Abstract
Spinal muscular atrophy (SMA) is a motoneuron disease caused by reduced levels of survival motoneuron (SMN) protein. Previous studies have assigned SMN to uridine-rich small nuclear ribonucleoprotein particle (U snRNP) assembly, splicing, transcription, and RNA localization. Here, we have used gene silencing to assess the effect of SMN protein deficiency on U snRNP metabolism in living cells and organisms. In HeLa cells, we show that reduction of SMN to levels found in SMA patients impairs U snRNP assembly. In line with this, induced silencing of SMN expression in Xenopus laevis or zebrafish arrested embryonic development. Under less severe knock-down conditions, zebrafish embryos proceeded through development yet exhibited dramatic SMA-like motor axon degeneration. The same was observed after silencing two other essential factors in the U snRNP assembly pathway, Gemin2 and pICln. Importantly, the injection of purified U snRNPs into either SMN- or Gemin2-deficient embryos of Xenopus and zebrafish prevented developmental arrest and motoneuron degeneration, respectively. These findings suggest that motoneuron degeneration in SMA patients is a direct consequence of impaired production of U snRNPs.
Keywords: Survival motor neurons (SMN), U snRNP assembly, motoneuron, spinal muscular atrophy, zebrafish
Spinal muscular atrophy (SMA) is a hereditary neuromuscular disease caused by reduced levels of the survival motoneuron (SMN) protein (Lefebvre et al. 1995). In humans, the SMN protein is encoded by two almost identical gene copies, SMN1 and SMN2. In both healthy and affected individuals, only small amounts of functional SMN protein are expressed from SMN2, due to the alteration of a splice regulatory sequence in exon 7 (Lorson et al. 1999; Lorson and Androphy 2000; Cartegni and Krainer 2002; Kashima and Manley 2003). The expression of functional SMN protein therefore largely depends on SMN1. Most pathogenic mutations identified so far map to this copy of the gene and result in reduced expression for functional SMN protein. It is widely accepted that this alone causes the degeneration of α-motoneurons in the spinal cord and the muscular atrophy symptomatic for SMA. Indeed, recent studies carried out in mice and zebrafish elegantly recapitulated this scenario, showing that induced reduction of SMN protein levels leads to motoneuron defects (Hsieh-Li et al. 2000; Monani et al. 2000; Jablonka et al. 2002; McWhorter et al. 2003). Opposed to SMN protein insufficiency, a complete lack of SMN protein in patients has never been reported and, since homozygous inactivation of SMN in mice causes embryonic death, may be presumed to be lethal also in humans.
The tissue-specific degeneration of cells caused by SMN protein deficiency is puzzling, given that SMN is expressed ubiquitously rather than restricted to motoneurons. To solve this paradox, several experimental approaches have been exploited to assign SMN to a specific function. These studies have revealed links of SMN to the assembly of spliceosomal and histone mRNA-processing uridine-rich small nuclear ribonucleoprotein particles (U snRNPs) (Meister and Fischer 2002; Pillai et al. 2003; Gubitz et al. 2004), to pre-mRNA splicing catalysis (Pellizzoni et al. 1998; Meister et al. 2000), to axonal transport (Jablonka et al. 2001; Rossoll et al. 2003; Zhang et al. 2003), and to transcription (Strasswimmer et al. 1999).
One function that is understood in some detail is SMN's contribution to the assembly of the spliceosomal U snRNPs U1, U2, U4, and U5. A key step in the formation of these particles takes place in the cytosol, where seven so-called Sm-proteins that are common to these snRNPs associate with a U snRNA (U1, U2, U4, or U5, respectively) in a stable, ring-shaped structure (Kambach et al. 1999). The functional state of a U snRNP is achieved once additional, particle-specific, proteins have bound to this Sm core particle. Ultimately, the cap structures of the U snRNAs undergo hypermethylation, and the resulting U snRNPs are imported into the nucleus, where they function in splicing (Will and Luhrmann 2001). Although the formation of the Sm core can occur spontaneously in vitro, we and others have shown that it requires the activity of SMN in a test system that recapitulates in vivo conditions (Meister et al. 2001a; Meister and Fischer 2002; Pellizzoni et al. 2002). SMN operates in a functional entity termed the SMN complex, whose major protein components have been grouped as Gemins 2-7, and a factor termed unrip (Meister and Fischer 2002; Gubitz et al. 2004). Immunodepletion of cell extracts showed that this entity is essential for U snRNP assembly. Furthermore, purified SMN complex alone was shown to be sufficient for the transfer of Sm proteins onto U snRNAs (Meister and Fischer 2002; Pellizzoni et al. 2002). Nonetheless, the activity of the SMN complex becomes greatly enhanced through the interaction with a second entity termed PRMT5 complex (Meister and Fischer 2002). In addition to its eponym, the type II arginine methyltransferase PRMT5, this complex contains the Sm protein-binding factor pICln and the WD repeat protein WD45/MEP50 (Branscombe et al. 2001; Friesen et al. 2001; Meister et al. 2001b). The PRMT5 complex symmetrically dimethylates arginines in the Sm proteins B/B′, D1, and D3, thereby enhancing their affinity for the SMN complex (Brahms et al. 2001; Friesen et al. 2001). In view of this, one may presume that the interplay between SMN and PRMT5 complexes occurs in coordinated, and most likely coregulated, steps, with SMN itself being a key player along the assembly line. Indeed, there is now evidence for the regulation of SMN's activity by a kinase that recognizes both SMN and pICln (Grimmler et al. 2005). This most recent study is one of many indicating that SMN is indispensable for the formation of U snRNPs in vivo.
Despite the progress in unraveling the cellular functions of SMN, neither its role in U snRNP assembly nor the additional functions assigned to SMN (splicing, transcription, axonal transport) have been convincingly linked to the pathology of SMA. We have therefore dedicated our efforts to this aspect and asked whether impaired U snRNP assembly, due to reduced levels of SMN, can elicit SMA-like symptoms in animal test systems.
Results
Induced reduction of SMN protein in HeLa cells impairs U snRNP assembly
Most studies carried out so far aimed at the depletion of the SMN complex from cell extracts (Meister et al. 2001a; Meister and Fischer 2002; Pellizzoni et al. 2002), others focused on site-specific inactivation of SMN (Pellizzoni et al. 1998; Grimmler et al. 2005). From these experiments, we learned that the SMN complex is essential for U snRNP assembly. Nonetheless, it was unclear whether U snRNP assembly was likewise impaired in patients and whether this, indeed, conferred the motoneuron defects associated with the SMA phenotype. As mentioned above, all patients, even those suffering from the most severe form of SMA (type I SMA), express SMN to some extent, with levels of functional SMN protein amounting up to 30%-40% of the normal level (Lefebvre et al. 1997). Thus, we first asked whether this extent of SMN protein reduction affected U snRNP assembly at all. One approach toward this could be to analyze U snRNP assembly in primary cells from patients, using the assay system described previously (Meister et al. 2001a). As opposed to HeLa cells or Xenopus egg extract, however, we did not succeed in preparing active extracts, neither from cells of patient origin nor from unaffected individuals. We therefore decided to elaborate experimental conditions in HeLa cells that could reflect the reduced SMN expression observed in patients.
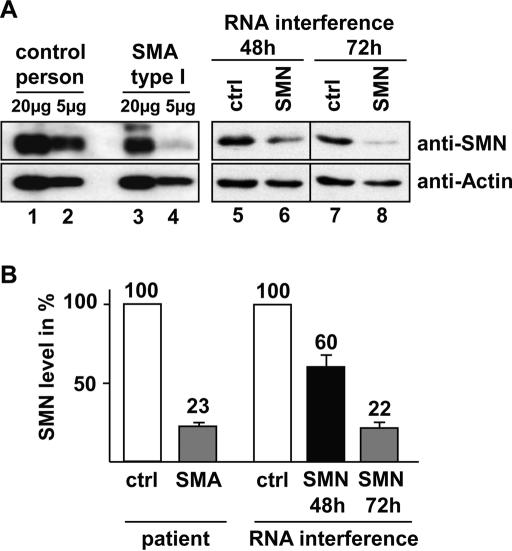
As a first approach, we tested in tissue culture whether RNA interference (RNAi) was suitable for adjusting SMN expression levels to that observed in patients. Primary fibroblasts from type I SMA patients were used as a standard for pathogenic conditions. Compared with fibroblasts from unaffected individuals, the expression of endogenous SMN protein in these cells was reduced to 23% of the normal level (Fig. 1A [cf. lanes 1,2 and 3,4], B). We then mimicked this situation in HeLa cells using a 22-nt-long duplex of small interfering RNAs (siRNAs) that corresponds to positions 183-205 of human SMN mRNAs. As shown in Figure 1A and B, transfection of HeLa cells with this duplex reduces the SMN protein level to ∼60% within 48 h, as compared with cells transfected with a control siRNA (Fig. 1A, lanes 5,6). After 72 h, expression of SMN was suppressed to ∼22% of SMN protein remaining (Fig. 1A, lanes 7,8). The transfection efficiency in these experiments was in the range of 95%, ruling out the possibility that a significant amount of the extract was derived from untransfected cells (see Supplementary Fig. 1). These findings showed that the amounts of SMN protein in our test system can be adjusted to pathological levels. Cells treated in this way proceed through several indicatory defects along the RNAi time course, such as the disappearance of SMN from nuclear structures termed gems, slowed proliferation, and eventually apoptosis (Supplementary Fig. 1A; data not shown).
Figure 1.
Reduction of SMN levels in HeLa cells. (A) Immunoblot analysis of SMN in fibroblasts from a healthy individual (lanes 1,2) or a SMA type I patient (lanes 3,4), and in HeLa cells subjected to RNAi (lanes 5-8). Twenty micrograms of total protein (lanes 1,3,5-8) or 5 μg (lanes 2,4) was analyzed for SMN protein content. For RNAi, extracts from cells treated with siRNAs complementary to SMN (lanes 6,8) or nonspecific control siRNAs (lanes 5,7) were analyzed either 48 h (lanes 5,6) or 72 h (lanes 7,8) after transfection. Lower panels show an immunodetection of actin on the same blot. (B) Densitometric quantification of the relative SMN levels versus actin levels shown in A.
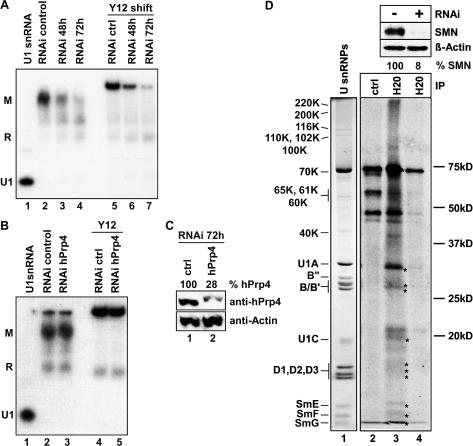
Having the RNAi tool at hand, we next applied a U snRNP assembly assay to extracts from RNAi-treated HeLa cells. This assay is based on the ability of HeLa cell extract to retard a radiolabeled U1 snRNA in native polyacrylamide gel electrophoresis (Meister et al. 2001a). Consistent with previous data, extracts prepared from control cells efficiently promoted the formation of a slowly migrating M complex (Fig. 2A, lane 2). We have previously shown that this complex contains the fully assembled Sm core domain of snRNPs (Meister and Fischer 2002). Consistent with this, the anti-Sm protein antibody Y12 retards this complex from migration (Fig. 2A, lane 5). Importantly, the formation of the M complex was significantly reduced in extracts from cells that had been treated with the SMN siRNA duplex for 48 h (Fig. 2A, lanes 3,6). Extracts prepared from cells harvested after 72 h of RNAi treatment were intensely inhibited in performing this reaction (Fig. 2A, lanes 4,7). Both conditions did not affect the formation of a group of bands (Fig. 2A, bands denoted R) that contain the U1-specific A protein but lack an Sm core domain (Meister and Fischer 2002). Importantly, an inhibition of assembly was not observed in extracts derived from cells with RNAi-induced reduction of the essential splicing factor hPrp4 (Fig. 2B,C). This suggests that the observed assembly defect in the extract derived from SMN-deficient cells is specific.
Figure 2.
Induced reduction of SMN protein in HeLa cells impairs the assembly of U snRNPs. (A) Assay for the assembly of the U1 snRNP in vitro. [32P]-labeled U1 snRNA (lane 1) was incubated with the RNAi-treated HeLa cell extracts described in Figure 1 (lanes 2-7) for 30 min, and assembly was monitored by native gel electrophoresis as described in Meister et al. (2001a). (Lanes 5-7) Supershifts with antibody Y12 were used to identify complexes that contain Sm proteins. (B) The same assay as described in A was performed with extracts from cells treated with control siRNAs (lanes 2,4) or siRNAs against the hPrp4 mRNA (lanes 3,5). (C) Immunoblot analysis of hPrp4 and β-actin in the HeLa cell extracts used in B. (D) Impaired synthesis of U snRNPs de novo upon SMN protein reduction. HeLa cells transfected with SMN siRNA (lane 4) or control siRNA (lanes 2,3) were pulsed with [35S]cysteine and [35S]methionine and, after exchanging the growth medium, extracts were prepared. Immunoprecipitations were carried out with anti-RNA cap antibody H20 (lanes 3,4) or an unrelated antibody (lane 2), and precipitated proteins were analyzed by SDS-PAGE and autoradiography. See lane 1 for Coomassie-stained U snRNP proteins. Upper panels display immunoblot analyses of SMN and actin in extracts of SMN siRNA-treated cells. The level of SMN protein quantified by densitometry is shown as a percentage value in relation to actin.
The above findings support the idea that the amount of functional SMN protein in cells is a bottleneck to efficient U snRNP assembly. To further validate this in vivo, we designed pulse-chase experiments to monitor U snRNP synthesis de novo. HeLa cells were treated for 72 h with SMN or control siRNAs, followed by a 1-h pulse with [35S]methionine to allow for the labeling of nascent proteins. Lysates from these cells were then subjected to immunoprecipitation using an antibody that is directed against the cap structure of U snRNAs (antibody H20) (Bringmann and Luhrmann 1986). An immunoprecipitate containing radiolabeled U snRNP proteins would therefore be indicatory of de novo production of U snRNPs. Indeed, upon comparison of autoradiographs with Coomassie-stained U snRNP proteins (Fig. 2D, lanes 1,3), we identified specific bands in the immunoprecipitate that correspond to the seven Sm proteins and the U1 snRNP-specific proteins 70K, U1A, and U1C (see asterisks in Fig. 2D; note that additional U snRNP-specific proteins became visible after extended exposure of the film). In contrast, much weaker signals were obtained after immunoprecipitation from SMN-deficient cell lysates (Fig. 2D, cf. lanes 4 and 3; also see upper panel for SMN protein deficiency). The only intense band remaining is nonspecific, as it also appears in the negative control (Fig. 2D, cf. lanes 4 and 2). Similar but less severe effects were observed in cells containing intermediate levels of SMN (Supplementary Fig. 2). Thus, data from both RNAi approaches, that is, the analysis of extracts in vitro and the pulse-chase study in vivo, provide strong support that insufficiency of SMN protein impairs U snRNP assembly in human cells.
Purified U snRNPs rescue the developmental arrest of SMN-deficient Xenopus laevis embryos
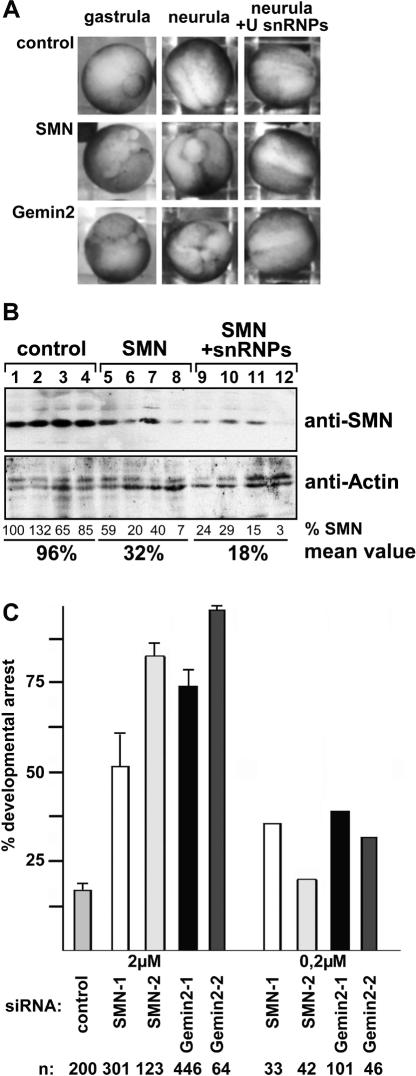
The observed effects of SMN deficiency on U snRNP assembly encouraged us to ask whether disturbances in this pathway are directly linked to the degeneration of cells. In this case, we should be able to protect SMN-deficient cells from degeneration through an exogenous supply of U snRNPs. Most somatic cell lines available from SMA patients do not exhibit dramatic phenotypes and are thus not suitable for this approach. Therefore, we devised an RNA-interference strategy in developing X. laevis embryos. SiRNAs complementary to X. laevis SMN mRNA were injected into both blastomeres of two-cell-stage embryos. The analysis of proteins from individual embryos on Western blots (Fig. 3B) revealed a significant reduction of SMN protein (Fig. 3B, cf. lanes 1-4 and 5-8). This became apparent only at gastrula stages (probably due to the large maternal pool of SMN), indicating that injection of the siRNAs does not impair the initial phase of embryonic development. However, a large fraction of embryos exhibited developmental arrest during gastrulation (typically between 52% and 80%, depending on the batch of eggs), whereas control embryos developed normally (Fig. 3A,C). At stage 11, the control RNAi-injected embryos begun to close the blastoporus, whereas the SMN-deficient embryos failed to finish gastrulation. Instead, detached white cells surrounding the blastoporus indicated massive developmental defects (Fig. 3A, first column). In neurula stages, the control-injected embryos formed the neural tube, while SMN-deficient embryos that survived until this stage did not close the blastoporus (Fig. 3A, second column). The same phenotype was observed when the SMN-interacting protein Gemin2 was targeted. In this case, up to 95% of injected embryos did not proceed through development beyond gastrulation (Fig. 3A,C). This effect was dose-dependent, since a 10-fold lower concentration of siRNAs (0.2 μM) had only marginal effects on development, whereas a higher concentration (2 μM) was toxic (Fig. 3C; data not shown).
Figure 3.
SMN and Gemin2 are essential for the development of Xenopus embryos. (A) SiRNAs complementary to SMN or Gemin2, or nonspecific siRNAs were injected into X. laevis embryos, and survival was scored at gastrula (left column) or neurula (middle and right columns). Dead or developmentally arrested embryos display an abnormal outgrowth of white cell mass. (B) Western blot analysis of the SMN level in embryos treated with control (lanes 1-4) or SMN-siRNAs (lanes 5-8) in comparison to actin levels. Analyses of embryos that received a coinjection of U snRNPs (see also legend for Fig. 4 for details) are shown in lanes 9-12. Each lane represents an individual embryo. Relative SMN expression levels were estimated by densitometry in comparison to actin levels. (C) Quantification of the injection studies shown in A. Results are shown for two siRNA concentrations (2 and 0.2 μM) and two different siRNAs per target. The numbers of examined embryos (n) are indicated below each row.
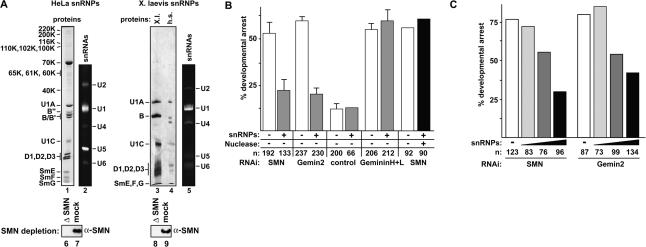
We next asked whether supplementation with purified U snRNPs could compensate for the loss of SMN or its direct binding partner, Gemin2. For this approach, we used U snRNPs that had been isolated either from HeLa nuclear extract or X. laevis cells (XTC) by H20 anti-m3G immunoaffinity chromatography at elevated ionic strength (420 mM KCl) (Bringmann and Luhrmann 1986). This preparation contains the Sm core particles of U1, U2, U4, and U5 snRNPs (see Fig. 4A for Sm proteins B/B′, D1, D2, D3, E, F, G, and for U snRNAs) and reflects the end products of the SMN-mediated assembly pathway. In addition to the U1 snRNP-specific proteins 70K, U1A, and U1C, this isolate also contains a subset of specific proteins from U2, U4, and U5 snRNPs (some of them indicated in Fig. 3A). As an approach to remove contaminating SMN complex associated with U snRNPs, these particles were passed over an anti-SMN immunoaffinity matrix before they were injected into the embryos. Indeed, SMN was not detected in either U snRNP preparation (Fig. 4A, lanes 6-9).
Figure 4.
Isolated U snRNPs protect SMN- and Gemin2-deficient Xenopus embryos from developmental arrest. (A) Characterization of U snRNPs used for injection experiments. U snRNP proteins were resolved by SDS-PAGE and analyzed either by Coomassie staining (lane 1) or by silver staining (lanes 3,4). (Lanes 2,5) U snRNAs were separated by a denaturing RNA-gel electrophoresis and visualized by ethidium bromide staining. The U snRNPs were passed over an anti-SMN column prior to injection to remove residual SMN protein. Immunoblot analysis of the SMN-depletion is shown in lanes 6 and 8; lanes 7 and 9 show the mock depletions. (B) X. laevis embryos were treated by injection with control siRNA or siRNAs directed against SMN or Gemin2, respectively, either alone or along with purified U snRNPs. As a first control for the specificity of the rescue, U snRNPs were coinjected into embryos with siRNAs against proteins whose function does not relate to U snRNP assembly (Geminins H and L). As a second control, embryos were coinjected with digested U snRNPs. Quantification of embryonic survival at stage 13 upon injection of siRNAs alone, siRNA together with U snRNPs, or siRNA together with RNase T1-digested U snRNPs. (C) X. laevis embryos were coinjected with siRNAs against either SMN or Gemin2 and increasing amounts of U snRNPs. Quantification of the survival rates is shown.
Along with siRNAs against SMN or Gemin2, 1 ng of purified human U snRNPs per embryo was injected. Given an approximate volume of the embryo of 0.5-1 μL, the amount injected roughly corresponds to a concentration of 1-2 nM. Strikingly, this coinjection efficiently prevented the developmental arrest observed in embryos injected with siRNAs alone (Fig. 4B; see also Fig. 3A [third column], B, lanes 9-12 for knock-down efficiency). Importantly, the rescue was dose-dependent as the injection of lower amounts (i.e., 0.1 ng or 0.01 ng of U snRNP) had less severe effects (Fig. 4C). In contrast, the injection of isolated U snRNPs per se did not alter the development of embryos treated with control siRNA (Figs. 3A [first row], 4B). These data were verified with purified U snRNPs isolated from Xenopus tissue culture (data not shown).
Two controls were performed to ensure the specificity of this rescue. First, siRNAs directed against a gene involved in an unrelated cellular pathway were used in conjunction with U snRNP injections. The siRNAs applied here correspond to the mRNA sequences of two endogenous inhibitors of DNA replication termed Geminin H and Geminin L (McGarry 2002). Geminins are unstable inhibitors of DNA replication that are degraded at the metaphase/anaphase transition. In agreement with data from McGarry (2002), the injection of siRNAs against either mRNA caused a strong developmental arrest at gastrulation (Fig. 4B). The developmental defects elicited by RNAi against Geminins H and L are thus qualitatively and temporally equivalent to the silencing of SMN or Gemin2 expression. In contrast to SMN- or Gemin2-deprived embryos, however, a coinjection of U snRNPs did not circumvent the developmental arrest of embryos treated with Geminin siRNAs (Fig. 4B). In a second control experiment, the RNA moiety of the U snRNPs was digested with RNase T1 prior to injection into SMN-deficient embryos. As seen in Figure 4B, in this case developmental arrest could not be prevented (see also Supplementary Fig. 3 for efficiency of RNase treatment). Taken together, these data indicate that isolated and intact U snRNPs supplied in trans can efficiently and specifically bypass the requirement for SMN and Gemin2 in the developing embryo.
Gene silencing of the snRNP assembly pathway causes motor axon degeneration in zebrafish
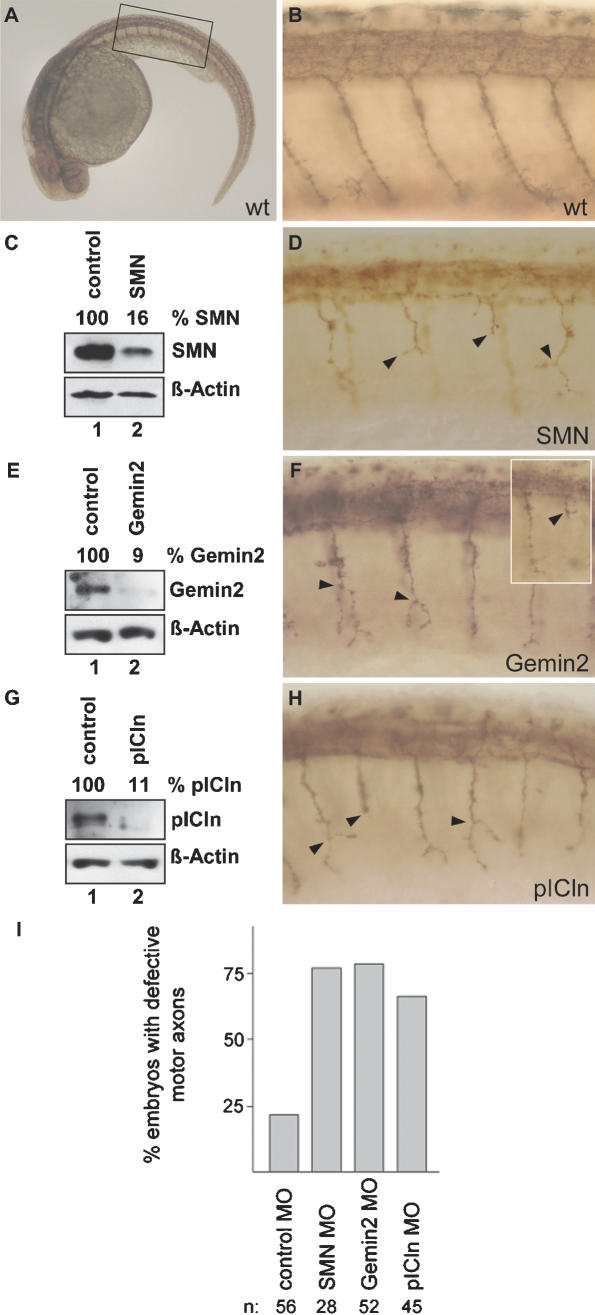
The rescue of SMN-deficient frog embryos by U snRNPs prompted us to ask whether a SMA-like motoneuron degeneration could likewise be linked to the U snRNP pathway. In this case, the impaired expression of any essential factor whose sole function is the assembly of U snRNPs should give rise to a motoneuron phenotype. To address this issue, we used a recently described approach to study motor axon degeneration in zebrafish (McWhorter et al. 2003). Consistent with our observations in X. laevis, the authors of this study showed that injection of antisense Morpholinos against SMN into zebrafish embryos terminates development in a dose-dependent manner (McWhorter et al. 2003). The transparency of these embryos also allowed for a close inspection of neurons, revealing specific defects in outgrowth and pathfinding of motoneurons. This was particularly evident at lower Morpholino concentrations, where survival rates of the embryos improved (McWhorter et al. 2003). We have adopted this approach and consistently observe abnormal morphologies of motoneurons after the expression of SMN had been knocked down to 16% of the control level (Fig. 5C, cf. lanes 2 and 1). Immunohistochemistry using the znp1 antibody (Melancon et al. 1997) revealed abnormal outgrowth of axons in the caudal primary (CaP) and variable primary (VaP) motoneurons, leaving most of these axons truncated and erroneously branched (Fig. 5 [cf. B and D], see A for specification of the examined region). More than 75% of the SMN-deficient embryos developed this phenotype at 31 h post-fertilization (Fig. 5I; Supplementary Table 2), similar to the situation described earlier (McWhorter et al. 2003). Injection of no or a control Morpholino had no significant effect on axon length and morphology (Fig. 5B,I; see also Supplementary Table 2).
Figure 5.
Motoneuron defects in zebrafish embryos upon reduced expression of SMN, Gemin2, or pICln. (A) Lateral view of a zebrafish embryo at 31 h post-fertilization (hpf). Motor axons are stained with the znp1 antibody. A rectangle marks the area at midtrunk level that contains 16 motor axons and was evaluated in all analyzed embryos. (B,D,F,H) Lateral views of trunk regions showing znp1 positive motor axons in a noninjected (B) and SMN (D), Gemin2 (F), and pICln (H) Morpholino-injected embryos. Arrowheads indicate truncated or branched axons. Inset in F shows truncated axon in embryo different from that shown in F. (C,E,G) Western blot analysis of extracts from zebrafish treated with Morpholino oligonucleotides against SMN (C, lane 2), Gemin2 (E, lane 2), or pICln (G, lane 2). Extracts from noninjected control embryos are always shown in lane 1. Actin was detected to control for equal loading and relative quantification of the respective target protein level. (I) Quantification of motor axon defects in zebrafish embryos after knock-down of SMN, Gemin2, and pICln. Embryos with bifurcated or more than one truncated motor axon were scored as defective.
We next asked whether similar phenotypes could likewise be observed upon knocking down other U snRNP assembly factors. For these studies we chose (1) Gemin2, a major component of the SMN complex, and (2) pICln, the Sm protein-binding factor of the PRMT5 complex (Meister et al. 2000, 2001b; Friesen et al. 2001). The zebrafish orthologs of human Gemin2 and human pICln were identified in the NCBI database and exhibit 68% identity (Gemin2) or 67% identity (pICln), respectively. When the amount of Gemin2- or pICln-Morpholino injected into embryos was adjusted to sublethal doses, a phenotype similar to that of SMN morphants was obtained (see Fig. 5E-I; see also Supplementary Tables 1, 2). Compared with noninjected controls, embryos injected with a sublethal dose of SMN-, Gemin2-, and pICln-Morpholinos showed slightly delayed development with shortened body axes and kinked tails, but otherwise developed normally (see Supplementary Table 2). The myotome regions, representing the targets of motoneuronal innervation, formed normally in these embryos (regular myoD expression in all embryos injected with Mo Gemin2 [n = 36] or Mo pICln [n = 21]) (data not shown). Also, the number of motoneuronal cell bodies was only slightly decreased in a subset of injected embryos (reduced number of isl2-positive cells in six out of 20 embryos injected with Gemin2 Mo and three out of 21 for pICln). The remaining expression in morphants for Gemin2 or pICln was 9% or 11%, respectively, of control levels (Fig. 5E,G). These observations indicate that the reduced expression of three factors with a proven role in the biogenesis of U snRNPs causes motor axon degeneration.
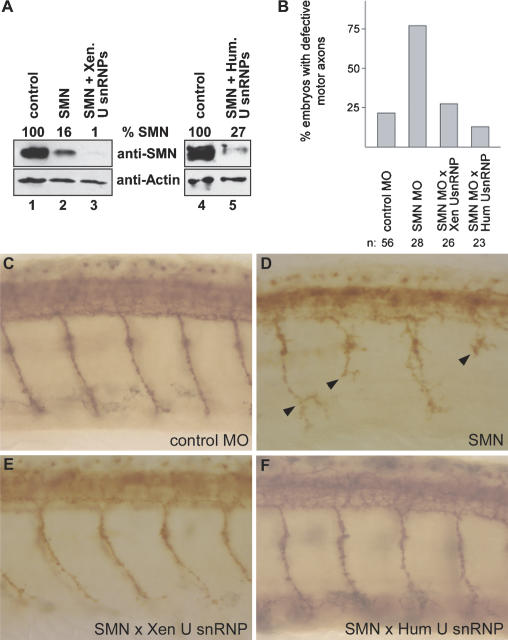
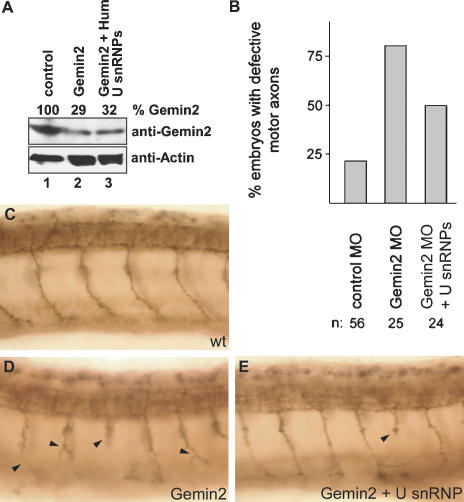
The results shown above encouraged us to analyze whether motor axon degeneration could be prevented upon coinjection of isolated U snRNPs. To test this possibility, the SMN-Morpholino was injected into zebrafish embryos either alone or in the presence of purified U snRNPs. As observed previously, embryos treated with SMN-Morpholino alone displayed reduced expression of SMN (Fig. 6A, lane 2) and erroneously truncated and branched motoneurons (Fig. 6D). Strikingly, these motor axon defects were efficiently prevented by coinjected U snRNPs (Fig. 6E,F). Due to the high degree of evolutionary conservation of Sm proteins, U snRNP isolates from HeLa cells or Xenopus were equally suitable for this rescue (Fig. 6, cf. E and F; see also Fig. 4A for U snRNP composition). The motoneurons of animals that had received a U snRNP injection at the embryonic stage developed normally and resembled the control group injected with a standard control Morpholino (Fig. 6, cf. E,F and C). We have evaluated defects of caudal primary motoneurons (CaP) in a statistically relevant number of animals (Fig. 6B) and find that 76.9% of motoneurons in SMN morphants are defective, whereas treatment with U snRNPs reduces this number to control levels (13%-31%). Since treatment with U snRNPs does not significantly elevate the level of SMN protein in morphants (Fig. 6A, cf. lanes 2 and 3,5), we consider it unlikely that this effect was mediated by potential trace amounts of SMN in the U snRNP preparation. Finally, we analyzed whether a similar strategy would also rescue Gemin2-deficient zebrafish embryos. Indeed, complementation with human U snRNPs rescued motor axonal defects (Fig. 7, cf. D,E and C). This rescue, however, was considerably less efficient than observed for SMN (cf. Figs. 6B and 7B). The number of embryos with motor axonal defects was reduced from 80% (n = 25) in Gemin2 Morpholino-injected embryos to 50% (n = 24) in embryos simultaneously coinjected with Gemin2 Morpholino and human U snRNPs. Thus, SMN- and Gemin2-deficient zebrafish develop normal motoneurons, provided that U snRNPs are supplied in trans. The animal model for SMA applied here provides the first evidence in vivo that interference with the RNP assembly pathway causes motor axon degeneration.
Figure 6.
Coinjection of U snRNPs and SMN Morpholinos prevents motor axon defects in zebrafish embryos. (A) Western blot analysis of SMN levels in noninjected control embryos (lanes 1,4) and in embryos injected with SMN Morpholino (lane 2) or SMN Morpholino in combination with isolated U snRNPs from X. laevis (lane 3) or HeLa cells (lane 5; see also Fig. 4A for Coomassie-stained proteins). Actin levels served as loading control and for relative densitometric quantification levels of SMN levels. (B) Quantification of motoneuron defects from SMN-deficient zebrafish embryos that had been treated with or without exogenous U snRNPs. (C-F) Lateral views of zebrafish trunk regions showing znp1-positive motor axons after immunostaining. Motor axon phenotypes in control Morpholino-injected embryos (C), SMN-deficient embryos (D), and embryos coinjected with SMN Morpholino and U snRNPs from X. laevis (E) and HeLa cells (F). Arrowheads indicate truncated or branched axons in SMN Morpholino-injected embryos in D.
Figure 7.
Coinjection of U snRNPs and Gemin2 Morpholinos prevents motor axon defects in zebrafish embryos. (A) Western blot analysis of Gemin2 levels in embryos injected with no Morpholino (lane 1), Gemin2 Morpholino (lane 2), or Gemin2 Morpholino in combination with isolated human U snRNPs. Actin levels served as loading control and for relative densitometric quantification levels of Gemin2 levels. (B) Quantification of motor axon defects in the injected zebrafish embryos. (C-F) Lateral views of zebrafish trunk regions showing znp1-positive motor axons after immunostaining. Motor axon phenotypes in control embryos (C), Gemin2-deficient embryos (D), and embryos coinjected with Gemin2 Morpholino and U snRNPs. Arrowheads indicate truncated or branched axons in Gemin2 Morpholino-injected embryos.
Discussion
Although the genetics of spinal muscular atrophy are well known, the search for causal links between cellular processes and the disease is still at its beginning. One obstacle to this endeavor is the confusing variety of functions assigned to the SMN protein. Here, we focused on a well-understood activity of SMN by analyzing, in live organisms, whether the reduction of SMN protein impairs U snRNP assembly and thereby elicits motor axon degeneration. The first indications that SMN levels affect the formation of U snRNP in vivo were: (1) the low efficiency of assembly in extracts from cells whose SMN content had been reduced by RNAi to pathological levels, and (2) impaired synthesis of U snRNPs de novo in these cells. Encouraged by this observation, we proceeded to animal models and showed that an induced SMN protein deficiency was lethal to embryos of X. laevis or zebrafish. When the reduction of SMN in zebrafish was less stringent, survival rates improved and an SMA-like degeneration of motor axons came to light (McWhorter et al. 2003; this study).
Although the above findings suggest that reduced snRNP assembly is a cause of SMA, this correlation would be speculative if no direct correlation between motoneuron defects and U snRNP insufficiency could be established. We therefore reasoned that the data need to be in accordance with two postulates: First, the reduced expression of any other U snRNP assembly factor should likewise lead to the degeneration of motor axons. Second, the developmental defects observed should be preventable upon supplementation with the end product of this pathway; that is, isolated U snRNPs.
To test whether insufficient supply of any other U snRNP assembly factor elicits SMA-like symptoms, we knocked down in zebrafish the expression of pICln or Gemin2. Indeed, these animals showed the same erroneous axonal outgrowth as seen in the SMN morphants. Neither Gemin2 nor pICln has so far been directly linked to other functions of SMN such as axonal transport, splicing catalysis, or transcription. In the case of axonal transport, a participation of Gemin2 has been ruled out (Jablonka et al. 2001). However, it is worth to note that Gemin2, like SMN, is found not only in the cytosolic complex that mediates U snRNP assembly, but also in a nuclear complex. We have recently provided evidence that this complex is an intermediate of the assembly pathway rather than a unit of different function (Grimmler et al. 2005). The functional interplay between SMN and Gemin2 may therefore be restricted to the formation of U snRNPs. As for pICln, whose initial identification as a modulator of chloride conductance (Krapivinsky et al. 1994) awaits further validation, the only cellular pathway it appears to have in common with SMN is U snRNP assembly (Pu et al. 1999; Friesen et al. 2001; Meister et al. 2001b; Meister and Fischer 2002). Thus, silencing the expression of genes encoding Gemin2 or pICln is presumed to specifically affect the U snRNP assembly pathway. In view of these considerations, we believe that the reduction of SMN leads to impaired U snRNP assembly and thereby causes motor axon defects in zebrafish embryos. This conclusion is further supported by genetic studies in Drosophila melanogaster, showing that deficiency for the U snRNA export factor PHAX induces a neuromuscular disorder (G. Matera, pers. comm.).
Our second and probably most striking support for the correlation between motor axon degeneration and U snRNP assembly was the finding that purified U snRNPs injected into Xenopus or zebrafish embryos can compensate for the loss of SMN and Gemin2. U snRNPs rescued only SMN- and Gemin2-deprived embryos but not knock-downs directed against proteins of other function (the regulators of replication, Geminin H or L). Furthermore, U snRNPs that had been digested with RNase T1 failed to rescue SMN-deficiency in X. laevis embryos. Therefore, we consider the possibility unlikely that this rescue was conferred by potential impurities in the U snRNP preparation.
The ability of U snRNPs to prevent the degeneration of SMN- and Gemin2-deficient motoneurons brings an unanswered question back to mind, that is: Why does the reduced expression of ubiquitous proteins (SMN and Gemin2) cause a tissue-specific phenotype? The second question we may now add is: Why can U snRNPs, essential for the life of every cell, compensate this phenotype? In pursuit of an answer, it is worth discussing another disease related to RNA metabolism, termed Retinitis pigmentosa (RP). Like SMA, RP is characterized by the tissue-specific degeneration of cells, in this case retinal photoreceptors. Among many loci linked to RP, recent studies have identified mutations in three integral U snRNP components (for review, see Faustino and Cooper 2003). Presuming that the mutated proteins are inactive, as opposed to a gain of function, the resulting haploinsufficiency would be strikingly similar to SMA. What makes this comparison between SMA and RP so remarkable is the fact that in each case the affected tissue exhibits a tremendous turnover of specific mRNAs. While development and homeostasis of photoreceptors require large quantities of opsin (Korenbrot and Fernald 1989), axonal pathfinding of motoneurons requires a plethora of receptors and neurotrophins (for reviews, see Chisholm and Tessier-Lavigne 1999; Tear 1999; Beattie 2000). It is easy to envisage that cells cannot meet the demand for such proteins if splicing of their encoding pre-mRNAs is inefficient. Faustino and Cooper (2003) recently raised the hypothesis that the majority of ubiquitously expressed pre-mRNAs are more rapidly spliced than tissue-specific pre-mRNAs. Thinking along this line, a tissue-specific pre-mRNA containing suboptimal splice elements may thus become more sensitive to low levels of splicing factors than others. The reduction of proteins involved in the biogenesis of U snRNPs (as in SMA) or of U snRNP components (as in RP) would, in both cases, evoke a specific phenotype.
Alternatively, inefficient U snRNP assembly may only weaken the motoneuron such that disturbance of another SMN-dependent reaction can impair the cell cumulatively. SMN's proposed activity in axonal transport (Jablonka et al. 2001; Rossoll et al. 2003; Zhang et al. 2003) could be one such function, since SMN-deficient motoneurons exhibit severe defects in pathfinding (McWhorter et al. 2003; this study), a process that intimately depends on efficient transport (Steward 2002). A detailed evaluation of SMN's other activities in the context of SMN-deficient cells will be required to address this properly.
An intriguing question that comes with motoneuron rescue by U snRNPs is, at what time during development must SMN-deficiency be compensated? Zebrafish motoneurons are known to pass checkpoints as their axon projects through tissue (for review, see Beattie 2000). These checkpoints occur in spatial and temporal order and involve a series of cytokines and receptor proteins presented at or to the growth cone of the axon (for reviews, see Tessier-Lavigne and Goodman 1996; Chisholm and Tessier-Lavigne 1999). The two types of α-motoneurons that are found sensitive to the reduction of SMN, caudal primary (CaP) and variable primary neurons (VaP), are among the first to develop in the zebrafish embryo. Their fate is determined at the end of gastrulation, which, transformed to our experimental setup, begins only a few hours after the coinjection of the anti-SMN-Morpholino and U snRNPs. Therefore, albeit speculative, we favor the idea that there is a short time frame within which U snRNP levels are critical to development. The work presented here also raises the question of how robust the compensation of a SMN-deficient phenotype by U snRNPs stays over time. To this end, we know that rescued SMN morphants survive well beyond 31 h post-injection, which extends to the event of hatching. Future studies are under way to assess the persistence of this rescue throughout zebrafish ontogenesis.
Materials and methods
Cell culture, RNAi, and pulse-chase analysis
HeLa cells and human fibroblasts (Jablonka et al. 2001) were cultured in complete DMEM (Dulbecco's modified Eagle's medium containing glucose and L-glutamin [PAA laboratories], 10% fetal calf serum [FCS; PAA laboratories], penicillin [100 U/mL; Invitrogen], and streptomycin (0.1 mg/mL; Invitrogen]). For RNAi experiments, a 22-nt-long RNA duplex corresponding to nucleotides 183-205 of the SMN cDNA was generated. Cells were transfected with siRNAs using Oligofectamine reagent (QIAGEN) according to the manufacturer's protocol, and incubated at 37°C for the time indicated. For pulse-chase studies, cells were washed twice with PBS 72 h after transfection and incubated for 1 h in starvation medium (GIBCO-BRL) containing 10% FCS, 2 mM L-alanyl-L-glutamine, and 25 mM HEPES-KOH (pH 7.4). Then, [35S]methionine/[35S]cysteine (ProMix; Amersham-Biosciences) was added for 45 min. Cells were washed again with PBS and incubated for 1 h in complete DMEM.
Preparation of cellular extracts and immunoblotting
For the preparation of extracts from HeLa cells, the cells were trypsinized, washed with PBS, resuspended in lysis buffer (150 mM NaCl, 1 mM EDTA, 10 mM HEPES · KOH at pH 7.6, 1.5 mM DTT, 0.5% NP-40, 0.5 U/μL RNAsin [Promega], Complete protease inhibitor [Roche]), and homogenized by shearing with a 26G needle. The lysate was centrifuged for 30 min at 16,000g, and the supernatant was used for follow-up experiments. Extracts from whole Xenopus embryos were made by boiling the embryos in SDS sample buffer. Zebrafish embryos were frozen in liquid nitrogen, resuspended in lysis buffer, and sheared with a 26G needle. The extract was separated from insoluble matter by two centrifugations at 16,000g. The amount of protein in each extract was quantified by Bradford colorimetry and equal loading onto SDS-polyacrylamide gels was verified via immunoblotting using monoclonal α-actin antibody (Sigma). The antibodies used for the detection of assembly factors were 7B10 mouse monoclonal antibody for human SMN (Meister et al. 2000), MANSMA 7 and MANSMA 21 for zebrafish SMN (a kind gift of G. Morris, Robert Jones and Agnes Hunt Orthopaedic Hospital, Oswestry, UK), and affinity-purified rabbit polyclonal antibodies against Xenopus SMN, human pICln and human Gemin2, respectively.
Transcription of U snRNAs in vitro and reconstitution of U snRNPs
For U snRNP assembly reactions in vitro, U1 snRNA was co-transcriptionally labeled using [α-32P]UTP (3000 Ci/mmol; Amersham) and gel-purified as described previously (Meister et al. 2000). In a standard reconstitution assay, 25 fmol of [32P]-labeled U snRNA was incubated with 10 μL of HeLa cell extract (adjusted with PBS to 2.5 mg total protein/mL) in the presence of 2.5 pmol of unlabeled tRNA for 30 min at 37°C. For supershift analyses of complex M, Y12 antibody (2 μg ascite fluid) was added and the incubation was continued for 10 min. Samples were mixed with 1 volume of sample buffer (16% glycerol, 10 mg/mL Heparin, 0.025% bromphenol blue, 0.025% xylene cyanole) and analyzed by native RNA gel-electrophoresis as described previously (Meister et al. 2001a).
Immunoprecipitations
Antibodies were covalently bound to protein G-Sepharose beads (Amersham Biosciences) using dimethylpimelimidate (PIERCE) according to the manufacturer's protocol. Immunoprecipitations were carried out by incubating extracts with the immobilized antibodies (15 μg antibody/20 μL bead volume) for 1 h at 4°C. Beads were washed four times with PBS (pH 7.5)/0.01% NP-40 before proteins were extracted with phenol, precipitated by trichloroacetate, and analyzed by 12% SDS-PAGE. H20 antibody directed against the RNA cap structure (a kind gift from R. Lührmann, Max-Planck-Institute of Biophysical Chemistry, Gottingen, Germany) was used as a positive control, and M5 α-Flag antibody (Sigma) served as negative control. Isolations of U snRNPs from HeLa as well as XTC cells were performed as described before (Bringmann and Luhrmann 1986). For the digestion of human U snRNPs, the preparations were incubated overnight with 5 × 10-4 U/μL RNase T1.
Injection of X. laevis embryos
Eggs from X. laevis were fertilized in vitro and cultivated as described (Kuhl et al. 1996). At the two-cell stage, siRNAs (final concentration 0.2 μM or 2 μM, respectively) were injected into both cells, either alone or in combination with affinity-purified U snRNPs (final concentration 0.5-1 μM; generously provided by R. Lührmann). Embryos were grown until stage 13 in 1/10 MBSH [8.8 mM NaCl, 0.1 mM KCl, 0.24 mM NaHCO3, 0.08 mM MgSO4, 0.03 mM Ca(NO3)2, 0.04 mM CaCl2, and 1 mM HEPES at pH 7.4] at 16°C and scored for survival. Sequences of siRNAs were as follows (sense strands listed only): Xl-SMN #1 (5′-GCAUAUGAUAAAGCAGUGUdTdT-3′); Xl-SMN #2 (5′-GAGAGCUCUGAAGAAUGAGdTdT-3′); Xl-Gemin2 #1 (5′-GCAGCACGUUGUCCUGAUGdTdT-3′); Xl-Gemin2 #2 (5′-G UUGCGAAAGAAACAGACCdTdT-3′); Xl-GemininH #1 (5′-UACCAACAAGAAGCAGAGAdTdT-3′); Xl-GemininH #2 (5′-UCUCUGCUUCUUGUUGGUAdTdT-3′); Xl-GemininL #1 (5′-TCTGCATCTGGGTGCCTTGdTdT-3′); and Xl-GemininL #2 (5′-CAAGGCACCCAGAUGCAGAdTdT-3′).
Silencing of gene expression and U snRNP injections in zebrafish
Zebrafish EST sequences encoding putative orthologs of Gemin2 (accession nos. CN510554, AI793714) and pICln (NM_131424, AAH66713) were identified in NCBI databases using human sequences as queries. NCBI BLAST shows 68% identity and 83% similarity for Gemin2 or 67% identity and 78% similarity for pICln, respectively. The ESTs contained full-length open reading frames and 5′-UTR sequences that were used for designing Morpholino antisense oligonucleotides (Gene Tools). Zebrafish embryos were injected at the one- or two-cell stage with ∼0.5 nL of the following Morpholino antisense oligonucleotide solutions: SMN (5′-CGACATCTTCTGCACCATTGGC-3′, 3.1 mg/mL); Gemin2 (5′-TCAACTCCTCGGCGTCTGACTTCAT-3′, 6.3 mg/mL); pICln (5′-GGTAAACTCTTCAACAGCACCATCC-3′, 6.3 mg/mL), and standard control (5′-CCTCTTACCTCAGTTACAATTTATA-3′, 6.3 mg/mL). For rescue experiments, SMN and Gemin2 Morpholinos (final concentration 3.1 and 6.3 mg/mL, respectively) were coinjected with purified Xenopus or human U snRNPs (0.1 mg/mL; provided by R. Lührmann) as described. Following injection, the embryos were reared at 28°C until the prim-15 stage (30-31 h post-fertilization) (see Kimmel et al. 1995) and then subjected to in situ hybridizations and immunostainings.
RNA in situ hybridizations and immunostainings
Whole-mount RNA in situ hybridizations for islet2 expression in motoneurons (Appel et al. 1995) and myoD in somites (Weinberg et al. 1996) were performed as described earlier (Winkler et al. 2003). For whole-mount immunostainings, embryos were fixed overnight in 4% paraformaldehyde/PBS/0.1% Tween, dechorionated in PBS/0.1% Tween, and stored in methanol. After rehydration, embryos were soaked in water for 1 h and subsequently incubated in PBDT blocking solution (1% DMSO, 1% BSA, 0.5% Triton X-100, 2.5% goat serum in PBS) for 1 h. The samples were then incubated with the znp1 antibody (diluted 1:100 in PBDT) as described by Melancon et al. (1997) and detected using the Vectastain Elite kit (Vector Laboratories) with diaminobenzidine as chromogenic substrate. The znp1 antibody was kindly provided by the Zebrafish International Resource Center ZIRC (NIH #RR12546). Trunk regions from stained embryos were manually dissected from the remaining embryo using razor blades and mounted flat in 100% glycerol for photography.
Acknowledgments
We are indebted to Reinhard Lührmann for providing purified U snRNPs and the H20 antibody and to Glenn Morris for antibodies against zebrafish SMN. We also thank Greg Matera for sharing unpublished data, Andreas Brandstätter and Cordula Neuner for help with immunostainings, and Ashwin Chari for critical readings of the manuscript. This work was funded by grants of the Deutsche Forschungsgemeinschaft (SFB581, GRK1048, FOR426) and Families of SMA.
Supplemental material is available at http://www.genesdev.org.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.342005.
References
- Appel B., Korzh, V., Glasgow, E., Thor, S., Edlund, T., Dawid, I.B., and Eisen, J.S. 1995. Motoneuron fate specification revealed by patterned LIM homeobox gene expression in embryonic zebrafish. Development 121: 4117-4125. [DOI] [PubMed] [Google Scholar]
- Beattie C.E. 2000. Control of motor axon guidance in the zebrafish embryo. Brain Res. Bull. 53: 489-500. [DOI] [PubMed] [Google Scholar]
- Brahms H., Meheus, L., de Brabandere, V., Fischer, U., and Luhrmann, R. 2001. Symmetrical dimethylation of arginine residues in spliceosomal Sm protein B/B′ and the Sm-like protein LSm4, and their interaction with the SMN protein. RNA 7: 1531-1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branscombe T.L., Frankel, A., Lee, J.H., Cook, J.R., Yang, Z., Pestka, S., and Clarke, S. 2001. PRMT5 (Janus kinase-binding protein 1) catalyzes the formation of symmetric dimethylarginine residues in proteins. J. Biol. Chem. 276: 32971-32976. [DOI] [PubMed] [Google Scholar]
- Bringmann P. and Luhrmann, R. 1986. Purification of the individual snRNPs U1, U2, U5 and U4/U6 from HeLa cells and characterization of their protein constituents. EMBO J. 5: 3509-3516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartegni L. and Krainer, A.R. 2002. Disruption of an SF2/ASF-dependent exonic splicing enhancer in SMN2 causes spinal muscular atrophy in the absence of SMN1. Nat. Genet. 30: 377-384. [DOI] [PubMed] [Google Scholar]
- Chisholm A. and Tessier-Lavigne, M. 1999. Conservation and divergence of axon guidance mechanisms. Curr. Opin. Neurobiol. 9: 603-615. [DOI] [PubMed] [Google Scholar]
- Faustino N.A. and Cooper, T.A. 2003. Pre-mRNA splicing and human disease. Genes & Dev. 17: 419-437. [DOI] [PubMed] [Google Scholar]
- Friesen W.J., Paushkin, S., Wyce, A., Massenet, S., Pesiridis, G.S., Van Duyne, G., Rappsilber, J., Mann, M., and Dreyfuss, G. 2001. The methylosome, a 20S complex containing JBP1 and pICln, produces dimethylarginine-modified Sm proteins. Mol. Cell. Biol. 21: 8289-8300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimmler M., Bauer, L., Nousiainen, M., Korner, R., Meister, G., and Fischer, U. 2005. Phosphorylation regulates the activity of the SMN complex during assembly of spliceosomal U snRNPs. EMBO Rep. 6: 70-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubitz A.K., Feng, W., and Dreyfuss, G. 2004. The SMN complex. Exp. Cell Res. 296: 51-56. [DOI] [PubMed] [Google Scholar]
- Hsieh-Li H.M., Chang, J.G., Jong, Y.J., Wu, M.H., Wang, N.M., Tsai, C.H., and Li, H. 2000. A mouse model for spinal muscular atrophy. Nat. Genet. 24: 66-70. [DOI] [PubMed] [Google Scholar]
- Jablonka S., Bandilla, M., Wiese, S., Buhler, D., Wirth, B., Sendtner, M., and Fischer, U. 2001. Co-regulation of survival of motor neuron (SMN) protein and its interactor SIP1 during development and in spinal muscular atrophy. Hum. Mol. Genet. 10: 497-505. [DOI] [PubMed] [Google Scholar]
- Jablonka S., Holtmann, B., Meister, G., Bandilla, M., Rossoll, W., Fischer, U., and Sendtner, M. 2002. Gene targeting of Gemin2 in mice reveals a correlation between defects in the biogenesis of U snRNPs and motoneuron cell death. Proc. Natl. Acad. Sci. 99: 10126-10131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kambach C., Walke, S., Young, R., Avis, J.M., de la Fortelle, E., Raker, V.A., Luhrmann, R., Li, J., and Nagai, K. 1999. Crystal structures of two Sm protein complexes and their implications for the assembly of the spliceosomal snRNPs. Cell 96: 375-387. [DOI] [PubMed] [Google Scholar]
- Kashima T. and Manley, J.L. 2003. A negative element in SMN2 exon 7 inhibits splicing in spinal muscular atrophy. Nat. Genet. 34: 460-463. [DOI] [PubMed] [Google Scholar]
- Kimmel C.B., Ballard, W.W., Kimmel, S.R., Ullmann, B., and Schilling, T.F. 1995. Stages of embryonic development of the zebrafish. Dev. Dyn. 203: 253-310. [DOI] [PubMed] [Google Scholar]
- Korenbrot J.I. and Fernald, R.D. 1989. Circadian rhythm and light regulate opsin mRNA in rod photoreceptors. Nature 337: 454-457. [DOI] [PubMed] [Google Scholar]
- Krapivinsky G.B., Ackerman, M.J., Gordon, E.A., Krapivinsky, L.D., and Clapham, D.E. 1994. Molecular characterization of a swelling-induced chloride conductance regulatory protein, pICln. Cell 76: 439-448. [DOI] [PubMed] [Google Scholar]
- Kuhl M., Finnemann, S., Binder, O., and Wedlich, D. 1996. Dominant negative expression of a cytoplasmically deleted mutant of XB/U-cadherin disturbs mesoderm migration during gastrulation in Xenopus laevis. Mech. Dev. 54: 71-82. [DOI] [PubMed] [Google Scholar]
- Lefebvre S., Burglen, L., Reboullet, S., Clermont, O., Burlet, P., Viollet, L., Benichou, B., Cruaud, C., Millasseau, P., Zeviani, M., et al. 1995. Identification and characterization of a spinal muscular atrophy-determining gene. Cell 80: 155-165. [DOI] [PubMed] [Google Scholar]
- Lefebvre S., Burlet, P., Liu, Q., Bertrandy, S., Clermont, O., Munnich, A., Dreyfuss, G., and Melki, J. 1997. Correlation between severity and SMN protein level in spinal muscular atrophy. Nat. Genet. 16: 265-269. [DOI] [PubMed] [Google Scholar]
- Lorson C.L. and Androphy, E.J. 2000. An exonic enhancer is required for inclusion of an essential exon in the SMA-determining gene SMN. Hum. Mol. Genet. 9: 259-265. [DOI] [PubMed] [Google Scholar]
- Lorson C.L., Hahnen, E., Androphy, E.J., and Wirth, B. 1999. A single nucleotide in the SMN gene regulates splicing and is responsible for spinal muscular atrophy. Proc. Natl. Acad. Sci. 96: 6307-6311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGarry T.J. 2002. Geminin deficiency causes a Chk1-dependent G2 arrest in Xenopus. Mol. Biol. Cell 13: 3662-3671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McWhorter M.L., Monani, U.R., Burghes, A.H., and Beattie, C.E. 2003. Knockdown of the survival motor neuron (Smn) protein in zebrafish causes defects in motor axon outgrowth and pathfinding. J. Cell Biol. 162: 919-931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meister G. and Fischer, U. 2002. Assisted RNP assembly from purified components: SMN- and pICln-complexes cooperate in the formation of spliceosomal U snRNPs. EMBO J. 21: 5853-5863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meister G., Buhler, D., Laggerbauer, B., Zobawa, M., Lottspeich, F., and Fischer, U. 2000. Characterization of a nuclear 20S complex containing the survival of motor neurons (SMN) protein and a specific subset of spliceosomal Sm proteins. Hum. Mol. Genet. 9: 1977-1986. [DOI] [PubMed] [Google Scholar]
- Meister G., Buhler, D., Pillai, R., Lottspeich, F., and Fischer, U. 2001a. A multiprotein complex mediates the ATP-dependent assembly of spliceosomal U snRNPs. Nat. Cell Biol. 3: 945-949. [DOI] [PubMed] [Google Scholar]
- Meister G., Eggert, C., Buehler, D., Brahms, H., Kambach, C., and Fischer, U. 2001b. Methylation of Sm proteins by a complex containing PRMT5 and the putative U snRNP assembly factor pICln. Curr. Biol. 11: 1990-1994. [DOI] [PubMed] [Google Scholar]
- Melancon E., Liu, D.W., Westerfield, M., and Eisen, J.S. 1997. Pathfinding by identified zebrafish motoneurons in the absence of muscle pioneers. J. Neurosci. 17: 7796-7804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monani U.R., Sendtner, M., Coovert, D.D., Parsons, D.W., Andreassi, C., Le, T.T., Jablonka, S., Schrank, B., Rossol, W., Prior, T.W., et al. 2000. The human centromeric survival motor neuron gene (SMN2) rescues embryonic lethality in Smn-/- mice and results in a mouse with spinal muscular atrophy. Hum. Mol. Genet. 9: 333-339. [DOI] [PubMed] [Google Scholar]
- Pellizzoni L., Kataoka, N., Charroux, B., and Dreyfuss, G. 1998. A novel function for SMN, the spinal muscular atrophy disease gene product, in pre-mRNA splicing. Cell 95: 615-624. [DOI] [PubMed] [Google Scholar]
- Pellizzoni L., Yong, J., and Dreyfuss, G. 2002. Essential role for the SMN complex in the specificity of snRNP assembly. Science 298: 1775-1779. [DOI] [PubMed] [Google Scholar]
- Pillai R.S., Grimmler, M., Meister, G., Will, C.L., Luhrmann, R., Fischer, U., and Schumperli, D. 2003. Unique Sm core structure of U7 snRNPs: Assembly by a specialized SMN complex and the role of a new component, Lsm11, in histone RNA processing. Genes & Dev. 17: 2321-2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pu W.T., Krapivinsky, G.B., Krapivinsky, L., and Clapham, D.E. 1999. pICln inhibits snRNP biogenesis by binding core spliceosomal proteins. Mol. Cell. Biol. 19: 4113-4120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossoll W., Jablonka, S., Andreassi, C., Kroning, A.K., Karle, K., Monani, U.R., and Sendtner, M. 2003. Smn, the spinal muscular atrophy-determining gene product, modulates axon growth and localization of β-actin mRNA in growth cones of motoneurons. J. Cell Biol. 163: 801-812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steward O. 2002. Translating axon guidance cues. Cell 110: 537-540. [DOI] [PubMed] [Google Scholar]
- Strasswimmer J., Lorson, C.L., Breiding, D.E., Chen, J.J., Le, T., Burghes, A.H., and Androphy, E.J. 1999. Identification of survival motor neuron as a transcriptional activator-binding protein. Hum. Mol. Genet. 8: 1219-1226. [DOI] [PubMed] [Google Scholar]
- Tear G. 1999. Neuronal guidance. A genetic perspective. Trends Genet. 15: 113-118. [DOI] [PubMed] [Google Scholar]
- Tessier-Lavigne M. and Goodman, C.S. 1996. The molecular biology of axon guidance. Science 274: 1123-1133. [DOI] [PubMed] [Google Scholar]
- Weinberg E.S., Allende, M.L., Kelly, C.S., Abdelhamid, A., Murakami, T., Andermann, P., Doerre, O.G., Grunwald, D.J., and Riggleman, B. 1996. Developmental regulation of zebrafish MyoD in wild-type, no tail and spadetail embryos. Development 122: 271-280. [DOI] [PubMed] [Google Scholar]
- Will C.L. and Luhrmann, R. 2001. Spliceosomal UsnRNP biogenesis, structure and function. Curr. Opin. Cell Biol. 13: 290-301. [DOI] [PubMed] [Google Scholar]
- Winkler C., Schafer, M., Duschl, J., Schartl, M., and Volff, J.N. 2003. Functional divergence of two zebrafish midkine growth factors following fish-specific gene duplication. Genome Res. 13: 1067-1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H.L., Pan, F., Hong, D., Shenoy, S.M., Singer, R.H., and Bassell, G.J. 2003. Active transport of the survival motor neuron protein and the role of exon-7 in cytoplasmic localization. J. Neurosci. 23: 6627-6637. [DOI] [PMC free article] [PubMed] [Google Scholar]