the inverse dosage model.
Materials and methods
Preparation of dsRNA
Target sequences were scanned to exclude any complete 21-mer homology with other genes. Primer sequences for generation of a GFP dsRNA template by PCR from pEGFP-N1(Clontech) (Boutros et al. 2004) were forward, 5′-TAATACGACTCACTATAGGGAGAGGTGAGCAAGGGCGAGGAGCT-3′, and reverse, 5′-TAATACGACTCACTATAGGGAGATCTTGAAGTTCACCTTGATGCCG-3′. An msl2 segment (-230 to +90 base pair [bp] relative to the ATG start codon) was inserted into pBlue-script to enable generation of an msl2-dsRNA template by PCR using M13 forward and reverse primers. The GFP-dsRNAs and msl2-dsRNAs were generated using an Ambion MEGAscript T7/T3 Kit. The final size of the msl2 and GFP dsRNAs was 500 bp, including flanking Bluescript sequences included in the msl2 construct. Transcription reactions were treated with DNase I for 15 minutes at 37°C to remove the template. After purification, equimolar amounts of sense and antisense msl2 RNAs were mixed, heated for 5 min to 85°C, and annealed by slowly cooling to room temperature.
RNAi
One day prior to the addition of dsRNA, 3 mL of cells per well were placed into six-well cell culture dishes at a final concentration of 1.0 × 106 per mL in Gibco Schneider Drosophila Incubation Media (supplemented with 5% fetal bovine serum and 1% antibiotic/antimycotic). Immediately prior to the addition of dsRNA, the old medium with serum was replaced with 1 mL of Schneider Drosophila Medium without fetal bovine serum. Thirty micrograms of either GFP-dsRNA or msl2-dsRNA was added per well. Cells were incubated at room temperature for 30 min, before the addition of 3 mL of Schneider Cell Growth Media with fetal bovine serum. Cells were incubated for 4 d prior to analysis of gene expression (Kiger et al. 2003; Boutros et al. 2004).
Quantitive real-time RT–PCR
Real-time RT–PCR was performed as described in Bai et al. (2004). The pka gene was used as the internal reference for normalization. The primer sequences for msl2 were as follows: forward, 5′-GCAAGTTGAGGAATCTGATG-3′, and reverse, 5′-GTTTGTGTAGGTGACTGTGAAG-3′. Relative quantification of msl2-mRNA was determined by the comparative CT method based on the manufacture's instruction (ABI Prism 7700 Sequence Detection System Use Bulletin #2, Applied Biosystems). Standard curves for pka- and msl2-primers were constructed using a serial dilution of cDNA to verify equal amplification efficiency of the two systems.
Western analysis
Crude lysates were prepared from cells 4 d after treatment with GFP-dsRNA or msl2-dsRNA. Samples were separated by SDS-PAGE using a 4%–12% Tris-Glycine gel (Invitrogen). Western blots were incubated with affinity-purified anti-MSL antibodies or anti-α tubulin antibody (Sigma) and detected by ECL plus Western blotting detection system (Amersham Biosciences) using HRP secondary antibodies.
Immunostaining
Immunolocalization of MSL proteins was performed as described in Copps et al. (1998) with minor modifications. The cells were centrifuged onto slides at 2000 rpm for 5 min in a Shandon Cytospin 3 cytocentrifuge, fixed by incubation in PBS plus 2% formaldehyde for 30 min. Fixed cells were washed twice in PBS, then dehydrated in acetone for 3 min at -20°C. After two more washes in PBS, slides were blocked by PBS plus 10% donkey serum for 30 min. Slides were treated with affinity-purified anti-MSL antibodies or anti-histone H4K16ac antibody (1:500 dilution, Serotec) overnight, washed two times in PBST (0.1% Tween20), and then incubated with an appropriate Texas Red secondary antibody (Jackson Laboratories) for 1 h. The stained samples were washed two times in PBST and mounted in Vectashield containing DAPI (Vector laboratories, Inc.).
Statistical analysis
The microarrays were processed using GeneChip Operating Software (GCOS) 1.1 from Affymetrix, Inc. The ratios in each experiment were computed from the probe level (rather than probe-set level) data using the same software for increased accuracy. The probe design was based on FlyBase version 3.1, and 18,369 probes were mapped to their locations on the genome (Affymetrix alignment, November 2004). There were two filtering criteria: (1) to eliminate the genes that are expressed in neither control nor RNAi, those with more than three Affymetrix “Absent” calls out of the six samples were removed; (2) the standard deviation of the log ratios were calculated for each gene and those with inconsistent fold ratios (mostly genes with low expression levels) in the triplicates were eliminated. The threshold standard deviation of 0.25 was used, but other values give qualitatively similar results. The estimated fold changes for the remaining 5436 genes were deemed reasonably accurate, with a relatively small number of false positives for high fold changes. The overall results are robust to various parameters in data analysis, as described further in Supplemental Material. This and the raw data are available from http://chip.tch.harvard.edu/~ppark/KurodaLab.
Acknowledgments
We are grateful to E. Larschan and A. Alekseyenko for many helpful discussions and members of the Kuroda lab for critical reading of the manuscript. We thank P. Becker and colleagues for sharing results prior to publication. This work was supported by the National Institutes of Health (GM45744 to M.I.K. and GM67825 to P.J.P.) and the Howard Hughes Medical Institute. M.I.K. is an HHMI Investigator.
Supplemental material is available at http://www.genesdev.org.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1343705.
References
- Akhtar A. and Becker, P.B. 2000. Activation of transcription through histone H4 acetylation by MOF, an acetyltransferase essential for dosage compensation in Drosophila. Mol. Cell 5: 367-375. [DOI] [PubMed] [Google Scholar]
- Bai X., Alekseyenko, A.A., and Kuroda, M.I. 2004. Sequence-specific targeting of MSL complex regulates transcription of the roX RNA genes. EMBO J. 23: 2853-2861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bashaw G.J. and Baker, B.S. 1995. The msl-2 dosage compensation gene of Drosophila encodes a putative DNA-binding protein whose expression is sex specifically regulated by Sex-lethal. Development 121: 3245-3258. [DOI] [PubMed] [Google Scholar]
- Belote J.M. and Lucchesi, J.C. 1980a. Control of X chromosome transcription by the maleless gene in Drosophila. Nature 285: 573-575. [DOI] [PubMed] [Google Scholar]
- ____. 1980b. Male-specific lethal mutations of Drosophila melanogaster. Genetics 96: 165-186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhadra U., Pal-Bhadra, M., and Birchler, J.A. 1999. Role of the male specific lethal (msl) genes in modifying the effects of sex chromosomal dosage in Drosophila. Genetics 152: 249-268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birchler J.A., Pal-Bhadra, M., and Bhadra, U. 2003. Dosage dependent gene regulation and the compensation of the X chromosome in Drosophila males. Genetica 117: 179-190. [DOI] [PubMed] [Google Scholar]
- Bone J.R., Lavender, J., Richman, R., Palmer, M.J., Turner, B.M., and Kuroda, M.I. 1994. Acetylated histone H4 on the male X chromosome is associated with dosage compensation in Drosophila. Genes & Dev. 8: 96-104. [DOI] [PubMed] [Google Scholar]
- Boutros M., Kiger, A.A., Armknecht, S., Kerr, K., Hild, M., Koch, B., Haas, S.A., Consortium, H.F., Paro, R., and Perrimon, N. 2004. Genome-wide RNAi analysis of growth and viability in Drosophila cells. Science 303: 832-835. [DOI] [PubMed] [Google Scholar]
- Breen T.R. and Lucchesi, J.C. 1986. Analysis of the dosage compensation of a specific transcript in Drosophila melanogaster. Genetics 112: 483-491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buscaino A., Kocher, T., Kind, J.H., Holz, H., Taipale, M., Wagner, K., Wilm, M., and Akhtar, A. 2003. MOF-regulated acetylation of MSL-3 in the Drosophila dosage compensation complex. Mol. Cell 11: 1265-1277. [DOI] [PubMed] [Google Scholar]
- Chiang P.W. and Kurnit, D.M. 2003. Study of dosage compensation in Drosophila. Genetics 165: 1167-1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemens J.C., Worby, C.A., Simonson-Leff, N., Muda, M., Maehama, T., Hemmings, B.A., and Dixon, J.E. 2000. Use of double-stranded RNA interference in Drosophila cell lines to dissect signal transduction pathways. Proc. Natl. Acad. Sci. 97: 6499-6503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cline T.W. and Meyer, B.J. 1996. Vive la difference: Males vs females in flies vs worms. Annu. Rev. Genet. 30: 637-702. [DOI] [PubMed] [Google Scholar]
- Copps K., Richman, R., Lyman, L.M., Chang, K.A., Rampersad-Ammons, J., and Kuroda, M.I. 1998. Complex formation by the Drosophila MSL proteins: Role of the MSL2 RING finger in protein complex assembly. EMBO J. 17: 5409-5417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorman M., Franke, A., and Baker, B.S. 1995. Molecular characterization of the male-specific lethal-3 gene and investigations of the regulation of dosage compensation in Drosophila. Development 121: 463-475. [DOI] [PubMed] [Google Scholar]
- Gu W., Szauter, P., and Lucchesi, J.C. 1998. Targeting of MOF, a putative histone acetyl transferase, to the X chromosome of Drosophila melanogaster. Dev. Genet. 22: 56-64. [DOI] [PubMed] [Google Scholar]
- Hiebert J.C. and Birchler, J.A. 1994. Effects of the maleless mutation on X and autosomal gene expression in Drosophila melanogaster. Genetics 136: 913-926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilfiker A., Hilfiker-Kleiner, D., Pannuti, A., and Lucchesi, J.C. 1997. mof, a putative acetyl transferase gene related to the Tip60 and MOZ human genes and to the SAS genes of yeast, is required for dosage compensation in Drosophila. EMBO J. 16: 2054-2060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley R.L., Solovyeva, I., Lyman, L.M., Richman, R., Solovyev, V., and Kuroda, M.I. 1995. Expression of msl-2 causes assembly of dosage compensation regulators on the X chromosomes and female lethality in Drosophila. Cell 81: 867-877. [DOI] [PubMed] [Google Scholar]
- Kiger A.A., Baum, B., Jones, S., Jones, M.R., Coulson, A., Echeverri, C., and Perrimon, N. 2003. A functional genomic analysis of cell morphology using RNA interference. J. Biol. 2: 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroda M.I., Kernan, M.J., Kreber, R., Ganetzky, B., and Baker, B.S. 1991. The maleless protein associates with the X chromosome to regulate dosage compensation in Drosophila. Cell 66: 935-947. [DOI] [PubMed] [Google Scholar]
- Meller V.H. and Kuroda, M.I. 2002. Sex and the single chromosome. Adv. Genet. 46: 1-24. [DOI] [PubMed] [Google Scholar]
- Mukherjee A.S. and Beermann, W. 1965. Synthesis of ribonucleic acid by the X-chromosomes of Drosophila melanogaster and the problem of dosage compensation. Nature 207: 785-786. [DOI] [PubMed] [Google Scholar]
- Muller H. 1932. Further studies on the nature and causes of gene mutations. In Proceedings of the 6th International Congress of Genetics, Vol. 1 (ed. D.F. Jones), pp. 213-215. Menasha, WI: Banta. [Google Scholar]
- Nusinow D.A. and Panning, B. 2005. Recognition and modification of seX chromosomes. Curr. Opin. Genet. Dev. 15: 206-213. [DOI] [PubMed] [Google Scholar]
- Oh H., Park, Y., and Kuroda, M.I. 2003. Local spreading of MSL complexes from roX genes on the Drosophila X chromosome. Genes & Dev. 17: 1334-1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal Bhadra M., Bhadra, U., Kundu, J., and Birchler, J.A. 2005. Gene expression analysis of the function of the MSL complex in Drosophila. Genetics 169: 2061-2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer M.J., Mergner, V.A., Richman, R., Manning, J.E., Kuroda, M.I., and Lucchesi, J.C. 1993. The male-specific lethal-one (msl-1) gene of Drosophila melanogaster encodes a novel protein that associates with the X chromosome in males. Genetics 134: 545-557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer M.J., Richman, R., Richter, L., and Kuroda, M.I. 1994. Sex-specific regulation of the male-specific lethal-1 dosage compensation gene in Drosophila. Genes & Dev. 8: 698-706. [DOI] [PubMed] [Google Scholar]
- Ryner L.C. and Baker, B.S. 1991. Regulation of doublesex pre-mRNA processing occurs by 3′-splice site activation. Genes & Dev. 5: 2071-2085. [DOI] [PubMed] [Google Scholar]
- Smith E.R., Pannuti, A., Gu, W., Steurnagel, A., Cook, R.G., Allis, C.D., and Lucchesi, J.C. 2000. The drosophila MSL complex acetylates histone H4 at lysine 16, a chromatin modification linked to dosage compensation. Mol. Cell. Biol. 20: 312-318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straub T., Gilfillan, G.D., Maier, V.K., and Becker, P.B. 2005. The Drosophila MSL complex activates the transcription of target genes. Genes & Dev. (this issue). [DOI] [PMC free article] [PubMed]
- Turner B.M., Birley, A.J., and Lavender, J. 1992. Histone H4 isoforms acetylated at specific lysine residues define individual chromosomes and chromatin domains in Drosophila polytene nuclei. Cell 69: 375-384. [DOI] [PubMed] [Google Scholar]
- Zhou S., Yang, Y., Scott, M.J., Pannuti, A., Fehr, K.C., Eisen, A., Koonin, E.V., Fouts, D.L., Wrightsman, R., Manning, J.E., et al. 1995. Male-specific lethal 2, a dosage compensation gene of Drosophila, undergoes sex-specific regulation and encodes a protein with a RING finger and a metallothionein-like cysteine cluster. EMBO J. 14: 2884-2895. [DOI] [PMC free article] [PubMed] [Google Scholar]