Abstract
Isolated soil DNA from an oak-hornbeam forest close to Cologne, Germany, was suitable for PCR amplification of gene segments coding for the 16S rRNA and nitrogenase reductase (NifH), nitrous oxide reductase (NosZ), cytochrome cd1-containing nitrite reductase (NirS), and Cu-containing nitrite reductase (NirK) of denitrification. For each gene segment, diverse PCR products were characterized by cloning and sequencing. None of the 16S rRNA gene sequences was identical to any deposited in the data banks, and therefore each of them belonged to a noncharacterized bacterium. In contrast, the analyzed clones of nifH gave only a few different sequences, which occurred many times, indicating a low level of species richness in the N2-fixing bacterial population in this soil. Identical nifH sequences were also detected in PCR amplification products of DNA of a soil approximately 600 km distant from the Cologne area. Whereas biodiversity was high in the case of nosZ, only a few different sequences were obtained with nirK. With respect to nirS, cloning and sequencing of the PCR products revealed that many false gene segments had been amplified with DNA from soil but not from cultured bacteria. With the 16S rRNA gene data, many sequences of uncultured bacteria belonging to the Acidobacterium phylum and actinomycetes showed up in the PCR products when isolated DNA was used as the template, whereas sequences obtained for nifH and for the denitrification genes were closely related to those of the proteobacteria. Although in such an experimental approach one has to cope with the enormous biodiversity in soils and only a few PCR products can be selected at random, the data suggest that denitrification and N2 fixation are not genetic traits of most of the uncultured bacteria.
Microbial life in soils is complex. It is estimated that 5 × 103 to 1 × 104 different bacterial ribotypes occur per g of soil (7, 41). Knowledge of the number of species (species richness) and the relative fraction of each of the different species (species evenness) within a soil is a prerequisite to understanding its biological activities, e.g., in the nitrogen cycle. Soils are known to be a significant sink and/or source for N2, N2O, and NO, and the last two gases have detrimental impacts on the atmosphere (9). The microbial basis for the production and consumption of N2O and NO is complicated (8). Both gases are formed in soils as side products in nitrification by either autotrophic or heterotrophic nitrifiers (42). In denitrification, nitrate serves as an electron acceptor alternative to O2 and is reduced to N2 via nitrite, NO, and N2O. The enzymes catalyzing the different steps (NO3− → NO2−, nitrate reductase; NO2− → NO, either cytochrome cd1- or Cu-containing nitrite reductase, depending on the organism; NO → N2O, cytochrome bc-containing nitric oxide reductase; N2O → N2, Cu-containing nitrous oxide reductase) seemingly do not completely use up their respective substrates. This might also result in a partial release of NO and N2O to the atmosphere. In addition, bacteria that do not possess the full set of denitrification enzymes are known. For instance, some reduce nitrate to N2O but do not possess the N2O reductase (47). Others involve only single steps; e.g., Escherichia coli performs nitrate ammonification and additionally converts N2O to N2 (16). In dinitrogen fixation, N2O is an alternative substrate to nitrogenase (6). Thus, NO and N2O fluxes between soil and atmosphere result from complex and little-understood microbial activities which proceed simultaneously in soils (8).
Molecular techniques provide relatively new avenues for studying bacterial communities participating in the biological N cycle. Since genes coding for the different enzymes of denitrification, N2 fixation, and nitrification have been sequenced from diverse organisms, 0.5- to 1.0-kb gene probes can be developed which recognize the target DNA from a broad range of organisms by Southern hybridization. Alternatively, oligomer primers can be developed from strongly conserved motifs which allow the amplification of gene segments from purified culture or soil DNA by PCR.
In initial studies, this laboratory (17, 19) performed hybridization experiments with probes of genes coding for different steps of denitrification and for nitrogenase reductase (nifH) to screen for the occurrence of these genes in bacteria isolated from different soils and enriched in media. These studies were only of limited value, because the number of published sequences for the genes was small when the studies were initiated. Therefore, universal primers for PCR and “broad-band” gene probes recognizing denitrification genes of bacteria from all taxonomic affinities could not be developed then. Moreover, only 0.1 to 5% of the total population of soil bacteria can be cultivated by current methods (2, 24). Earlier studies (17, 19) showed that N2-fixing and denitrifying bacteria occur abundantly in the upper soil layer (∼5-cm depth), but the majority of bacteria, which are still uncultured, could not be assessed then. Others employed the terminal restriction fragment length polymorphism (t-RFLP) pattern technique, which allows the determination of the number and abundance of numerically different ribotypes within the total amplified DNA (20) and can be taken to monitor gradients of denitrifying, N2-fixing, nitrifying, or other bacteria within aqueous sediments and soils (3, 14, 27, 34) but not their species identity. In a different approach, our laboratory isolated and purified DNA from different soil layers and also assessed the relative distribution of denitrifying and N2-fixing bacteria by hybridization with the now improved DNA probes (25, 26). The quantification of the signal strengths obtained also indicated that denitrifying and N2-fixing bacteria predominantly occur in the upper soil layer and that their abundance decreases with the soil depth. Any information about the identities of the species of uncultured bacteria in soils can, however, come only from efforts to sequence their DNA.
Although we were well aware of the fact that only a rather limited range of bacteria can be identified by such a sequencing approach, the present study focused on amplifying segments of genes coding for nitrogenase reductase and for different steps of denitrification. To have an estimate of the total bacterial life in this soil, a gene segment coding for the 16S rRNA was also amplified from DNA of this soil by PCR. The PCR products obtained were cloned and characterized by sequencing. This is seemingly the first attempt to assess the biodiversity of denitrifying and N2-fixing microorganisms in parallel with the total bacterial population in an environmental sample. It provided unexpected results compared to data published for denitrifying bacteria in marine sediments (5, 32, 33).
MATERIALS AND METHODS
Location of the soil sampling sites.
Samples were taken at depths of 5 and 25 cm from cores of an alluvial, acid soil of an oak-hornbeam forest (Chorbusch Forest) located about 20 km west of the city of Cologne. This forest (51°02′50″N, 06°48′40″E) carries a largely undisturbed vegetation. Its soil is rich in nitrogen but low in phosphorus. Details of the soil parameters have been described in a preceding publication (26). For comparison, some bacterial PCR products of a strongly acid spruce stand taken at a 5-cm depth at Villingen, Black Forest (48°02′30″N, 08°23′00″E) were also analyzed. This soil has also been characterized previously (25).
Isolation of bacteria and extraction of DNA from the soil samples.
For the isolation of bacteria, 100 mg of soil was vigorously mixed on a magnetic stirrer for 3 h in 50 ml of phosphate-buffered saline to get a homogeneous suspension. Then 50 μl was plated on agar containing either yeast extract mannitol medium (YEM) or heterotrophic mineral medium (17, 25) and incubated at 30°C. Single colonies were obtained by isolating and repeatedly plating them on agar containing the media.
For the extraction of DNA from the different soils, samples were taken to the Cologne laboratory and used for the isolation not more than 1 day after sampling. The extraction was performed by conventional steps, exactly as described previously (26). The pretreatment step is worth mentioning: soil samples (5 g) were suspended in 10 ml of 0.1% Na4P2O7 in 10 mM Tris-HCl buffer (pH 8.0)-1 mM EDTA, incubated at room temperature (10 min), and centrifuged (10 min, 6000 × g). This step removed most of the humic acids, free DNA, and other interfering substances but left the soil bacteria intact.
Primers used for PCR amplifications.
Table 1 contains the information about the primers used to amplify the different gene segments encoding 16S rRNA, cytochrome cd1-containing nitrite reductase (nirS gene), Cu-containing nitrite reductase (nirK), nitrous oxide reductase (nosZ), and dinitrogenase reductase (nifH). The nosZ-F-nosZ-R and nifH-F-nifH-R primer pairs were newly developed and were, therefore, tested with DNA from laboratory cultures. Distinct PCR products of the expected sizes were obtained with DNA from the organisms listed in Table 2. In these, the correctness of the amplification products listed in boldface was confirmed by sequencing. As expected, no PCR product for nosZ was obtained in the cases of Escherichia coli K-12 and Pseudomonas chlororaphis (12).
TABLE 1.
PCR primers used in the present study
| Gene | Primer | Sequence (5′ → 3′)a | Derivation sequence | Organism (GenBank accession no.) | Reference for the derivation sequence |
|---|---|---|---|---|---|
| 16S rRNA | EUB338-F | ACT CCT ACG GGA GGC AGC | N338-N355 | Escherichia coli (7415865) | 43 |
| RM2-R | GAT CTC TAC GCA TTT CAC CGC TAC | N685-N708 | |||
| nirS | KA3-F | CAC GGY GTB CTG CGC AAG GGC GC | N292-N314 | Paracoccus denitrificans (U05002) | 10 |
| KA25-R | CGC CAC GCG CGG YTC SGG GTG GTA | N967-N990 | |||
| nirK | K15-F | GGC ATG GTA CCT TGG CAC GTA ACC TCG GGC | N526-N555 | Alcaligenes faecalis (D13155) | 28 |
| K16-R | CAT TAG ATC GTC GTT CCA ATC ACC GGT | N1075-N1101 | |||
| nosZ | nosZ-F | CGY TGT TCM TCG ACA GCC AG | N1211-N1230 | Paracoccus denitrificans (398932) | 13 |
| nosZ-R | CAT GTG CAG NGC RTG GCA GAA | N1897-N1917 | |||
| nifH | nifH-F | AAA GGY GGW ATC GGY AAR TCC ACC AC | N34-N59 | Sinorhizobium meliloti (46285) | 40 |
| nifH-R or | TTG TTS GCS GCR TAC ATS GCC ATC AT | N466-N491 | |||
| K07-F | GCG TTC TAC GGT AAG GGC GGT ATC GGN AAR | N22-N51 | |||
| AMR-R | GCT ACT ACY TCG CCS GA | N457-N473 |
Boldface letters denote degenerate positions. B, G + T; M, A + C; N, A + C + G + T; R, A + G; S, G + C; Y, C + T; W, A + T.
TABLE 2.
The occurrence of genes coding for denitrification enzymes and nitrogenase reductase in laboratory cultures assessed by PCR amplification
| Bacterial strain | PCR result for indicated gene (primer pair)
|
||||
|---|---|---|---|---|---|
| nirS (KA3-KA25) | nirK (K15-K16) | nosZ (nosZ-F-nosZ-R) | nifH (nifH-F-nifH-R) | nifH (K07-AMR) | |
| Proteobacteria, α subdivision | |||||
| Azospirillum amazonense Y-1T | − | − | − | + | + |
| Azospirillum brasilense Sp7030 (nirS mutant) | − | − | + | + | + |
| Azospirillum brasilense Sp7T | + | − | + | + | + |
| Azospirillum doebereinerae GSF 71T | − | + | + | + | + |
| Azospirillum halopraeferens Au4T | + | − | + | + | + |
| Azospirillum irakense KA3 | − | + | + | + | + |
| Azospirillum irakense KBCIT | − | + | + | + | |
| Azospirillum largomobile ACM 2041 | + | − | + | + | |
| Azospirillum lipoferum Sp59bT | + | − | + | + | + |
| Azospirillum strain SpA1-3 | + | + | + | + | |
| Azorhizobium caulinodans ORS 571T | − | + | + | + | |
| Blastobacter denitrificans IFAM 1005T | − | − | − | − | |
| Bradyrhizobium japonicum LMG 6137 | − | + | + | − | |
| Gluconacetobacter diazotrophicus PAL3 | + | − | − | + | + |
| Ochrobactrum anthropi LMG 3331 | − | + | + | − | − |
| Paracoccus denitrificans Pd1222 | + | + | − | − | |
| Rhizobium trifolii ANV 845 | − | − | + | + | |
| Rhizobium tropici CIAT 899 | − | + | + | + | |
| Rhodospirillum rubrum S1T | + | − | + | + | |
| Sinorhizobium meliloti Rm1021 | − | + | + | + | + |
| Proteobacteria, β subdivision | |||||
| Achromobacter ruhlandii DSM 653T | − | − | + | − | |
| Achromobacter xylosoxidans NCIMB 11015 | − | + | − | − | |
| Aquaspirillum autotrophicum SA32T | − | + | + | + | |
| “Burkholderia brasilensis” M130 | + | − | − | + | |
| Herbaspirillum frisingense GSF 30T | − | + | + | + | + |
| Herbaspirillum rubrisubalbicans LMG 1278 | − | + | − | + | + |
| Herbaspirillum seropedicae Z67T | + | − | + | + | + |
| Herbaspirillum seropedicae Z78 | − | + | − | + | + |
| Ralstonia eutropha H16 | + | + | + | − | |
| Proteobacteria, γ subdivision | |||||
| Azomonas agilis DSM 375 | − | + | + | + | |
| Azorhizophilus paspali DSM 2283 | − | + | + | ||
| Azotobacter vinelandii DSM 2289 | − | + | + | ||
| Enterobacter cloacae NCIMB 11463 | − | − | + | + | |
| Escherichia coli K-12 | − | − | − | − | |
| Frateuria aurantia DSM 6220T | − | + | + | + | |
| Pseudomonas aeruginosa DSM 6195T | + | + | − | − | |
| Pseudomonas chlororaphis NCIMB 9030 | − | − | − | − | |
| Pseudomonas stutzeri (= Alcaligenes faecalis) A15 | + | + | + | + | |
| Pseudomonas stutzeri ZoBell 632 | + | + | + | ||
| Firmicutes, Actinomycetales | |||||
| Frankia alni ArI3 (DSM 44251) | − | − | + | + | |
| Frankia strain DSM 43829 | − | + | + | ||
| Frankia strain DSM 44263 | − | + | + | ||
| Kocuria varians LTH 1429 | − | − | − | − | |
| Micrococcus luteus DSM 20030T | − | − | − | − | |
| Nocardioides simplex DSM 20130T | − | − | − | − | |
| Rhodococcus erythropolis (from H. Papen, Garmisch- Partenkirchen) | − | − | − | + | |
| Firmicutes, Bacillus/Clostridium group | |||||
| Clostridium pasteurianum DSM 525T | − | − | + | + | |
| Paenibacillus validus DSM 3037T | − | − | + | + | |
| Staphylococcus carnosus LTH 55 | − | − | − | − | |
| Cyanobacteria | |||||
| Anabaena variabilis ATCC 29413 | + | ||||
| “Chroococcidiopsis thermalis” PCC 7203 | + | ||||
| Microcystis strain PCC 7806 | − | ||||
| Nostoc muscorum Agardh CCAP 1453/12 | + | ||||
| Plectonema boryanum PCC 6306 | + | ||||
| Synechococcus strain PCC 7942 | − | ||||
+, PCR product of the right size obtained; −, no PCR product obtained; boldface +, PCR product was cloned and sequenced and the sequence was deposited in the data banks.
PCR, sequencing reactions, and sequence comparison.
The following steps were employed in the PCR: a 4-min hot start at 97°C before Taq DNA polymerase (MasterTaq kit; Eppendorf, Hamburg, Germany) was added; 1 cycle of denaturation at 96°C for 20 s, annealing at 65°C for 30 s, and elongation at 72°C for 30 s; 2 cycles of denaturation at 96°C for 20 s, annealing at 62°C for 30 s, and elongation at 72°C for 35 s; 3 cycles of denaturation at 96°C for 20 s, annealing at 59°C for 30 s, and elongation at 72°C for 40 s; 4 cycles of denaturation at 96°C for 20 s, annealing at 56°C for 30 s, and elongation at 72°C for 45 s; 5 cycles of denaturation at 96°C for 20 s, annealing at 53°C for 30 s, and elongation at 72°C for 50 s; 25 cycles of denaturation at 94°C for 20 s, annealing at 50°C for 45 s, and elongation at 72°C for 60 s; and an extension at 72°C for 10 min. The products obtained were purified with a Qiaquick gel extraction kit (QIAGEN, Hilden, Germany) and cloned into the pGEM-T-Easy vector (Promega, Madison, Wis.) according to the protocols of the manufacturers. The plasmids were transformed into competent E. coli XL1-Blue by the heat shock method described in reference 15. Sequencing was performed on an ABI sequencer using the ABI PRISM dye terminator cycle sequencing reaction kit (Perkin-Elmer, Foster City, Calif.). Sequence data were compared with sequences in the National Center for Biotechnology Information data bank using the BLAST program (1). Alignment of DNA sequences was done with the CLUSTAL X program (39), and phylograms were edited with TreeView (29). Sequences of the PCR products were always analyzed by editing out primer sequences. Phylograms were constructed by the neighbor-joining method with 1,000 replicate trees (31). The method of Kimura was employed to correct for multiple substitutions (see reference 39).
Nucleotide sequence accession numbers.
Nucleotide sequence accession numbers were AF315402 to AF315425 and AY072242 to AY072262 for the 16S rRNA, AF315434 to AF315457 and AY072227 to AY072241 for nosZ; AY072278 to AY072282 for nirS, AY072271 to AY072277 for nirK, and AF315426 to AF315433 and AY072265 to AY072270 for nifH.
RESULTS
DNA contents in the different layers of a forest soil.
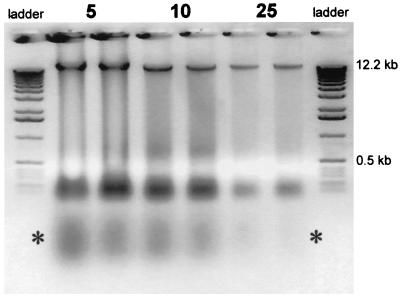
DNA isolation from the 5-, 10-, and 25-cm layers of the acid soil of an oak-hornbeam forest close to Cologne, Germany, was performed as described by Mergel et al. (26). The typical banding pattern on a 0.7% agarose gel is shown in Fig. 1. The DNA band at ≥12.2 kb showed little smearing, indicating that the isolation procedure did not degrade the DNA into small fragments. Two independent DNA isolations from the same soil sample gave approximately the same band intensities (Fig. 1). Signal strength obtained for the isolated DNA decreased with the depth of the soil, which might reflect the lower number of bacteria in the deeper soil layers. The preparation at this stage contained a lot of RNA (band below 0.5 kb) and also other interfering material (below this band, marked with an asterisk in Fig. 1) which showed a yellow-greenish autofluorescence (probably mostly humic acids). When the DNA band was cut out and eluted, the ratio of the values at 260 and 280 nm was 1.4 ± 0.2 (n = 3). The yields from two different preparations were 15.1 and 15.4 μg/g (fresh weight) of soil at the 5-cm layer and twice 2.1 μg/g at 25 cm in the case of the Chorbusch soil. With the Villingen samples, the preparation yielded 9.7 ± 1.2 μg of DNA/g of soil (n = 3) of the same purity in the 5-cm layer. Approximately 30% of the total DNA preparations were amenable to DNA amplification by PCR, irrespective of the soil used. Alternatively, when the DNA was purified with a UltraCleanSoil DNA kit (MoBio Laboratories, Solana Beach, Calif.), about 50% of the preparations could successfully be used for DNA amplifications but the yield was lower, e.g., only 0.7 ± 0.1 μg (n = 3) per g (fresh weight) with the Chorbusch soil from the 5-cm layer and thus ∼5% of that obtained by the method developed in our laboratory and described in reference 25. It must be kept in mind that the DNA quality might have an impact on the following sequence recovery efforts.
FIG. 1.
The DNA contents in different soil layers of an oak-hornbeam forest were extracted as described in Materials and Methods. The DNA extracted from 0.2 g (wet weight) of soil was blotted onto each lane. Samples were taken from 5-, 10-, and 25-cm depths. Parallels on these 0.7% agarose gels are for two independent DNA extractions from the same soil sample. The ∗ denotes the smear under the RNA band, due probably to humic acids and other interfering substances.
16S rRNA gene sequences of PCR products obtained with DNA isolated either from soils or from cultured bacteria.
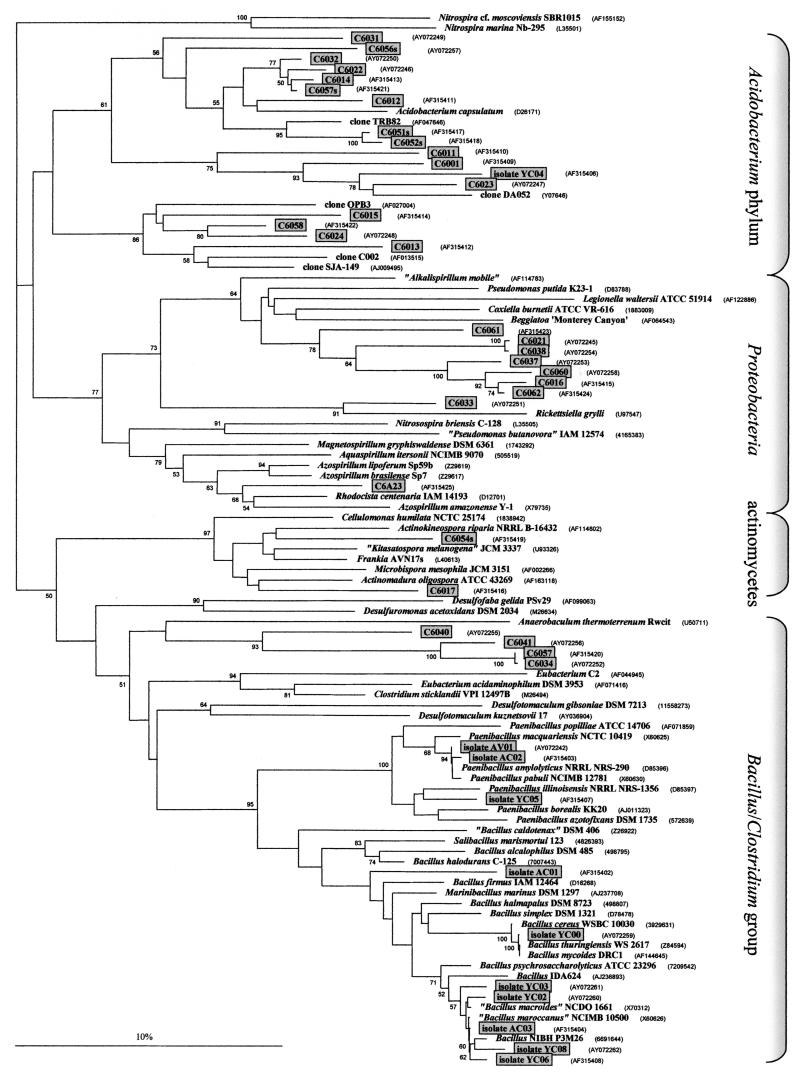
The region of the 16S rRNA gene amplified contained two hypervariable stretches (nucleotide positions N450 to N480, helix H17, and positions N600 to N640, helix H21), whereas the region from N500 to N530 located in H18 was strongly conserved (numbers refer to the nomenclature of Thermus thermophilus [45]). The 350-bp gene segment was short enough to be sequenced in one single step but sufficient to provide the necessary information for phylogenetic analyses. All together, 45 randomly selected PCR products were sequenced: 31 of the amplification products obtained with DNA from the 5-cm layer of the Chorbusch soil, 10 from cultured bacteria from the same layer, and 4 from cultured bacteria from the Villingen soil. Sequences of all cultured bacteria except one were of monophyletic origin and belonged to the Bacillus/Paenibacillus clade (Fig. 2 ). The one exception (isolate YC04) showed 77.2% identity to Acidobacterium capsulatum. Isolates AV01 to AV04 were 99.7% identical and thus might come from the same bacterial species since a difference of 0.3% might reflect a PCR amplification or a sequencing error in one position.
FIG. 2.
Phylogram of the 16S rRNA gene sequences. The sequences of two Nitrospira spp. served as an outgroup to root the phylogram. The clones that are framed and shaded were sequenced and deposited by us. The abbreviations used for the clones are explained in Materials and Methods. The alignment was done with 328 nt. The dendrogram was derived from 261 nt because of deletions and/or insertions in some sequences. The 59 GenBank sequences included in the phylogram were next most closely related to the clones obtained by us. These 59 were selected by employing the Ribosomal Database Project Phylip interface, followed by a BLASTN search. The scale bar indicates 10% nucleotide substitutions. Numbers at the branches indicate the percentages of the occurrence of the respective nodes in a bootstrap analysis of 1,000 resamplings. Only values above 50% are shown. Abbreviations (for all subsequent figures): A, cultured bacterium grown in Azospirillum medium; Y, cultured bacterium grown in YEM; C, amplification product of soil DNA or of a cultured bacterium from Chorbusch soil; V, amplification product of soil DNA or of a cultured bacterium from Villingen; F, amplification product from nifH; S, amplification product from nirS; K, amplification product from nirK; Z, amplification product from nosZ; 6, 16S rRNA; F, forward primer; R, reverse primer. The small letters behind the numbers are internal codes.
The sequenced PCR products obtained with isolated soil DNA showed a different picture. Out of a total of 31 PCR products, 16 belonged to the Acidobacterium phylum, as did isolate YC04. These 16 sequences could be divided into three different subgroups. One of these was apart from the other two. Eight other sequences bore a weak resemblance to those from γ-proteobacteria in the proximity of Coxiella burnetii, Legionella waltersii, and Pseudomonas putida, whereas clones C6017 and C6054s belonged to the actinomycetes. Clones C6034, C6040, C6041, and C6057 stood apart but were distantly related to Anaerobaculum thermoterrenum (75.8 to 80.7% identity). However, the sequence identity to Desulfotomaculum spp., Nostoc spp., actinomycetes, or clostridia was nearly as high (73 to 79%). Thus, these clones might belong to an unknown bacterial clade. Clone C6A23 revealed the sequence of an α-proteobacterium with relatedness to Azospirillum. It should be stressed that none of the sequences, either from cultured bacteria or from the PCR products of the 16S rRNA gene, obtained with isolated DNA as the template, was identical with any sequence deposited in the data banks.
Sequences obtained both from bacteria isolated from the soils and from the clones of the PCR products from the soil DNA were compared with 59 GenBank sequences of the organisms listed in Fig. 2. Each of the new sequences and its next most closely related sequence in the data banks were compared in pairs. The cultured bacteria from the Chorbusch soil had an average relatedness to their next most closely related neighbors of 98.6 ± 1.8%, with maximal and minimal sequence identities of 99.7 and 93.6%, respectively (however, isolate YC04 was not considered in this calculation). Corresponding values for the clones from the isolated soil DNA were 85.7% ± 5.0%, with a minimum of 76.5% and a maximum of 92.7%. Thus, in the 16S rRNA gene segment analyzed, the cultured bacteria were much more closely related to bacteria described in the data banks than were the PCR clones of the soil DNA, which might be mainly as-yet-uncultured bacteria.
Some control experiments must be mentioned here. In the absence of template DNA, no PCR products were obtained under the conditions employed and with the primers for the 16S rRNA gene segment (also for the other genes mentioned below). The test for chimeric artifacts (23) indicated that all PCR products had nonchimeric structures, with the exception of clone AC01, which may consist of Paenibacillus macquariensis (nucleotides [nt] 1 to 130) and Bacillus macroides/Bacillus simplex (nt 131 to 323).
Sequences obtained for the nosZ gene encoding nitrous oxide reductase.
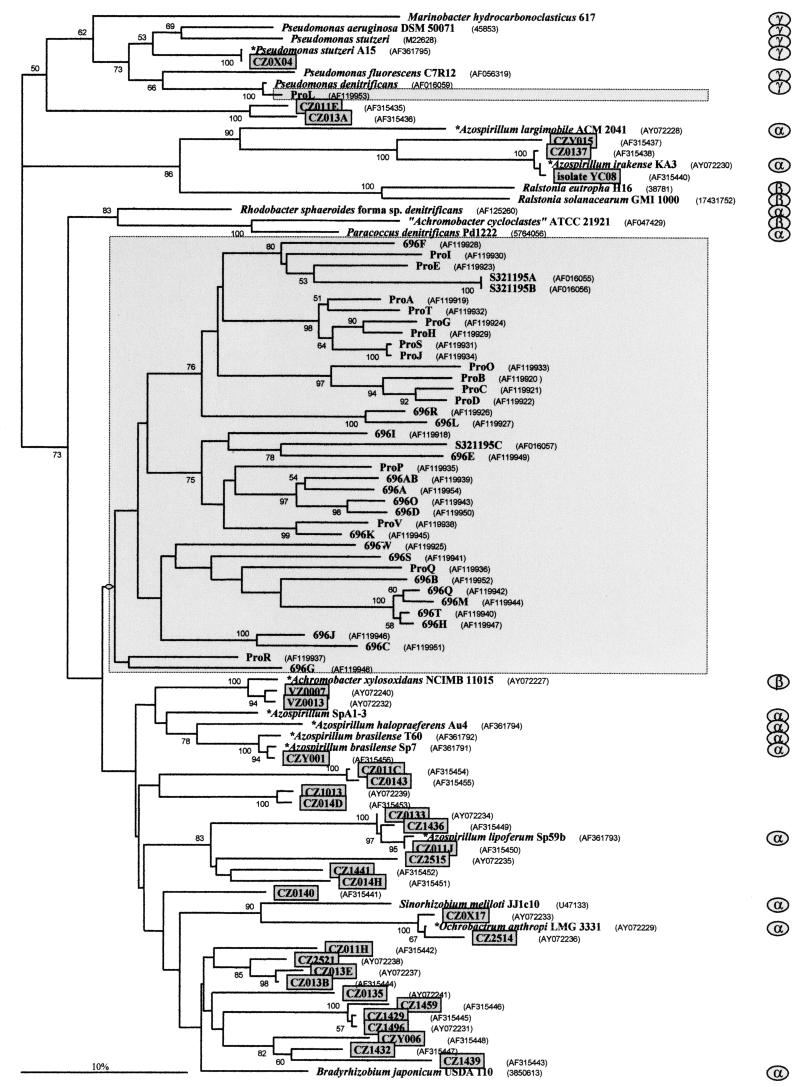
The newly developed primer pair nosZ-F and nosZ-R allowed the amplification of a 700-bp segment coding for part of the Cuz and the CuA catalytic domains and the Cu ligands His376, His437, Cys561, Cys565, and His569. All together, 69 cloned PCR products of the Chorbusch soil were examined, resulting in 32 different sequences. Of these, 20% gave unique sequences and 26% occurred twice. Sequences almost identical to those of CZ011C and CZ0143 were detected 10 times (maximal divergence, 4 nt), but these clones had no close relative among the cultured bacteria (Fig. 3). A sequence closely related to that of Azospirillum irakense (98.6% identity with the total 700 bp sequenced) showed up seven times (with a maximal divergence of 9 nt). The phylogram (Fig. 3) for nosZ utilizes only 230 bp out of the 700-bp segment, since these 230 bp overlapped those used by Scala and Kerkhof (32, 33). The phylogram contains all sequences available in the data banks and the data obtained from nine cultured bacteria and from 32 different PCR clones of soil DNA. None of the new sequences was completely identical with any already known. However, almost all sequences could be assigned to the Rhizobium, Azospirillum, or Pseudomonas group. Three different sequences shown in Fig. 3 clustered with Azospirillum irakense, which has already been found to be far apart from the other azospirilla also in its 16S rRNA gene sequence (18). The sequences of CZ013A and CZ011E apparently were separated from that of any described bacterium. The primers nosZ-F and nosZ-R provided no amplification product with soil DNA from Villingen. Therefore, primer K11, located 4 bp upstream of nosZ-F (18), was used. The primer pair K11-nosZ-R resulted in 12 clones, all of whose sequences were related to that of Achromobacter xylosoxidans. Surprisingly, the primer pair K11-nosZ-R did not provide amplification products with DNA from the Chorbusch soil. It is not clear whether such problems in the amplifications are due to the DNA quality.
FIG. 3.
Phylogram for nosZ, which encodes the dissimilatory nitrous oxide reductase. This phylogram is an alignment of only 230 positions to allow a comparison with the sequences for nosZ genes from marine bacteria published by Scala and Kerkhof (32, 33). Their data are framed and shaded in light grey. The sequences of pseudomonads were used as an outgroup. The clone designations that are framed and shaded in dark grey are our sequences. Data for other items (scale bar, numbers at the branches) are as described in the legend to Fig. 2.
None of the 32 nosZ segments was more than 90% identical to their next most closely related neighbor in the data banks. The mean relatedness within the sequences from all cultured bacteria (including those deposited by us) was only 68% ± 8%. This value was 71% ± 11% for the clones obtained by PCR of soil DNA. These data refer to the approximately 700 bp of the nosZ-F-nosZ-R segment; the corresponding identities for the 230-bp part analyzed in Fig. 3 were 73% ± 8% for the cultured bacteria and 78% ± 8% for the clones from the Chorbusch DNA. This relatively low sequence identity might reflect the high diversity of the N2O reductase genes of the organisms of the Chorbusch soil. It is noteworthy that sequencing of two of the PCR clones with isolated DNA from the Chorbusch soil revealed non-N2O reductase sequences.
Sequences for nirS and nirK.
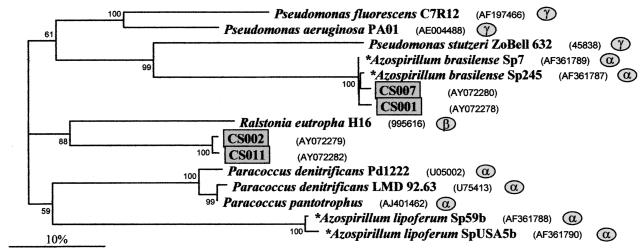
Only a few sequences were obtained for nirS, which encodes the cytochrome cd1-containing nitrite reductase, and for nirK, which encodes the Cu-containing nitrite reductase. The nirS primers KA3 and KA25 gave a 700-bp gene segment containing the cytochrome d1 binding domain and the strictly conserved His207. PCR products of the expected sizes were obtained with DNA of diverse laboratory cultures which were known to possess this enzyme (17, 25, 28) (see also Table 2). Using isolated DNA from the 5-cm layer of the Chorbusch soil, 15 out of the 40 clones obtained were sequenced. Surprisingly, 10 of them gave false, non-nitrite reductase sequences. Of the remaining five, a 357-bp-long sequence within the segment was analyzed. Three of the sequences showed distinct homology to that of Azospirillum brasilense, and the other two showed homology to Ralstonia eutropha (Fig. 4). None of these partial sequences were found to have been deposited in the data banks.
FIG. 4.
Phylogram for nirS, which encodes the cytochrome cd1-containing nitrite reductase. The alignment was performed on 357 positions. The sequences of three pseudomonads served as an outgroup. Otherwise, see the legend to Fig. 2.
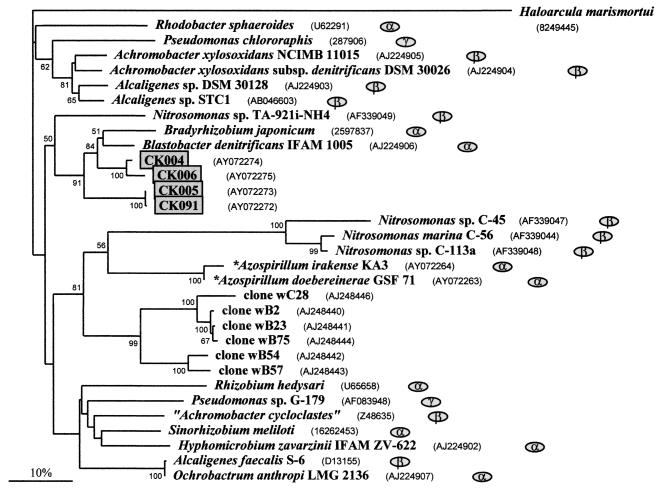
The primers K15-F and K16-R (18), spanning a segment of 520 bp, were used for nirK. As positive controls, PCR products of the nirK segment were obtained with DNA from Azospirillum irakense KA3, Ochrobactrum anthropi LMG3331, Sinorhizobium meliloti Rm1021, and others (Table 2). DNA from either the Chorbusch or the Villingen soil could be amplified with these primers. All together, 12 clones (10 from Chorbusch and 2 from Villingen) were sequenced. Of them, 10, including the 2 Villingen clones, provided remarkably similar sequences with 99.6 to 99.8% identity. Therefore, only clones CK091, CK090, and CK005 out of these 10 are listed in Fig. 5. The sequences of the other two clones were 85.2% identical to each other, around 86% identical to the other 10 clones, and approximately 89% identical to sequences in the data banks, in which the closest score is for Bradyrhizobium japonicum (Fig. 5).
FIG. 5.
Phylogram for nirK, which encodes the Cu-containing nitrite reductase. Three hundred forty-one positions were utilized for the alignment, and the sequence of Haloarcula marismortui was the outgroup. Otherwise, see the legend to Fig. 2.
Evenness of the sequences obtained for nifH, which encodes nitrogenase reductase of N2 fixation.
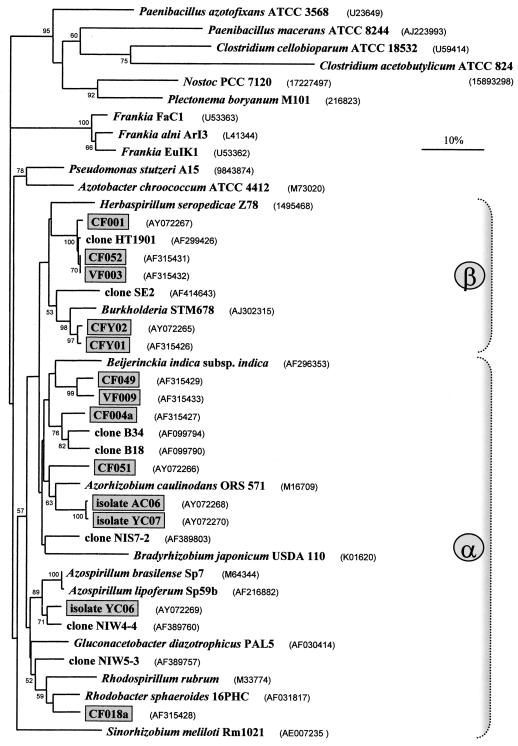
Among the nitrogenase proteins, the NifH subunit is the most conserved. The primers nifH-R and nifH-F amplify the target nifH gene segment in a wide range of N2-fixing organisms (Table 2). These include three different Frankia isolates (Fig. 6). Out of the PCR products obtained, 16 originating from DNA of the 5-cm layer of the Chorbusch soil, 5 from DNA of the same layer from Villingen, and 5 from DNA of Chorbusch isolates from an enrichment in YEM were characterized by cloning and sequencing. The sequences showed that the nifH DNA segment had been amplified in all 26 cases. The clones CF001 (from Chorbusch) and VF003 (from Villingen) were identical (Fig. 6). A sequence similar to that of CF001/VF003 was found in 12 clones, and the maximal divergence between any two sequences of this group was 0.6%. As a sequence divergence of 0.2% reflects an exchange of a single nucleotide in this 400-bp-long segment (without the primers), a difference of 0.6% or even less may be due to PCR amplification and/or sequencing errors but may not indicate a novel bacterial strain. The closely related sequences of CFY01 and CFY02 occurred six times, CF049 occurred three times in Chorbusch samples, and VF009 occurred twice in Villingen samples. Three sequences only, CF051, CF004a, and CF018a, were unique. The comparison by pairs revealed that the new different sequences had an average identity of 83.4% ± 4.0%. They could be subdivided into three groups. Clone CF051 clustered with Azorhizobium caulinodans ORS 571; CF049 and VF009 clustered with Beijerinckia indica; and CF052, VF003, CFY02, and CFY01 bore some resemblance to β-proteobacteria. A single isolated clone, CF018a, grouped with the Rhodospirillum/Azospirillum cluster. It should be mentioned that none of the new sequences totally matched any deposited in the data banks. Two further, only partial sequences of YEM-grown isolates from the Chorbusch soil clustered with Azorhizobium caulinodans and Bradyrhizobium japonicum, respectively (not documented in Fig. 6).
FIG. 6.
Phylogram for nifH, which encodes dinitrogenase reductase (NifH). The alignment was performed with 293 nt (gaps excluded) or 310 nt (gaps included). The outgroup sequences were from Paenibacillus spp., clostridia, and cyanobacteria. α and β denote arbitrary classifications. Otherwise, see the legend to Fig. 2.
DISCUSSION
In any attempt to characterize the microbial soil community, one is immediately faced with its enormous biodiversity. Apart from the fact that only an indeterminable percentage of the DNA from soil bacteria may be suitable for amplification by PCR, only a small fraction of the PCR products can be characterized by sequencing due to limitations with respect to time and funds. Therefore, clones can be selected only at random. In any approach like the present one investigators are immediately faced with the problem of the general validity of the results. This investigation attempted to characterize the microbial flora of a soil by the use of primers for PCR amplifications of the nitrogenase gene, three denitrification genes, and the 16S rRNA gene side by side. Although the bacterial population could not be characterized comprehensively, some unexpected results, partly apparently of general validity, emerged.
All 31 sequenced 16S rRNA gene clones and even some of the isolates from Chorbusch soil came from bacteria that have not been described, as their sequences were not identical with any of the approximately 30,000 deposited in the data banks (including 20,000 sequences from cultured bacteria). The divergence in sequences of the 31 clones compared to those of the 59 selected from the database (Fig. 2) was 7.3% at a minimum and 26.5% at a maximum. This is far beyond PCR amplification and sequencing errors.
Each of the new 16S rRNA gene sequences was unique. However, some sequences were rather similar. Clones C6021 and C6038 had a sequence identity of 99.0%; thus, only 3 bases were exchanged. Such differences may be due to errors in PCR amplification and the cloning of closely related 16S rRNA gene sequences, which can cause up to a 1.2% divergence within 16S rRNA gene clone libraries (36). If one assumes that the error in the sequence determination amounts to even 3%, 24 out of the 31 clone sequences were still different. Clones C6014, C6032, C6022, and C6057s were between 93 and 95% identical when analyzed in pairs. Such small differences may reflect that bacteria of such a habitat may be composed of a genetic continuum where isolates can hardly be differentiated into clearly separated species (37). It was not expected that exactly the same 16S rRNA gene sequence would show up twice. Even though a PCR with universal DNA primers amplifies the segment with different levels of efficacy in bacteria (38), the biodiversity of the PCR products would still be enormous. If only 10% of the gene segments were amplified under the conditions employed, the complete analysis would require sequencing of 103 different PCR products of the ribotypes per g of soil. The high biodiversity in soils is also reflected by findings that 200 clones from a sandy loam soil yielded 73 different total-RFLP signals and 154 different RFLP patterns (11).
The 16S rRNA gene sequences of the cultured isolates all belonged to bacteria of the Bacillus/Paenibacillus group. Bacteria of this group are common among culturable soil bacteria (22). Sequencing did not reveal the occurrence of either rhizobia or pseudomonads and revealed the sequence of only one of the azospirilla in this soil; thus, their numbers may be small at the Chorbusch location. Their occurrence in this soil was, however, documented by employing primers specific for either azospirilla or rhizobia (C. Rösch, unpublished data). The major part of the PCR products obtained with the universal bacterial primers used in the present study belonged to members of the Acidobacterium phylum, to unknown γ-proteobacteria, and to actinomycetes. The sequences of the PCR products around those of C6040 and C6057 were only distantly related to actinomycetes or to deep-rooted members of the Bacillus/Clostridium group. Members of the Acidobacterium phylum and many of the actinomycetes are as yet uncultured or grow at best with a long generation time (30). A similar abundance of bacteria of the Acidobacterium phylum and of actinomycetes in the soil microbial community of a wheat field in The Netherlands has recently been described (35).
In contrast to the apparent richness in the 16S rRNA gene sequences, the PCR products from DNA of the Chorbusch soil were surprisingly uniform for nifH. Only one clone provided a unique sequence, whereas all of the other 25 could be subdivided into three groups with very similar sequences in each. The two PCR clones obtained from Villingen had sequences almost identical to those of Chorbusch samples, indicating a low level of species richness in the population of soil bacteria in the case of nifH. In the data banks, approximately 1,000 sequences are available for nifH, among which are also some from the actinomycete Frankia spp. The primers developed might allow PCR amplifications of nifH from all organisms, including Frankia spp. Thus, it is tentatively concluded that the actinomycetes and the members of the Acidobacterium phylum are non-N2 fixers in such a soil unless sequences were present in the DNA but not recovered by PCR. A previous study (26) showed that N2-fixing bacteria predominantly occur in the upper soil layer and are estimated to make up ∼5% of the total bacterial population. A similar low level of diversity in nifH-positive bacteria was also found in a soil of a Douglas fir forest in Oregon, where 64 nifH clones provided only 13 RFLP patterns (44).
Two more specific observations with nifH are worth mentioning. Isolate YC06 grown in YEM is most closely related to the noncultured clone NIW 4-4 (21), both of which cluster with Azospirillum (Fig. 6). Four nearly identical sequences have now been reported for clones CF001, CF052 (both from Chorbusch), VF003 (from Villingen), and HT1901 (from the North Pacific Ocean close to Hawaii [46]), suggesting a broad distribution of some N2-fixing species.
With nosZ, the biodiversity was higher. Out of the 69 sequences analyzed from the Chorbusch soil, 32 were different. However, the nine from the Villingen soil were almost identical. All sequences were related to those from proteobacteria and did not cluster in groups apart, in contrast to the situation with the 16S rRNA gene segment. In line with this, for nosZ, the data banks do not contain any sequence next to actinomycetes and the Acidobacterium phylum. Thus, these groups of organisms might not be significant contributors to denitrification and N2 fixation in such soils unless they possess unusual genes or have acquired proteobacterial genes by lateral gene transfer. Our 700-bp PCR product overlapped by 230 bp those of the Atlantic or Pacific coast (32, 33). The sequence information in these common 230 bp was sufficient to combine the data for a joint phylogram for nosZ (Fig. 3). The phylogram derived from the longer (700-bp) segment closely matched the one constructed from the 230-bp sequences shown in Fig. 3, with the exception that the clusters for Azospirillum and Rhizobium were more distinctly resolved in the 700-bp comparison (C. Rösch, unpublished data). In the case of the marine sediments (32, 33), the sequences clustered monophyletically, with the exception of ProL. The sequences from the Chorbusch soil were more diverse and remarkably did not overlap those of any from the marine sediments of the Pacific and Atlantic coasts. Thus, the occurrence of bacteria possessing the N2O reductase gene is seemingly habitat specific, although the geographical distance might not to be neglected.
Previous DNA-DNA hybridization experiments with probes for either the cytochrome cd1- or Cu-containing nitrite reductase showed that bacteria with these genes occur in both the Chorbusch and the Villingen soil, with a possible enrichment of bacteria with the Cu enzyme in the bulk, root-free soil (25, 26). For both genes, the DNA probes hybridized and the primers provided amplification products with target DNA of a whole range of culturable bacteria (17, 18, 25, 26). Unexpectedly, in the case of nirS, false PCR products were obtained for 10 out of 15 clones. The rest, however, showed strong sequence homologies to nirS, and all were next most closely related to the gene from Ralstonia eutropha and Azospirillum brasilense. Such a problem with false amplification products occurred occasionally in the case of nosZ but not with other genes. Primers recently developed by others (4, 5) may well be more suited for assessing the occurrence of nirS in bacteria and possibly also in soil DNA. In the case of nirK, the 10 clones from Chorbusch and 2 clones from Villingen soil DNAs showed strong sequence identities among them, which casts doubt as to whether the population of bacteria with nirK has been assessed representatively under the conditions employed. However, the biodiversity of bacteria with this gene was reported to be low in marine habitats as well (3).
The unexpected outcome of this study is that the sequences obtained here were new for each gene, but this reflects the enormous biodiversity in such soils. At the beginning of this study in the spring of 2000 we were faced with the problem that only a few sequences of the denitrification genes were available in the data banks, e.g., in the cases of both nitrite reductases. Thus, it was difficult to find universal primers recognizing the target genes in all organisms. As the new sequences obtained here were deposited into the data banks and as the sets for the genes are currently increasing almost monthly, it should be possible to develop primers of motifs conserved in all organisms which might allow us to more widely screen for the distribution of functional genes in diverse habitats in the near future. Studies similar to the present attempt for soils will help us to understand structure-function relationships in environments with complex biodiversities.
Acknowledgments
This work was kindly supported by a grant from the Gas- und Elektrizitätswerke Stiftung in Cologne, Germany, and by the Deutsche Bundesstiftung Umwelt in Osnabrück, Germany.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amann, R. I., W. Ludwig, and K. H. Schleifer. 1995. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 59:143-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Braker, G., H. L. Ayala-del-Rio, A. H. Devol, A. Fesefeldt, and J. M. Tiedje. 2001. Community structure of denitrifiers, bacteria, and archaea along redox gradients in Pacific Northwest marine sediments by terminal restriction fragment length polymorphism analysis of amplified nitrite reductase (nirS) and 16S rRNA genes. Appl. Environ. Microbiol. 67:1893-1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Braker, G., A. Fesefeldt, and K. P. Witzel. 1998. Development of PCR primer systems for amplification of nitrite reductase genes (nirK and nirS) to detect denitrifying bacteria in environmental samples. Appl. Environ. Microbiol. 64:3769-3775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Braker, G., J. Zhou, L. Wu, A. H. Devol, and J. M. Tiedje. 2000. Nitrite reductase genes (nirK and nirS) as functional markers to investigate diversity of denitrifying bacteria in Pacific Northwest marine sediment communities. Appl. Environ. Microbiol. 66:2096-2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burns, R. C., and R. W. F. Hardy. 1975. Nitrogen fixation in bacteria and higher plants, p. 46-60. Springer, Berlin, Germany. [DOI] [PubMed]
- 7.Chatzinotas, A., R. A. Sandaa, W. Schonhuber, R. Amann, F. L. Daae, V. Torsvik, J. Zeyer, and D. Hahn. 1998. Analysis of broad-scale differences in microbial community composition of two pristine forest soils. Syst. Appl. Microbiol. 21:579-587. [DOI] [PubMed] [Google Scholar]
- 8.Conrad, R. 1996. Soil microorganisms as controllers of atmospheric trace gases (H2, CO, CH4, OCS, N2O, and NO). Microbiol. Rev. 60:609-640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Crutzen, P. J. 1979. The role of NO and NO2 in the chemistry of the troposphere and stratosphere. Annu. Rev. Earth Planet Sci. 7:443-472. [Google Scholar]
- 10.De Boer, A. P., W. N. Reijnders, J. G. Kuenen, A. H. Stouthamer, and R. J. van Spanning. 1994. Isolation, sequencing and mutational analysis of a gene cluster involved in nitrite reduction of Paracoccus denitrificans. Antonie Leeuwenhoek 66:111-127. [DOI] [PubMed] [Google Scholar]
- 11.Dunbar, J., L. O. Ticknor, and C. R. Kuske. 2000. Assessment of microbial diversity in four Southwestern United States soils by 16S rRNA gene terminal restriction fragment analysis. Appl. Environ. Microbiol. 66:2943-2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Greenberg, E. P., and G. E. Becker. 1977. Nitrous oxide as end product of denitrification by strains of fluorescent pseudomonads. Can. J. Microbiol. 23:903-907. [DOI] [PubMed] [Google Scholar]
- 13.Hoeren, F. U., B. C. Berks, S. J. Ferguson, and J. E. McCarthy. 1993. Sequence and expression of the gene encoding the respiratory nitrous-oxide reductase from Paracoccus denitrificans. New and conserved structural and regulatory motifs. Eur. J. Biochem. 218:49-57. [DOI] [PubMed] [Google Scholar]
- 14.Horz, H.-P., J.-H. Rotthauwe, T. Lukow, and W. Liesack. 2000. Identification of major subgroups of ammonia-oxidizing bacteria in environmental samples by T-RFLP analysis of amoA PCR products. J. Microbiol. Methods 39:197-204. [DOI] [PubMed] [Google Scholar]
- 15.Huff, J. P., B. J. Grant, C. A. Penning, and K. F. Sullivan. 1990. Optimization of routine transformation of Escherichia coli with plasmid DNA. BioTechniques 9:570-577. [PubMed] [Google Scholar]
- 16.Kaldorf, M., K. H. Linne von Berg, U. Meier, U. Servos, and H. Bothe. 1993. The reduction of nitrous oxide to dinitrogen by Escherichia coli. Arch. Microbiol. 160:432-439. [DOI] [PubMed] [Google Scholar]
- 17.Kloos, K., U. M. Hüsgen, and H. Bothe. 1998. DNA-probing for genes coding for denitrification, N2-fixation and nitrification in bacteria isolated from different soils. Z. Naturforsch. C 53:69-81. [DOI] [PubMed] [Google Scholar]
- 18.Kloos, K., A. Mergel, C. Rösch, and H. Bothe. 2001. Denitrification within the genus Azospirillum and other associative bacteria. Aust. J. Plant Physiol. 28:991-998. [Google Scholar]
- 19.Linne von Berg, K. H., and H. Bothe. 1992. The distribution of denitrifying bacteria in soils monitored by DNA-probing. FEMS Microbiol. Ecol. 86:331-340. [Google Scholar]
- 20.Liu, W.-T., T. L. Marsh, H. Cheng, and L. J. Forney. 1997. Characterization of microbial diversity by determining terminal restriction fragment length polymorphisms of genes encoding 16S rRNA. Appl. Environ. Microbiol. 63:4516-4522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lovell, C. R., M. J. Friez, J. W. Longshore, and C. E. Bagwell. 2001. Recovery and phylogenetic analysis of nifH sequences from diazotrophic bacteria associated with dead aboveground biomass of Spartina alterniflora. Appl. Environ. Microbiol. 67:5308-5314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Madigan, M. T., J. M. Martinko, and J. Parker. 2000. In Brock biology of microorganisms, 9th ed. Prentice-Hall, New York, N.Y.
- 23.Maidak, B. L., G. L. Olsen, N. Larsen, R. Overbeck, M. J. McCaughey, and C. R. Woese. 1997. The RDP (Ribosomal Database Project). Nucleic Acids Res. 25:109-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mayr, C., A. Winding, and N. B. Hendriksen. 1999. Community level physiological profile of soil bacteria unaffected by extraction method. J. Microbiol. Methods 36:29-33. [DOI] [PubMed] [Google Scholar]
- 25.Mergel, A., K. Kloos, and H. Bothe. 2001. Seasonal fluctuations in the population of denitrifying and N2-fixing bacteria in an acid soil of a Norway spruce forest. Plant Soil 40:315-321. [Google Scholar]
- 26.Mergel, A., O. Schmitz, T. Mallmann, and H. Bothe. 2001. Relative abundance of denitrifying and dinitrogen-fixing bacteria in layers of a forest soil. FEMS Microbiol. Ecol. 36:33-42. [DOI] [PubMed] [Google Scholar]
- 27.Moeseneder, M. M., J. M. Arrieta, G. Muyzer, C. Winter, and G. J. Herndl. 1999. Optimization of terminal-restriction fragment length polymorphism analysis for complex marine bacterioplankton communities and comparison with denaturing gradient gel electrophoresis. Appl. Environ. Microbiol. 65:3518-3525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nishiyama, M., J. Suzuki, M. Kukimoto, T. Ohnuki, S. Horinouchi, and T. Beppu. 1993. Cloning and characterization of a nitrite reductase gene from Alcaligenes faecalis and its expression in Escherichia coli. J. Gen. Microbiol. 139:725-733. [DOI] [PubMed] [Google Scholar]
- 29.Page, R. 1996. TreeView: an application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12:357-358. [DOI] [PubMed] [Google Scholar]
- 30.Rheims, H., and E. Stackebrandt. 1999. Application of nested polymerase chain reaction for the detection of as yet uncultured organisms of the class Actinobacteria in environmental samples. Environ. Microbiol. 1:137-143. [DOI] [PubMed] [Google Scholar]
- 31.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogentic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 32.Scala, D. J., and L. J. Kerkhof. 1998. Nitrous oxide reductase (nosZ) gene-specific PCR primers for detection of denitrifiers and three nosZ genes from marine sediments. FEMS Microbiol. Lett. 162:61-68. [DOI] [PubMed] [Google Scholar]
- 33.Scala, D. J., and L. J. Kerkhof. 1999. Diversity of nitrous oxide reductase (nosZ) genes in continental shelf sediments. Appl. Environ. Microbiol. 65:1681-1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Scala, D. J., and L. J. Kerkhof. 2000. Horizontal heterogeneity of denitrifying bacterial communities in marine sediments by terminal restriction fragment length polymorphism analysis. Appl. Environ. Microbiol. 66:1980-1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Smit, E., P. Leeflang, S. Gommans, J. van den Broek, S. van Mil, and K. Wernars. 2001. Diversity and seasonal fluctuations of the dominant members of the bacterial soil community in a wheat field as determined by cultivation and molecular methods. Appl. Environ. Microbiol. 67:2284-2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Speksnijder, A. G. C. L., G. A. Kowalchuk, S. de Jong, E. Kline, J. R. Stephen, and H. J. Laanbroek. 2001. Microvariation artifacts introduced by PCR and cloning of closely related 16S rRNA gene sequences. Appl. Environ. Microbiol. 67:469-472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Staley, J. T. 1997. Biodiversity: are microbial species threatened? Curr. Opin. Biotechnol. 8:340-345. [DOI] [PubMed] [Google Scholar]
- 38.Suzuki, M. T., and S. J. Giovannoni. 1996. Bias caused by template annealing in the amplification of mixtures of 16S rRNA genes by PCR. Appl. Environ. Microbiol. 62:625-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Torok, I., and A. Kondorosi. 1981. Nucleotide sequence of the R. meliloti nitrogenase reductase (nifH) gene. Nucleic Acids Res. 9:5711-5723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Torsvik, V., J. Goksoyr, and F. L. Daae. 1990. High diversity in DNA of soil bacteria. Appl. Environ. Microbiol. 56:782-787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tortoso, A. C., and G. L. Hutschinson. 1990. Contributions of autotrophic and heterotrophic nitrifiers to soil NO and N2O emissions. Appl. Environ. Microbiol. 56:1799-1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wallner, G., R. Amann, and W. Beisker. 1993. Optimizing fluorescent in situ hybridization with rRNA-targeted oligonucleotide probes for flow cytometric identification of microorganisms. Cytometry 14:136-143. [DOI] [PubMed] [Google Scholar]
- 44.Widmer, F., B. T. Shaffer, L. A. Porteous, and R. J. Seidler. 1999. Analysis of nifH gene pool complexity in soil and litter at a Douglas fir forest site in the Oregon cascade mountain range. Appl. Environ. Microbiol. 65:374-380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wimberly, B. T., D. E. Brodersen, W. M. Clemons, Jr., R. J. Morgan-Warren, A. P. Carter, C. Vonrhein, T. Hartsch, and V. Ramakrishnan. 2000. Structure of the 30S ribosomal subunit. Nature 407:327-339. [DOI] [PubMed] [Google Scholar]
- 46.Zehr, J. P., J. B. Waterbury, P. J. Turner, J. P. Montoya, E. Omoregie, G. F. Steward, A. Hansen, and D. M. Karl. 2001. Unicellular cyanobacteria fix N2 in the subtropical North Pacific Ocean. Nature 412:635-638. [DOI] [PubMed] [Google Scholar]
- 47.Zumft, W. G. 1992. The denitrifying procaryotes, p. 443-582. In A. Balows, H. G. Trüper, M. Dworkin, W. Harder, and K.-H. Schleifer (ed.), The procaryotes. Springer, Heidelberg, Germany.