Abstract
Structural changes (deletions, insertions, and inversions) between human and chimpanzee genomes have likely had a significant impact on lineage-specific evolution because of their potential for dramatic and irreversible mutation. The low-quality nature of the current chimpanzee genome assembly precludes the reliable identification of many of these differences. To circumvent this, we applied a method to optimally map chimpanzee fosmid paired-end sequences against the human genome to systematically identify sites of structural variation ≥12 kb between the two species. Our analysis yielded a total of 651 putative sites of chimpanzee deletion (n = 293), insertions (n = 184), and rearrangements consistent with local inversions between the two genomes (n = 174). We validated a subset (19/23) of insertion and deletions using PCR and Southern blot assays, confirming the accuracy of our method. The events are distributed throughout the genome on all chromosomes but are highly correlated with sites of segmental duplication in human and chimpanzee. These structural variants encompass at least 24 Mb of DNA and overlap with >245 genes. Seventeen of these genes contain exons missing in the chimpanzee genomic sequence and also show a significant reduction in gene expression in chimpanzee. Compared with the pioneering work of Yunis, Prakash, Dutrillaux, and Lejeune, this analysis expands the number of potential rearrangements between chimpanzees and humans 50-fold. Furthermore, this work prioritizes regions for further finishing in the chimpanzee genome and provides a resource for interrogating functional differences between humans and chimpanzees.
Sites of structural variation (SVs) have considerable potential to impart both functional and irreversible difference between evolving species. In particular, the whole or partial deletion of genes has been proposed as one of the primary forces responsible for human evolution (Olson 1999). While cytogenetic comparisons of human and chimpanzee karyotypes have been effective in detecting large-scale (>5 Mb) SVs (Lejeune et al. 1973; Dutrillaux 1980; Yunis et al. 1980; Yunis and Prakash 1982), they are insensitive to submicroscopic changes. At the sequence level, single-base-pair nucleotide substitutions have been surveyed between these primate genomes and estimated to account for a 1.2% nucleotide difference between humans and chimpanzees (Kumar and Hedges 1998; Eichler et al. 2004b; The Chimpanzee Sequencing and Analysis Consortium 2005). The extent of variation affecting sequences larger than a few kb but too small to identify cytogenetically (<∼5Mb) has been difficult to resolve strictly by cytogenetic, microarray-based, or sequence-based methods. Comparative primate studies of segmental duplications (Jackson et al. 1999; Johnson et al. 2001; Stankiewicz et al. 2001; Samonte and Eichler 2002; Horvath et al. 2003) as well as comparisons between finished chimpanzee and human BACs (Britten 2002; Liu et al. 2003) suggest that such variation is common between the species. To date, however, there has been no systematic whole-genome assessment of such variation. Assessment of these intermediate-sized insertion, deletion, and inversion events is critical as variation of this size has great potential to affect the structure and genic complement in each species (Albertson et al. 2000; Snijders et al. 2001; Stankiewicz et al. 2001, 2004; Enard et al. 2002; Locke et al. 2003a,b, 2005; Lupski 2004; Sharp et al. 2005; Tuzun et al. 2005).
An understanding of such structural and functional differences is required to provide a more balanced perspective of the seemingly disparate phenotypic differences that distinguish humans and our closest primate relatives. Structural variation can lead to duplication or deletion of sequence elements, thereby creating species-specific exons, genes, or regulatory regions. A comparison of the mouse and human genome estimated that as much as 400 Mb of genetic material has been deleted in the mouse genome since the divergence of these two mammals 70–90 million years ago (Mouse Genome Sequencing Consortium (MGSC) 2002). The rate and impact of deletion/insertion/inversion between more closely related species has not been systematically addressed. Moreover, both gene duplication and gene loss have been proposed as important forces driving the evolution of the human lineage, but the relative importance of each with respect to human evolution has not been established (Ohno 1970; Olson 1999; Samonte and Eichler 2002). A complete catalog of all structural variation between humans and chimpanzees provides the framework to enable a better evaluation of the relative importance of each process.
The whole genome shotgun sequencing method (WGS) used for construction of the current chimpanzee assembly does not allow reliable detection of structural variation for two reasons. First, the current chimpanzee assembly contains a gap, on average, once every 8 kb (The Chimpanzee Sequencing and Analysis Consortium 2005). Second, the chimpanzee genome is still in draft form and, thus, contains many errors where the sequence has been fragmented, misassembled, or collapsed (The Chimpanzee Sequencing and Analysis Consortium 2005). Both gaps and improper assembly can create artifacts in pairwise genome alignments leading to unacceptable false discovery rates. Various attempts to identify a subset of chimpanzee deletions using the chimpanzee draft assembly have been made (The Chimpanzee Sequencing and Analysis Consortium 2005), including an analysis that characterized deletions (>15 kb in size) based on paired-end sequence analysis. A systematic analysis that considers insertions, deletions, and inversions, however, has not been performed.
Recently we developed a method for the systematic characterization of intermediate-sized structural variation (ISV) by optimal placement of fosmid paired-end sequences against the human genome reference sequence (Tuzun et al. 2005). The power of this approach stems from the stability and packaging constraints of the fosmid vector. These properties result in both genomic fidelity of inserts as well as a tight distribution of insert size around the mean. Given sufficient coverage, the presence of multiple fosmid pairs discordant by size or by orientation provides a useful metric to identify sites of structural variation. This method has been used to reliably identify insertions, deletions, and inversions between a single human individual and the human reference assembly with high (>8 kb) resolution (Tuzun et al. 2005).
In this study, we perform a similar analysis in which we initially ignore the chimpanzee genome assembly and instead use a library of chimpanzee fosmid end sequences to compare the genome of a single chimpanzee individual against the human reference sequence. During the chimpanzee genome sequencing project, ∼1.8 million fosmids were end-sequenced, providing ∼10-fold physical coverage of the genome. Because the forward and reverse sequence reads from each fosmid are physically linked in the chimpanzee genome, and capillary sequencing has essentially eliminated tracking errors, placement of these reads to the high-quality finished human assembly provides comparable power to detect structural variation between the two species (Eichler et al. 2004a; Tuzun et al. 2005). Implementation of this approach with chimpanzee data allowed us to double the number of putative large deletions (>12 kb) and provide one of the first comprehensive maps of structural variation between the two genomes.
Results
We initially mapped ∼1.8 million high-quality paired-end sequence reads from the chimpanzee fosmid library against the finished human genome reference sequence to identify discrepant regions (putative ISVs). To reduce the effect of sequencing errors, each fosmid end-sequence read was rescored based on trace quality, and only fosmids with high-quality reads (Phred ≥30) were retained for mapping (see Methods). In addition, during mapping we selected reads that unambiguously represented the “best match” for a particular region of the human genome. This “best match” criteria biased our set of mapped fosmid paired-end reads to regions where there was sufficient sequence divergence to unambiguously discern orthology—excluding many duplicated regions. We further excluded 137,110 clones either with sequence at only one end or with duplicated entries. Using these criteria we successfully mapped 976,000 (55%) of the ∼1.8 million chimpanzee fosmid sequences on the human assembly. These mapped pairs represent ∼20 Gb of DNA and therefore span ∼6.8× physical coverage of the genome (see Methods).
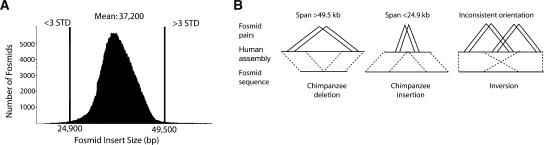
Putative ISVs were identified by mapping each pair of chimpanzee fosmid end sequences to the human genome and recording locations where the distance between the two ends in the human assembly was “larger” or “smaller” than expected, based on the average span of mapped fosmid insert sizes across the genome as a whole (Fig.1A). We also considered regions where multiple fosmid pairs showed consistent orientation differences with respect to the human genome (putative inversions). For each pair of chimpanzee fosmid end sequences that mapped to a “best” location against the human genome, we calculated the insert size based on the human reference sequence. We established length thresholds of at least three standard deviations beyond the mean of computed insert size of chimpanzee fosmid end sequences against the human genome (37.2 ± 4.2 Kb) as well as finished chimpanzee chromosome 22 (37.0 ± 4.1 kb) (Sakaki et al. 2003). When compared with a recent analysis of human fosmid paired-end sequence versus human genome sequence, the chimpanzee fosmid insert sizes were more widely distributed, possibly due to differences in library construction and/or genome architecture between the two species (Tuzun et al. 2005).
Figure 1.
Methodology. (A) Size distribution of 555,929 chimpanzee fosmids mapped unambiguously to the human genome assembly (build34). The distance between two end sequences was determined based on the coordinates within the human genome reference. A length threshold greater than or less than three SD beyond the mean (37.2 kb) was used to classify length discordancy. (B) A schematic depicting chimpanzee “deletions” (two or more fosmids showing a span >49.5 kb), “insertions” (two or more fosmids spanning <24.9 kb), and inversions in DNA (two or more fosmids with an inconsistent orientation of the end sequences with respect to the human genome for each breakpoint).
For the purpose of this study, we operationally defined all discordant sites with respect to the chimpanzee genome. Regions which showed two or more fosmids that were >49.5 kb were classified as “chimpanzee deletions.” Similarly, chimpanzee fosmids for which multiple fosmid pairs mapped too closely (<24.9 kb) based on the human reference genome were termed “chimpanzee insertions.” It should be noted, however, that such events could also, in principle, represent human-specific insertions and deletions, respectively (see below). These thresholds allowed us to detect putative insertion/deletion events >12 kb in size. All regions were graphically visualized (parasight software) and hand-curated based on additional criteria (see Methods).
Chimpanzee deletion events
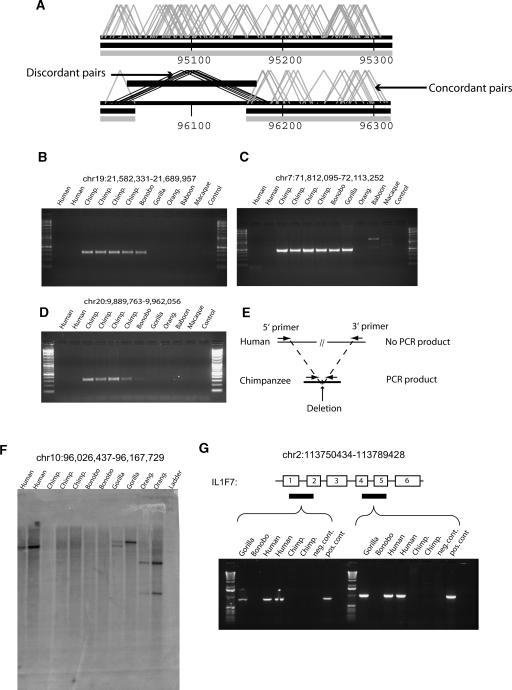
We initially identified ∼550 putative “chimpanzee deletions,” where two or more independent chimpanzee fosmid pairs predicted an insert size >49.5 kb when compared with the human genome (Fig. 1B). To reduce potential polymorphic variants, we further required that a region delineated by these mapped discordant end-pairs bracket a segment wherein no concordant chimpanzee paired sequences mapped. These interior discontinuities or “gaps” in physical coverage combined with two or more discordant fosmids significantly increased our power to detect a fixed structural variant between the two genomes. Figure 2A shows an example of a ∼123 kb deletion detected on chromosome 10. Using these criteria, we report 293 “chimpanzee deletions” ranging in size from 12.5 kb (the lower limit of detection based on the distribution in Fig. 1A) to 815 kb. In total, we estimate that these correspond to ∼21.1 Mb of human sequence that is missing in chimpanzee (Supplemental Table 1). As one measure of validation, we examined the corresponding regions within the chimpanzee assembly (The Chimpanzee Sequencing and Analysis Consortium 2005). Based on BLASTZ alignment between the human and chimpanzee assembly (http://genome.ucsc.edu/goldenPath/help/chain.html), we found corresponding deletions in the assembly >12 kb in length for ∼64% (187/293) of these paired-end sequence detected events. Twenty of these 187 regions mapped to scaffold gaps within the assembly, leaving 56% of the 293 events verified by comparison with the chimpanzee assembly.
Figure 2.
Detection and validation of “chimpanzee deletions.” (A) An example of a chimpanzee deletion event mapped to its corresponding position on human chromosome 10 (build34 coordinates in kb). Two criteria were used to identify chimpanzee deletions: multiple discordant (>49.5 kb) fosmid pairs (black angled lines covered by the black bar) and the absence of concordant fosmid pairs (gray lines) within the region. (B–D) Oligonucleotide sequences (Supplemental Table 5) were designed in regions of conserved human–chimpanzee sequence flanking each deletion breakpoint (see schematic in panel E). PCR products corresponding to the expected size were detected in chimpanzee but not human due to the increased distance between annealing oligonucleotides in the human genome. Results from other closely related apes and Old World monkeys provide outgroup information regarding lineage-specificity of the event. Bands of unexpected size are products of non-specific binding in more distant species. Panel C shows the deletion of a region on chromosome 7 that contains four human genes; POM121, WBSCR20C, TRIM50C, and FKBP6. (E) A schematic of the PCR primer design in chimpanzee and human. (F) Probes for Southern hybridization were developed based on human sequence corresponding to the predicted site of the deletion (see Methods; Supplemental Table 5) and hybridized against a primate panel of restriction-digested primate DNA. The probes successfully hybridized to human genomic DNA but not chimpanzee genomic DNA. Bands of different sizes and lighter intensity in more distant species likely show mutations in restriction enzyme sites. This panel shows a region that contains the human gene CYP2C18 on chromosome 10. (G) The results of an RT-PCR amplification of peripheral blood RNA from exons 1–2 and 3–4 in the IL1F7 gene on chromosome 2 in primates, and putatively deleted in chimpanzee. The primers successfully amplified the exons in humans and gorillas but yielded no products in chimpanzee, providing strong supporting evidence of the deletion.
As a second measure of validation, and in order to assess the lineage-specificity of these events, we experimentally characterized nine chimpanzee deletion events. First, six PCR assays were designed based on flanking conserved sequences adjacent to the chimpanzee deletion such that PCR amplification would readily amplify the deleted variant (Fig. 2E). Human, chimpanzee, bonobo, gorilla, orangutan, baboon, and macaque were then tested by PCR. Five assays verified the putative chimpanzee deletion events, and one showed a product of the expected size in human but not in chimpanzee, suggesting amplification of DNA other than our intended target (Fig. 2B–D; Supplemental Fig. 1A,B). In each of the five successful cases, a PCR product consistent with the size of the deleted allele was detected in chimpanzee (no products in human, Fig. 2B–D; Supplemental Fig. 1A,B). Four of the five PCR experiments show patterns of PCR amplification among the human/ape panel consistent with deletion events occurring specifically within the chimpanzee lineage (rather than an insertion event on the human lineage): three before chimpanzee/bonobo speciation (chromosomes 19 and 20, Fig. 2B,D; and chromosome 11, Supplemental Fig. 1A), and one specific to common chimpanzees only (chromosome 4, Supplemental Fig. 1B). In the remaining PCR experiment (chromosome 7, Fig. 2C) the pattern of PCR amplification among the apes suggests a human-specific insertion event. This region contains four human genes (POM121, WBSCR20C, TRIM50C, and FKBP6) that are not found at this location in chimpanzee. In addition, shared chimpanzee and human duplications, as well as human-specific segmental duplications, were found in this region, implying that duplicate copies of these genes may exist at other locations in both genomes.
As a more direct test, we designed hybridization probes specific to the deleted sequence for an additional three sites and performed Southern hybridization experiments against a primate panel of genomic DNA. All three of the experiments (chromosome 10, Fig. 2F, and chromosomes 22 and 6, Supplemental Fig. 1C,D) showed clear hybridization signals in human, gorilla, and orangutan, but not chimpanzee and bonobo, implying a deletion event specific to the chimpanzee/bonobo lineage of evolution. Each of these regions contains a gene found in humans: CYP2C18 on chromosome 10, ENPP3 on chromosome 6, and APOL4 on chromosome 22. In one case (chromosome 6, Supplemental Fig. 1D), the human population appeared to be polymorphic for the presence of this sequence, revealing a potentially ancient polymorphism or a site of recurrent rearrangement. We also assayed the expression potential of the IL1F7 gene in a putative deletion region on chromosome 2 using RT-PCR. Reverse transcriptase expression analysis of peripheral blood RNA samples from four species confirmed that the IL1F7 transcript exists in gorilla and human but neither bonobo nor chimpanzee (Fig. 2G). While expression of the IL1F7 gene could be lacking in both chimpanzee and bonobo for unrelated reasons, the lack of expression evidence provides supporting evidence that the gene is deleted in both species.
It is unlikely that all 293 putative chimpanzee deletion regions are fixed differences between humans and all chimpanzees. SNP data suggests that ∼14%–22% of single nucleotide differences between human and chimpanzee genomes are actually polymorphic within chimpanzee populations (Chen and Li 2001; Ebersberger et al. 2002). We evaluated this expectation for ISVs by examining the human sequence internal to the deletion regions (between discordant pairs and lacking concordant pair coverage) against the sequence libraries of two other western and three central chimpanzees (The Chimpanzee Sequencing and Analysis Consortium 2005). By retaining sequences of ≥95% identity to chimpanzee sequences >500 bp or more, and further requiring that ≥1000 bp of the internal coordinates of the deletion region aligned, we identified 97 (an upper bound) regions that did match sequence in at least one other chimpanzee individual. If we assume these regions are polymorphic in the chimpanzee population, it suggests that as much as 33% of the sites that vary between human and chimpanzee also vary within chimpanzee populations. However, this analysis cannot distinguish between false positives and polymorphisms and as such may be an overestimate. A second, more direct approach was to identify polymorphisms within the two haplotypes of the chimpanzee individual's genome. In our initial analysis we excluded deletion polymorphisms by focusing on regions that showed multiple fosmids that were discordant by size (“too large”) and the absence of sequence read data underlying the region of putative structural variant. If we eliminate the second criterion, we identify a comparable number of putative deletion regions where there is both discordancy and concordancy when compared with the human genome (n = 266). These data suggest that the ratio of fixed to polymorphic events is ∼1:2 (196:363), and is much lower than similar estimates for SNPs (2:1). It is possible that these differences may be attributed to the strong association of structural variation with segmental duplications (sites of recurrent rearrangement) between the two species.
We examined all 293 “chimpanzee deletions” with respect to annotation of the human genome assembly. Similar to structural variation in humans (Iafrate et al. 2004; Sebat et al. 2004; Sharp et al. 2005; Tuzun et al. 2005), the sequence between the breakpoints of 41% (120/293) of the chimpanzee deletions overlaps with human segmental duplication (SD) sequence (Supplemental Table 1). There are 10 chimpanzee deletion events whose breakpoints fall within 80 kb (the combined bounds of resolution for the results of both analyses) of the coordinates bounding human SVs (Supplemental Table 6).
Among the 178 RefSeq gene regions that intersect with these deletion regions (Supplemental Table 2), we found representatives of many duplicated gene families, including drug-detoxification (glycosyltransferase family, cytochrome P450 genes), immunity (chemokine, cytokine, MLC, HLA, and defensin families), and pregnancy-related proteins. We specifically compared all possible human RefSeq exons (n = 1001) underlying these fixed sites of structural variation to both the chimpanzee genome assembly and chimpanzee WGS. One hundred fifty exons, corresponding to 78 RefSeq genes, matched no chimpanzee sequence with ≥50 bp of ≥95% identity, suggesting that true orthologs of these 150 exons are not present in the genome of chimpanzees. However, only two of these 150 exons showed no sequence identity to other human gene models, indicating that the majority of exons within in these SVs arise from duplicate gene families and have paralogs elsewhere in the chimpanzee genome.
We tested whether these genes (n = 78) lacking exons might show an altered pattern of gene expression between the two species due potentially to altered reading frames, premature stop codons, and nonsense-mediated mRNA decay. We obtained human–chimpanzee expression data for 40 genes from a recently published microarray study from five tissues (brain, heart, liver, kidney, and testis; Khaitovich et al. 2005). Forty-two percent (17/40) of the genes showed reduced levels of expression in chimpanzee, while 15% (6/40) showed higher levels of expression in the chimpanzee (Supplemental Table 3). The remaining 17 genes did not report any significant differences in the expression assay. The number of genes (17, or 42%) with reduced chimpanzee expression was shown to be significantly (p < 0.01) higher than expected by chance from randomly sampling 40 genes from the total dataset 10,000 times (see Methods). In the majority of the cases (35/40), the probe sets map outside of the deletion region in question (Khaitovich et al. 2005). In four of the five remaining cases, the probe sets map at the periphery (<10 kb) of the predicted boundaries of the deletion. The correlation, thus, seems to be the result of lowered gene expression rather than absence of reporting probe sets. We found no evidence of relaxed selection operating on these particular genes when 1:1 orthologs were examined between human and mouse. For example, the average dN/dS value for a subsample of 15 genes was 0.49 and the median was dN/dS = 0.114, very similar to the median Ka/Ks value of 0.115 found for a set of ∼12,000 human–mouse orthologs (MGSC 2002).
Chimpanzee insertions
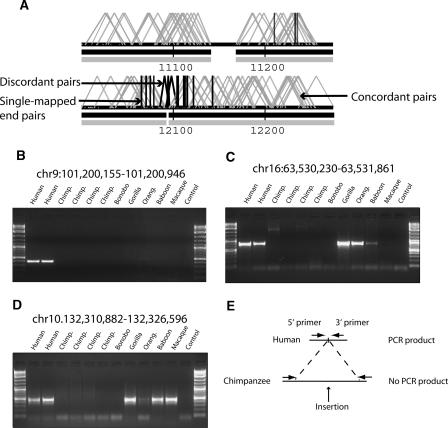
Similar to our deletion analysis, we required two separate criteria to classify potential insertions. First, we identified regions (n = 350) marked by two or more discordant fosmids with an insert size <24.9 kb. Since true insertions would create disruptions in paired-end continuity, we searched for the presence of singletons flanking each of these sites—fosmids for which both end sequences were high-quality but only one end of the pair mapped to the human reference sequence both at this site and in the orientation of the discontinuity. From the 350 discordant regions, we identified 164 regions with at least two discordant pairs (<24.9 kb) that were also flanked by multiple singletons orientated toward the discontinuity. Because the even distribution of unambiguously mapped end sequences can be disrupted by the presence of repeats or duplications at the breakpoints of insertions, we required the presence of singletons flanking each insertion event. An example of a ∼27-kb putative chimpanzee insertion event on chromosome 1 is shown in Figure 3A. This approach yielded 164 chimpanzee-specific regions (29 with flanking singleton clusters on both sides), which range in size from 12.4 kb to 36 kb, corresponding to 2.7 Mb of sequence.
Figure 3.
Detection and validation of “chimpanzee insertions.” (A) A chimpanzee insertion mapped to its corresponding position on human chromosome 1 (positions mapped in kb units from p arm). Two criteria were used to identify insertions: (1) two or more chimpanzee fosmids with an in silico insert size <24.9 kb (black angled lines) and (2) the presence of two or more “singletons” (vertical black lines) oriented toward the insertion. Concordant fosmid pairs are shown in gray. (B–D) PCR verification of three chimpanzee insertion events. Oligonucleotide sequences (Supplemental Table 5) were designed in regions of conserved human–chimpanzee sequence flanking each insertion breakpoint (see schematic in panel E). PCR products corresponding to the expected size were detected in human but not chimpanzees due to the increased distance between annealing oligonucleotides in the chimpanzee genome. Results from other closely related apes and Old World monkeys provide outgroup information regarding lineage-specificity of the event. Bands of unexpected size are products of non-specific binding in more distant species. (E) A schematic of the PCR primer design in chimpanzee and human.
Unlike deletion detection, an important caveat to our approach is that insertion events >40 kb cannot be readily captured due to packaging constraints of the fosmid cloning system. We therefore performed a separate analysis in which we identified clusters of “singletons” (see Methods) bracketing a discontinuity in clonal coverage (both discordant and concordant clones). We identified 20 additional putative chimpanzee-specific insertions in which a clone discontinuity was flanked on either side by at least two singletons. Although the precise length of these 20 insertions is unknown, this raises the total number of putative “chimpanzee insertions” to 184 (Supplemental Table 1) and identifies regions for more targeted sequence and assembly. Similar to the deletion analysis, we compared these regions to the chimpanzee assembly and confirmed 54% (100/184) of the insertions in which the chimpanzee contained >12 kb of unalignable sequence when compared with the human at that position. Seven of these corresponded to sites of chimpanzee-specific retroviral insertions (PTERV1) (Yohn et al. 2005).
We tested 13 regions with PCR in human, chimpanzees, and other primates to experimentally verify putative chimpanzee insertion events. PCR assays were designed to amplify the sequence spanning the site of structural variation in human but not the longer insertion site in chimpanzee (Fig. 3E). Of the 13 pairs tested, 10 supported our findings of structural variation between the two species (Fig. 3B–D; Supplemental Fig. 2A–F). Two showed a product of the wrong size in chimpanzee, suggesting amplification of a site other than our target DNA, and one showed a product of the expected size in human but also a product in chimpanzee, thus failing to verify the putative chimpanzee insertion. Parsimonious reconstruction of the event in each of the 10 successful experiments, based on products in other primates, suggests that four of the events are insertions on the chimpanzee and bonobo lineage, five are human-specific deletions, and the history of one event is ambiguous (Fig. 3B–D; Supplemental Fig. 2A–F). None of these 10 regions overlap with annotated human genes.
In stark contrast to the high percentage of chimpanzee deletions overlapping human SDs, the breakpoints of only 7.6% (14/184) of the chimpanzee insertions overlap with human segmental duplication sequence (Supplemental Table 1). We find three chimpanzee insertion events that map within 80 kb of the coordinates of human SVs (Supplemental Table 6). Only 54 of these insertion sites intersected with coordinates for human Ref-Seq genes (Supplemental Table 2), including the genes SPAG6 (important for spermatic flagellum development), SOX5 (associated with SRY function), and BARD1 (forms a heterodimer with BRCA1 required for proper apoptotic function). Thirty-three genes contained in this set were also tested for expression differences between humans and chimpanzee (Khaitovich et al. 2005). Five showed significant under-expression and four showed significant over-expression in chimpanzee (Supplemental Table 3), which was not significantly different than expected by simulation (see Methods). The remaining 24 genes showed no significant change in expression between the species.
Inversions
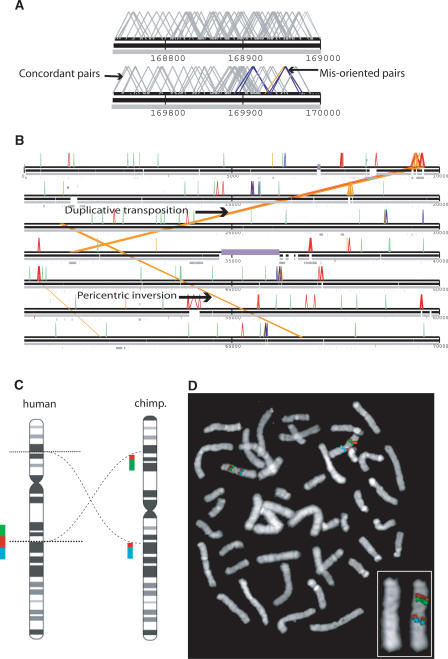
We identified 174 regions where two or more chimpanzee fosmid paired-end sequences showed an inconsistent orientation with respect to the human genome assembly (Fig. 1B). Such orientation inconsistencies may arise by either a conventional inversion of sequence in the reference genome assembly or a duplicative transposition event that transfers a copy of sequence to a new location but in an inverted orientation. Indeed, bicolor FISH analysis with probes flanking a subsample of 13 of these putative inversions showed that four were consistent with conventional inversions of intervening sequence, while eight showed the presence of segmental duplications at one or both boundaries (Supplemental Table 4). Sequence analysis of large insert-containing clones that traverse the duplicated and unique regions will be required to confirm whether these represent de novo duplications that are inverted between the two species. We noticed that the fosmid paired-end sequence signatures of conventional inversion events would present themselves as clusters of misoriented fosmids at either end of the inversion breakpoints (assuming both ends can be unambiguously detected), while duplicative transposition would be demarcated by a cluster of misoriented fosmids mapping at only one breakpoint. Forty-one of the regions show clear evidence of having captured reciprocal breakpoints (Supplemental Table 1) and are classified as conventional inversions, rather than duplicative transposition, by this second criterion. An example of such an inversion on chromosome 1 is shown in Figure 4A. The remaining 133 events may be either type of inversion. The smallest inversion event detected in the set of 41 inversions with reciprocal breakpoints is 1.5 kb, and the largest is 41 Mb (Supplemental Table 1).
Figure 4.
Detection and verification of inversions. (A) Inversion with discordant fosmid data spanning both breakpoints. Multiple chimpanzee fosmids that are discrepant by orientation (blue lines) demarcate the two ends of a hypothetical inversion breakpoint on chromosome 3. (B) A large (44-Mb) inversion on human chromosome 12 identified by our analysis. Orange lines correspond to a known pericentric inversion (ideogram in C) (Yunis et al. 1980; Yunis and Prakash 1982). The breakpoints depicted map within 40 kb of recently characterized sequence breakpoints (Kehrer-Sawatzki et al. 2005b). A second “putative” inversion shown on chromosome 12 is due to a lineage-specific transposition of a segmental duplication in the chimpanzee genome. (C) A schematic of FISH probe design using three BAC probes, one distal to a putative inversion breakpoint (green box, RP11–1007O22F), one spanning the breakpoint (red box RP11–348G10), and one proximal to the breakpoint (blue box, RP11–959B13C5). The expected pattern of probe colors in chimpanzee should split the red probe into two hybridization signals on either side of the centromere. (D) The tricolor FISH results confirm the presence of an inversion on chimpanzee chromosome 12.
Fifteen of the events span >20 Mb of distance and, thus, if they are conventional inversions rather than inverted duplications, they should have been clearly visible at the cytogenetic level. An example of a known pericentric inversion on chromosome 12 is shown in Figure 4B. The breakpoints of this event have been subsequently verified by FISH (Fig. 4C; Supplemental Table 4). Indeed, seven of these 15 large-scale events (human chromosomes 4, 5, 9, 12, 15, 17, and 18) do correspond precisely to chimp–human pericentric inversion breakpoints initially described by Yunis and coworkers (Yunis et al. 1980; Yunis and Prakash 1982) and subsequently refined at the molecular level (Table 1; Kehrer-Sawatzki et al. 2002, 2005a,c; Locke et al. 2003b; Dennehey et al. 2004; Goidts et al. 2004; Nickerson et al. 2005) including analysis of the chimpanzee genome assembly (The Chimpanzee Sequencing and Analysis Consortium 2005). Breakpoints for one additional known inversion on chromosome 1 were not identified in corresponding positions, but our set of 15 large-scale inversions does identify a centromere-spanning inversion on chromosome 1 that may represent the cytogenetic inversions on these chromosomes (Supplemental Table 1).
Table 1.
Summary of cytogenetically verified inversion events
|
Fosmid coordinates, build 34a
|
Breakpoints defined in literature
|
|||||
|---|---|---|---|---|---|---|
| Chr. | Bp. 1 | Bp. 2 | Chr. | Bp. 1 | Bp. 2 | Reference |
| chr1 | 87,288,446-87,328,446 | 145,375,657-145,415,657 | chr1 | unpublished | ||
| chr4 | 44,709,174-44,749,174 | 86,393,839-86,434,839 | chr4 | 44,730,692-44,751,795 | 86,275,393-86,461,364 | (Kehrer-Sawatzki et al. 2005a) |
| chr5 | 18,582,661-18,622,611 | 95,031,126-96,011,126 | chr5 | 18,443,766-18,614,471 | 95,891,549-96,072,074 | (Kehrer-Sawatzki et al. 2005c) |
| chr9 | 40,749,944-40,789,944 | 84,288,147-84,328,147 | chr9 | unmapped | 84,256,135-84,387,819 | (Kehrer-Sawatzki et al. 2005c) |
| chr12 | 20,845,308-20,885,308 | 66,631,594-66,671,594 | chr12 | 20,833,482-21,009,087 | 66,627,151-66,740,912 | (Kehrer-Sawatzki et al. 2005b) |
| chr15 | 20,702,019-20,742,019 | 26,722,088-26,762,088 | chr15 | unmapped | 28,025,787-28,486,050 | (Locke et al. 2003a) |
| chr17 | 8,123,673-8,163,673 | 48,068,346-48,108,346 | chr17 | 8,128,215-8,139,694 | 48,037,665-48,224,281 | (Kehrer-Sawatzki et al. 2002) |
| chr18 | 121,769-161,769 | 16,735,019-16,775,019 | chr18 | 102,251-103,561 | 16,762,886-16,898,525 | (Goidts et al. 2004) |
(Chr.) Chromosome, (Bp) breakpoint.
Fosmid coordinates are a range based on the genomic distances between two fosmid ends that determine the breakpoint
We note a very strong association of the inversion events with the locations of human segmental duplications. Of the 174 putative inversions, 78% overlap with human SDs. Notably, the putative inversion events identified by our method also overlap with chimpanzee SDs in 112 cases (64%). As discussed, this overlap with SDs significantly decreases the ability of our method to differentiate between duplicative transposition of material and more conventional inversions such as the large pericentric events. We identified 16 chimpanzee inversion events whose breakpoints map within 80 kb of a known human SV event (Supplemental Table 6). The breakpoints of the 41 double-ended conventional inversion events overlap with the coding region from 14 RefSeq genes (Supplemental Table 2). Given that the gene structure described is based on the human reference sequence, the coding regions of these 14 genes are possibly discontinuous in the chimpanzee genome. These 14 genes include a chemokine protein and a homeobox protein as well as several zinc fingers and hypothetical proteins. Five genes also correspond to genes tested for expression differences between human and chimpanzee by Khaitovich et al. (2005) (Supplemental Table 3). However, only one of these five genes reports any hybridization signal in any of the five tissues tested, and does not show a difference in expression between the two species (Supplemental Table 3).
Discussion
We have performed the first genome-wide assay of intermediate-scale structural variation between humans and chimpanzees by mapping chimpanzee fosmid paired-end sequences against the human reference sequence and identifying discordant regions by size and/or orientation. The method we have developed takes advantage of the high-quality reference of the human genome assembly and properties of the fosmid cloning system. We have demonstrated its potential to characterize interspecific structural variation in the absence of a genome assembly. Although we limited our analysis to the human and chimpanzee genomes, our approach to detect structural variation could be readily applied to any pair of genomes for which the genetic distance is relatively short (i.e., nucleotide divergence <10%) and one of the two genomes exists as a high-quality reference. Various species, subspecies, or strains of Drosophila, yeast, or mouse could be characterized in this fashion without the need to generate independent WGS assembly for each sibling species.
While this approach offers exquisite precision and resolution over other array-based approaches (Locke et al. 2003b; Fortna et al. 2004), it also suffers a number of limitations. First, proper placement of clone sequence ends requires a high-quality reference genome. Regions of incorrect assembly will yield discordant clones that represent false positives. Likewise, the human reference genome is incomplete (Eichler et al. 2004a) and sequence exists in the chimpanzee genome that is not represented in the human reference. Structural variation within these regions cannot be readily captured, leading to false negatives in the analysis. Second, this approach is expensive compared with techniques such as arrayCGH, as it requires considerable up-front investment in creating clone libraries and generating 0.3- to 0.4-fold sequence coverage of a genome. In the absence of significant cost reductions in sequencing and clone storage, it is currently not practical to apply this technique to screening large numbers of individuals. Finally, at the most stringent level, this method utilizes only those clones that map unambiguously to the reference genome, creating a significant bias against analysis of regions with recent or highly similar repeats and duplications.
In this analysis, we identified 651 regions of putative structural variation between the human genome assembly and a single chimpanzee individual (293 chimpanzee deletions, 184 chimpanzee insertions, and 174 inversions/duplicative transpositions; Table 2). Because these data were generated from a single chimpanzee individual, as much as ∼1/4 of these sites may be polymorphic within the chimpanzee population (The Chimpanzee Sequencing and Analysis Consortium 2005). Future interrogation of these sites in multiple chimpanzee individuals is required to discriminate between interspecific and intraspecific variation. Notwithstanding polymorphism, this analysis potentially increases the number of known structural variants between our two species by a factor of 50 beyond what was originally documented by cytogenetic techniques (Lejeune et al. 1973; Dutrillaux 1980; Yunis et al. 1980; Yunis and Prakash 1982). Details concerning the location of these structural variants mapped against the finished human genome may be found at http://humanparalogy.gs.washington.edu/CSV.
Table 2.
Summary of structural variants between human and chimpanzee
|
Insertions
|
Deletions
|
Inversions
|
||||
|---|---|---|---|---|---|---|
| Size | No. | Mb | No. | Mb | No. | Mb |
| 12-36 kb | 164 | 2.7 | 174 | 3.9 | 11 | 0.264 |
| 36-100 kb | 20a | - | 70 | 4.1 | 17 | 1.1 |
| 100-1000 kb | 0 | 0 | 49 | 13.1 | 65 | 24.8 |
| >1000 kbc | 0
|
0
|
0
|
0
|
7b
|
271
|
| Total | 184 | 2.7 | 293 | 21.1 | 100 | 297 |
The size of these events cannot be estimated from current assembly
These events represent seven of the nine pericentric inversions identified by Yunis et al. (1980)
Other events >1000 kb are not tallied here but can be found in Supplemental Table 1
These data serve two purposes. First, they provide a road map of regions of structural variation for further attention during the second phase of the chimpanzee genome assembly. Many of these regions were not properly assembled in the published version of the genome and we now have identified the specific fosmid clones for further characterization. Second, our set of disrupted or deleted genes provides a resource for interrogating differences between human and chimpanzee species at a functional level.
An important question that remains unaddressed is whether deletion and insertion events are symmetric or asymmetric with respect to frequency or abundance between human and chimpanzee lineages of evolution (Olson 1999; Locke et al. 2003a,b, 2004; Fortna et al. 2004). At first blush, it may appear that chimpanzee deletions outpace insertions (1.6:1 by count or 8:1 by bp in our analysis; Supplemental Table 1). However, with the exception of a small subset (n = 20) we have not determined the lineage-specificity of the majority of the events. Additionally, it is important to note that our fosmid-based approach creates a considerable bias against detecting large (>40 kb) chimpanzee insertions versus deletions, partially explaining the differences in event numbers and base pairs involved. If we limit our analysis to events estimated between 12.5–36.5 kb, we find that the margin narrows. One hundred sixty-four chimpanzee “insertion” events (2.7 Mb), were identified at this range, compared with 174 chimpanzee “deletion” events (3.9 Mb of DNA).
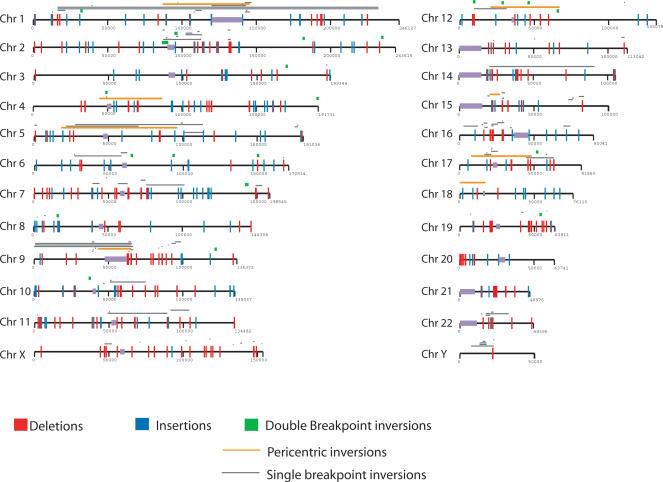
At the chromosomal level, the pattern of deletions, insertions, and inversion events mapped to the human reference assembly does not indicate any obvious genome-wide bias for the location of structural variants (Fig. 5). The three categories are intermixed and distributed across all chromosomes, with the possible exception of chromosome Y, which contains only one ISV (a chimpanzee deletion event). Although the Y chromosome may be the most rearranged chromosome between human and chimpanzee (Lahn and Page 1999; Ali and Hasnain 2002), it also contains a very high percentage of (lineage-specific) repetitive sequences, which our method specifically avoids because of the lack of reliable paired-end placement in such regions (Ali and Hasnain 2002). Thus, this method's ability to detect rearrangements in regions with the repetitive characteristics of the Y chromosome is low.
Figure 5.
Summary of structural variation between chimpanzee and human. A diagram of the location of all 651 structural variants between humans and chimpanzee mapped to the human reference assembly. Chimpanzee deletions (n = 293) are shown in red; insertions (n = 184) are shown in blue. Inversions/duplicative transpositions (n = 174) are classified into three groups: confirmed pericentric cytogenetic inversions from Yunis and Prakash (1982) (orange); double breakpoint inversions, if both of the breakpoints were captured (green); and single breakpoint inversions, if only one end was captured (gray). A significant fraction of the latter corresponds to duplicative transpositions of segmental duplications as opposed to bona fide inversions. The complete coordinate list for all sites of structural variants is provided in Supplemental Table 1. Supplemental Figures 3–26 provide a detailed map of all variation at the kb level for each chromosome.
At the regional level, certain areas show local hotspots for one or more types of variation. For example, the probability of observing four or more insertion or deletion events within a 1-Mb region by chance is <<0.001 (see Methods), suggesting that the events we predict in three regions (on chromosome 1, chromosome 2, and chromosome 14) may represent genomic hotspots of variation (see Methods). At the sequence level, a strong association emerges with respect to segmental duplications. The strongest association is observed for chimpanzee inversions and deletions. On average, 78% and 41% of the chimpanzee inversion and deletion events, respectively, overlap with SDs despite the fact that SDs make up only 5% of the human genome. The finding that 78% of the inversions overlap human SDs is not unexpected, since our algorithm cannot distinguish between conventional inversion events and duplicative transposition of sequence material. The enrichment seen in chimpanzee deletions extends and corroborates recent findings from human variation studies that show a similar structural bias to such regions (Fredman et al. 2004; Iafrate et al. 2004; Sebat et al. 2004; Sharp et al. 2005; Tuzun et al. 2005). Surprisingly, the enrichment is much less pronounced for chimpanzee insertion events, where only 8% map to sites of segmental duplication.
In total, we have identified 245 separate RefSeq genes that may be potentially affected by structural differences between chimpanzee and human (http://www.ncbi.nlm.nih.gov/RefSeq/). These 245 genes include members of a vast array of functional groups including those related to drug-detoxification, receptors, and reproduction. It is tempting to speculate that these functional groups have been good candidates for adaptive evolution since the divergence of humans and chimpanzees (Hollox et al. 2001; Gonzales et al. 2003). Over 71% (184/257) of these genes are in regions of chimpanzee deletions, and 78 genes contain 150 exons in humans that lack a corresponding high percent-identity match in the chimpanzee WGS sequences. More importantly, 23 of these genes show expression differences between humans and chimpanzees. Most of these genic regions are not well assembled in the current draft assembly. We recommend that such regions be prioritized for high-quality, clone-based sequencing.
In summary, our data establish the fosmid paired-end mapping strategy as a robust and accurate method for detecting mid-, and large-scale structural variation between humans and other primates. This method gives a high-resolution estimate of coordinates within ∼40 kb of the breakpoints of duplications, deletions, and inversion that are both too small to be detected by traditional cytogenetic analyses or too large to be reliably ascertained by comparisons of unfinished, low-quality, or low-coverage genomes to the human assembly (Pinkel et al. 1998; Snijders et al. 2001; Locke et al. 2003b). Our technique is also capable of detecting large-scale SVs and has yielded results that correspond well to all nine of the previously identified macro-inversions between humans and chimpanzee (Yunis et al. 1980; Yunis and Prakash 1982). In addition, our analysis identifies ∼245 genes that are potentially rearranged or deleted between the two species. Extensive future experimental study is required to demonstrate functional significance of any genes and their role in contributing to phenotypic difference between humans and chimpanzees.
Methods
Fosmid paired-end sequence placement
During the sequencing of the chimpanzee genome, a fosmid library (CHORI-1251) was constructed from peripheral blood obtained from the male chimpanzee genome sequence donor (Clint). The fosmid vector was chosen because of the insert stability, tight distribution of insert size, and the relatively low frequency of propagation errors when compared with other conventional cloning vectors (Kim et al. 1992). We obtained both the sequence and corresponding base quality for all traces from Washington University (http://www.ncbi.nlm.nih.gov/Traces/trace.cgi?), which yielded 1,788,428 end sequences (1,839,144,838 bp excluding “N”s) representing 866,328 nonredundant clones. Of these, we found 729,218 clones with trace sequences for both fosmid ends. All fosmid end sequences were optimally aligned and paired against both the reference human genome sequence and against chimpanzee chromosome 22 as part of a four-step process to detect putative rearrangements: (1) initial recruitment, (2) optimal realignment with quality rescoring, (3) determination of paired-end read placements, and (4) rearrangement detection.
Initial recruitment
During the recruitment phase all fosmid end sequences were aligned using NCBI Megablast (-p 80 -s 90 -v 7 -b 7 -w 12 -t 21) to the finishing reference human genome assembly (build34, July 2003). The score threshold (-s 90) was set to detect all alignments of ≥150 bp and ≥90% identity. A score cutoff allowed for the flexibility to detect shorter alignments with higher similarity or longer alignments with lower sequence identity, such as those due to base-calling errors in poor-quality traces. Additionally, an 80% identity threshold (-p 80) was set to avoid recruiting numerous pairwise alignments representing related transposable/repetitive elements. To capture all truly orthologous alignments while decreasing noise associated with more recently transposed repetitive sequences, only the alignments from the top seven scoring genomic reference fragments (and up to eight alignments within each genomic fragment) were retained. In total, 698,559 of the 866,328 clones (80.6%) with trace sequence for both ends were also high-quality sequence at both ends (30 bases of Phred Q 30). Of these 698,559 possible clones, 689,403 had recruitment of both ends with each end having one or more alignments. The remaining clones (<1%, or 9156/698,559) failed to align to human sequence at either end.
Optimal realignment with quality rescoring
All recruited alignments were then optimally realigned using an in-house Needleman-Wunsch implementation (match = +10, mismatch = -8, gap opening = -20, gap extension = -1, no penalty for terminal gaps) (Needleman and Wunsch 1970). Global realignment improved the treatment of insertions, deletions, and substitutions. The percent identity for each global alignment was then recalculated, base by base, including only those aligned bases where fosmid-end nucleotides were high-quality (Phred Q score 30, which equals a sequencing error rate of 10-3 per base) (Ewing and Green 1998). All reference genome sequence was considered high-quality, as published reports demonstrate extremely low error rates of <10-4 to 10-5 per base (International Human Genome Sequencing Consortium [IHGSC] 2001, 2004). A new alignment score, weighted for orthologous levels of identity, was then calculated based on the number of aligned bases and fraction identity (Global Alignment Score = base pairs × [2 × identity - 20 - [1 - identity]]). Alignments for each fosmid end were filtered to remove relatively small, lower-scoring, low-identity alignments, which do not likely represent orthologous locations.
Determination of best paired-end placements
We examined all pairwise combinations of end sequences that passed our criteria. In order to establish appropriate length thresholds, we initially examined the distribution of in silico insert sizes based on the mapping of 6172 chimpanzee fosmid paired-end sequences against the unique portions of human chromosome 21 and chimpanzee chromosome 22. We determined that the insert size was tightly distributed around the mean (PTR 22: 37.2 ± 4.1 kb; HSA 22: 37.2 ± 4.3 kb). This distribution was maintained after alignment of 555,929 high-quality clones to the whole human genome (Fig. 1A). Based on this distribution we chose a concordant insert size range of 24.9–49.5 kb (within three standard deviations of the mean of the chimpanzee chromosome 22 distribution), making it unlikely that size-discordant clones deviating outside of this range would represent chance occurrences or differences in the assembly rather than true rearrangements. For each fosmid, all paired-end alignment combinations were scored for placement. The placement score was essentially a four-point ordinal scale weighted for the longest (+1 per end) and most identical (+1 per end) end sequences, thus helping to avoid false-positive rearrangements due to recent segmental duplications or gene conversion events between extant or nonorthologous sites within the genome. Fosmids were retained for further analysis if they had a high-quality best placement (only one pair of end alignments having a highest placement score and both ends had 30 bp of Phred Q 30). To add additional stringency for the detection of putative rearrangements, we required discordant alignments (insert size <24.9 kb or >49.5 kb and/or misoriented ends) to have 96.5% identity, be at least 400 bp in length, and contain 150 bp of unique sequence (RepeatMasker-detected genomic elements with a sequence divergence <2% from consensus). In total, 488,887 of the 689,403 high-quality fosmids (with at least one end mapped in the human genome) demonstrated “best” placements at both ends, which represents a physical coverage of the human genome of ∼6.8-fold (40 kb × 488,887 fosmids / 2.85 Gb of euchromatic genome). Of these best placements, 484,322 (99%) and 4555 (1%) were concordant and discordant, respectively. The remaining 200,516 are high-quality clones that have one end that is a best placement, but the other end places non-optimally either on the same or a different chromosome, one or both ends place optimally or sub-optimally at multiple locations, or one end does not place in the human assembly at all (“singletons”). The high-quality discordant pairs (n = 4555) were classified as those in which the insert size was predicted to be too large (n = 3369) or too small (n = 849). Some of these discordant pairs (n = 337) also showed an incorrect orientation of ends with respect to the human.
Detection of rearrangements
Putative rearrangements were first identified computationally when two or more independent discordant fosmid clones supported the same type of rearrangement at an overlapping genomic position. Specifically, relative to the reference genome, multiple discordant fosmids supported an insertion when their insert size was too small, a deletion when the insert size was too large, and an inversion when the ends were directly oriented, rather than inverted. The minimal region containing the rearrangement on the reference genome was defined for each rearrangement by the position of the most juxtaposed/interior end sequences of the discordant clones overlapping the genomic region. For each minimal region of rearrangement, we calculated the amount of gap sequence, segmental duplication, and coverage of concordant fosmids. We used separate secondary criteria for insertion/deletion events to reduce the rate of false positives. To verify deletions, we required a break in concordant coverage (i.e., the bases spanned by concordant clones) to provide support for the configuration represented in the human reference sequence. We also removed 10 regions from our final set because they contained fosmids that spanned >1 Mb of DNA but failed to meet the second criteria with a sufficient gap in concordant fosmid coverage (i.e., <50% of the total length of the span). For insertion regions, we required the presence of at least two flanking singletons (clones in which one end is a best match in the human genome and the other is unaligned), in the appropriate orientation, for verification. Sequence annotation, discrepant clones, and putative regions of rearrangement, based on the two primary and secondary requirements described above, were displayed together for each chromosome using parasight (http://humanparalogy.gs.washington.edu/parasight/; Supplemental Figs. 3–26). During our analysis of discordant fosmids, we identified 37 regions where fosmid pairs span gaps within the sequence assembly. While such clones may be informative in directing gap closure, it is less likely that they represent true sites of structural variation due to the difficulties of accurately estimating gap sizes (IHGSC 2004). During this analysis we also identified 163 sites where the beginning and end positions of two different fosmids mapped within 20 bp of one another. We conservatively classified these as library amplification events (i.e., clonal propagates) and excluded these from further analysis. After elimination of clonal propagation and other assembly artifacts, we identified 651 sites of putative structural variation, corresponding to 293 chimpanzee deletions, 184 insertions, and 174 inversions (Fig. 5; Table 2; Supplemental Table 1). For each site, two statistics are calculated. To estimate the size of the structural variation, we first compute the “average discordance,” based on the difference of discordant fosmid pairs from the expected mean insert size, then added or subtracted the mean insert size (37.2 kb) as appropriate for regions with large or small clones. The minimum distance between clustered paired-end sequences, termed “genomic span,” provides precision on the map location of the variation (Supplemental Table 1). The largest deletion with respect to the chimpanzee genome that we detect is 815 kb in size, while the largest insertion is 36.5 kb (Supplemental Table 1, estimated insert size). It should be noted that while there is no upper bound for detecting deletions, there is a theoretical limit on insertion length since we cannot detect insertions that exceed the length of a fosmid insert (∼40 kb). Based on the genomic span, we can estimate inversion sizes from 1.5 kb to 215 Mb. As expected, inversion signatures occur frequently in pairs marking either end of the inversion breakpoint, especially in the case of larger events. Correcting for this effect, we estimate that 174 independent inversions are detected.
Permutation testing and generation of random distributions
We randomly sampled 40 genes from the set of ∼35,000 genes tested for expression differences between humans and chimpanzees (Khaitovich et al. 2005). We repeated this sampling procedure (n = 10,000), recording the proportion of genes showing increased or decreased expression in chimpanzee. The mean percentages for all 10,000 iterations showing under- or overexpression in chimpanzee were 22% and 14%, consistent with the entire data set (n = 35,000 genes). The proportion of genes overlapping with chimpanzee deletions showing reduced expression (17/40) was significantly increased (p <0.003) when compared with the total gene set.
To test the distribution of our insertion and deletion events across the genome, we randomly simulated the placement of 447 insertion and deletion events of equivalent size within the human reference assembly and recorded the distances between each event and its closest neighbor. We replicated this simulation of randomly placed events 10,000 times. We compared the distribution of these distances to the distribution of distances between closest neighbors found in the observed data set of 447 events and established a probability distribution (Poisson) for the number of events occurring by chance within a given amount of sequence.
Chimpanzee WSSD comparison
We implemented the WSSD duplication detection strategy, which measures the depth of coverage of random WGS sequence data against the human reference sequence to identify duplicated sequence in chimpanzee (>94% and >20 kb in length) (Cheng et al. 2005).
Microarray comparison
Gene expression differences between human and chimpanzee were assessed as described (Khaitovich et al. 2005). Briefly, five tissues (heart, brain, liver, kidney, and testis) were compared among five chimpanzee and six human individuals using Affymetrix® HG U133plus2 arrays. Eleven probes for each gene were chosen. All probes with significant difference in hybridization efficiency between humans and chimpanzees were excluded by first estimating the relative binding efficiency for each probe in the probe set by comparing the signal intensity of this probe to the intensities of all other probes within a probe set. We then compared the calculated binding efficiencies of the probes between all human and all chimpanzee samples using a t-test. If the binding efficiency of a probe differed significantly between human and chimpanzee samples (p <0.001), the probe was masked. Since this algorithm does not rely on actual sequence comparison, probes with different binding efficiencies caused by sequence differences in any copy of the gene will be masked (Khaitovich et al. 2004). Differentially expressed transcripts were defined as those which met the following criteria: (1) The corresponding probe set had to be expressed in all individuals from at least one species (detection p-value <0.065), and (2) the corresponding probe set had to show a change in expression in the same direction in all 30 pairwise comparisons. These cut-offs correspond to a false discovery rate ≤1.0% in all five tissues, estimated from 10,000 random permutations of sample labels.
PCR analyses and genomic hybridization
Oligonucleotides were designed within conserved sequence flanking sites of structural variation. PCR amplification conditions were as follows: Initial denaturation for 5 min at 95°C, “touchdown” from 65°Cto55°C, (60 sec 95°C, 60 sec 65°C, 60 sec 72°C, decreasing 1°C/cycle for 10 cycles), followed by 35 additional cycles of 60 sec 95°C, 60 sec 55°C, 60 sec 72°C (oligonucleotide sequences are shown in Supplemental Table 5). DNA samples (in the order of the gel): JK1051A (Homo sapiens), GM17015 (Homo sapiens), CO551 (Pan troglodytes), SFBR-4X0396 (Pan troglodytes), SFBR-4X0430 (Pan troglodytes), SFBR-4X0429 (Pan troglodytes), NG05253 (Pan paniscus), NG05251 (Gorilla gorilla), EEE-0002PPY (Pongo pygmaeus), SFBR-8320 (Papio hamadryas), NAO363446 (Macaca mullata). For Southern hybridizations, primate DNA (human [Homo sapiens], ELGP18; common chimpanzee [Pan troglodytes], AG16618, NA03448, NA03450, and NG06939; bonobo [Pan paniscus], LB501A and LB502A; gorilla [Gorilla gorilla], EEE0001GG0 and NG05251; orangutan [Pongo pygmaeus], EEE0003PPY and EEE0004PPY) was restriction enzyme-digested, transferred to nylon membrane, and hybridized as described previously (Yohn et al. 2005). Human PCR amplicons (see Supplemental Table 5 for PCR oligonucleotide sequence and conditions) corresponding to indels were used as radioactive probes.
RT-PCR validation of IL1F7
RNA purification was performed on peripheral blood samples according to standard protocol of the TriZol purification kit (Invitrogen Life Technologies, #155 96–026). Synthesis of cDNA from RNA was performed according to standard protocols of the ProtoScript cDNA synthesis kit (New England Biolabs, #E6500S). PCR amplification conditions were as follows: Initial denaturation for 2 min at 94°C, followed by 35 additional cycles of 60 sec 94°C, 30 sec 60°C, 30 sec 72°C (oligonucleotide sequences shown in Supplemental Table 5). Samples tested (in order of appearance on gel): gorilla (Gorilla gorilla, 465); bonobo (Pan paniscus, LB502); human (Homo sapiens, EEE0007HSA and EEE0008HSA); and chimpanzee (Pan troglodytes, BC450 and BC449).
FISH
Fluorescent in situ hybridization was used to validate potential inversions between human and chimpanzee. Human RP11-BACs (based on end-sequence map positions within the July 2004 UCSC human genome browser) were selected corresponding to non-duplicated human sequence on either side of the inversion breakpoint (Supplemental Table 3). Metaphase and interphase nuclei were hybridized (Horvath et al. 2000), and bicolor and tricolor FISH experiments were compared between chimpanzee and human chromosomes. A disruption in continuity and order of probes between the two species was taken as evidence of a true inversion. FISH experiments that showed colinearity of the markers and additional interphase or metaphase nuclei were scored as chimpanzee segmental duplications. At least 10 metaphases were examined for each experiment, and chromosome identity was established using standard DAPI staining according to the guidelines of the International Standard for Cytogenetic Nomenclature (ISCN 1985).
Supplementary Material
Acknowledgments
The authors would like to thank Dr. Jeff Bailey, Dr. Andy Sharp, Dr. Devin Locke, and Sierra Hansen for helpful comments and discussion regarding this manuscript. Non-human primate materials used in the research were provided in part by the Southwest National Primate Research Center (P51-RR013986). We thank the Chimpanzee Sequencing and Analysis Consortium for access to the chimpanzee genome assembly, WGS trace, and finished sequence data prior to publication. We are grateful to Jerilyn Pecotte, Steve Warren, and Jeffrey Rogers for providing some of the primate material used in this study. E.E.E. is an investigator of the Howard Hughes Medical Institute. This work was supported in part by NIH grants GM58815 and HG002385 to E.E.E. In addition, the authors gratefully acknowledge CEGBA (Centro di Eccellenza Geni in campo Biosanitario e Agroalimentare), MIUR (Ministero Italiano della Università e della Ricerca; Cluster C03, Prog. L.488/92), the European Commission (INPRIMAT, QLRI-CT-2002–01325), and the BMBF (Bundesministerium für Bildung und Forschung) for financial support.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.4338005. Article published online before print in September 2005.
Footnotes
[Supplemental material is available online at www.genome.org. The following individuals kindly provided reagents, samples, or unpublished information as indicated in the paper: The Southwest National Primate Research Center, The Chimpanzee Sequencing and Analysis Consortium, Jerilyn Pecotte, Peter Parham, Steve Warren, and Jeffrey Rogers.]
References
- Albertson, D.G., Ylstra, B., Segraves, R., Collins, C., Dairkee, S.H., Kowbel, D., Kuo, W.L., Gray, J.W., and Pinkel, D. 2000. Quantitative mapping of amplicon structure by array CGH identifies CYP24 as a candidate oncogene. Nat. Genet. 25: 144-146. [DOI] [PubMed] [Google Scholar]
- Ali, S. and Hasnain, S.E. 2002. Molecular dissection of the human Y-chromosome. Gene 283: 1-10. [DOI] [PubMed] [Google Scholar]
- Britten, R.J. 2002. Divergence between samples of chimpanzee and human DNA sequences is 5%, counting indels. Proc. Natl. Acad. Sci. 99: 13633-13635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, F.C. and Li, W.H. 2001. Genomic divergences between humans and other hominoids and the effective population size of the common ancestor of humans and chimpanzees. Am. J. Hum. Genet. 68: 444-456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, Z., Ventura, M., She, X., Khaitovich, P., Graves, T., Osoegawa, K., Church, D., DeJong, P., Wilson, R.K., Pääbo, S., et al. 2005. Nature 437: 88-93. [DOI] [PubMed] [Google Scholar]
- The Chimpanzee Sequencing and Analysis Consortium. 2005. Initial sequence of the chimpanzee genome and comparison with the human genome. Nature 437: 69-87. [DOI] [PubMed] [Google Scholar]
- Dennehey, B.K., Gutches, D.G., McConkey, E.H., and Krauter, K.S. 2004. Inversion, duplication, and changes in gene context are associated with human chromosome 18 evolution. Genomics 83: 493-501. [DOI] [PubMed] [Google Scholar]
- Dutrillaux, B. 1980. Chromosomal evolution of the great apes and man. J. Reprod. Fertil. Suppl. Suppl 28: 105-111. [PubMed]
- Ebersberger, I., Metzler, D., Schwarz, C., and Pääbo, S. 2002. Genomewide comparison of DNA sequences between humans and chimpanzees. Am. J. Hum. Genet. 70: 1490-1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichler, E.E., Clark, R.A., and She, X. 2004a. An assessment of the sequence gaps: Unfinished business in a finished human genome. Nat. Rev. Genet. 5: 345-354. [DOI] [PubMed] [Google Scholar]
- Eichler, E., Hillier, L., Warren, W., Mardis, E., and Wilson, R. 2004b. “Additional sequencing of the chimpanzee genome”, pp. 6. Washington University School of Medicine and Case Western Reserve University School of Medicine.
- Enard, W., Khaitovich, P., Klose, J., Zollner, S., Heissig, F., Giavalisco, P., Nieselt-Struwe, K., Muchmore, E., Varki, A., Ravid, R., et al. 2002. Intra- and interspecific variation in primate gene expression patterns. Science 296: 340-343. [DOI] [PubMed] [Google Scholar]
- Ewing, B. and Green, P. 1998. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 8: 186-194. [PubMed] [Google Scholar]
- Fortna, A., Kim, Y., MacLaren, E., Marshall, K., Hahn, G., Meltesen, L., Brenton, M., Hink, R., Burgers, S., Hernandez-Boussard, T., et al. 2004. Lineage-specific gene duplication and loss in human and great ape evolution. PLoS Biol. 2: E207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fredman, D., White, S.J., Potter, S., Eichler, E.E., Den Dunnen, J.T., and Brookes, A.J. 2004. Complex SNP-related sequence variation in segmental genome duplications. Nat. Genet. 36: 861-866. [DOI] [PubMed] [Google Scholar]
- Goidts, V., Szamalek, J.M., Hameister, H., and Kehrer-Sawatzki, H. 2004. Segmental duplication associated with the human-specific inversion of chromosome 18: A further example of the impact of segmental duplications on karyotype and genome evolution in primates. Hum. Genet. 115: 116-122. [DOI] [PubMed] [Google Scholar]
- Gonzales, M.J., Delwart, E., Rhee, S.Y., Tsui, R., Zolopa, A.R., Taylor, J., and Shafer, R.W. 2003. Lack of detectable human immunodeficiency virus type 1 superinfection during 1072 person-years of observation. J. Infect. Dis. 188: 397-405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollox, E.J., Poulter, M., Zvarik, M., Ferak, V., Krause, A., Jenkins, T., Saha, N., Kozlov, A.I., and Swallow, D.M. 2001. Lactase haplotype diversity in the Old World. Am. J. Hum. Genet. 68: 160-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath, J., Viggiano, L., Loftus, B., Adams, M., Rocchi, M., and Eichler, E. 2000. Molecular structure and evolution of an α/non-α satellite junction at 16p11. Hum. Molec. Genet. 9: 113-123. [DOI] [PubMed] [Google Scholar]
- Horvath, J.E., Gulden, C.L., Bailey, J.A., Yohn, C., McPherson, J.D., Prescott, A., Roe, B.A., De Jong, P.J., Ventura, M., Misceo, D., et al. 2003. Using a pericentromeric interspersed repeat to recapitulate the phylogeny and expansion of human centromeric segmental duplications. Mol. Biol. Evol. 9: 1463-1479. [DOI] [PubMed] [Google Scholar]
- Iafrate, A.J., Feuk, L., Rivera, M.N., Listewnik, M.L., Donahoe, P.K., Qi, Y., Scherer, S.W., and Lee, C. 2004. Detection of large-scale variation in the human genome. Nat. Genet.. 9: 949-951. [DOI] [PubMed] [Google Scholar]
- International Human Genome Sequencing Consortium (IHGSC). 2001. Initial sequencing and analysis of the human genome. Nature 409: 860-921. [DOI] [PubMed] [Google Scholar]
- ———. 2004. Finishing the euchromatic sequence of the human genome. Nature 431: 931-945. [DOI] [PubMed] [Google Scholar]
- ISCN. 1985. Report of the standing committee on human cytogenetic nomenclature. Birth Defects 21: 1-117. [PubMed] [Google Scholar]
- Jackson, M.S., Rocchi, M., Thompson, G., Hearn, T., Crosier, M., Guy, J., Kirk, D., Mulligan, L., Ricco, A., Piccininni, S., et al. 1999. Sequences flanking the centromere of human chromosome 10 are a complex patchwork of arm-specific sequences, stable duplications, and unstable sequences with homologies to telomeric and other centromeric locations. Hum. Mol. Genet. 8: 205-215. [DOI] [PubMed] [Google Scholar]
- Johnson, M.E., Viggiano, L., Bailey, J.A., Abdul-Rauf, M., Goodwin, G., Rocchi, M., and Eichler, E.E. 2001. Positive selection of a gene family during the emergence of humans and African apes. Nature 413: 514-519. [DOI] [PubMed] [Google Scholar]
- Kehrer-Sawatzki, H., Schreiner, B., Tanzer, S., Platzer, M., Muller, S., and Hameister, H. 2002. Molecular characterization of the pericentric inversion that causes differences between chimpanzee chromosome 19 and human chromosome 17. Am. J. Hum. Genet. 71: 375-388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kehrer-Sawatzki, H., Sandig, C., Chuzhanova, N., Goidts, V., Szamalek, J.M., Tanzer, S., Muller, S., Platzer, M., Cooper, D.N., and Hameister, H. 2005a. Breakpoint analysis of the pericentric inversion distinguishing human chromosome 4 from the homologous chromosome in the chimpanzee (Pan troglodytes). Hum. Mutat. 25: 45-55. [DOI] [PubMed] [Google Scholar]
- Kehrer-Sawatzki, H., Sandig, C.A., Goidts, V., and Hameister, H. 2005b. Breakpoint analysis of the pericentric inversion between chimpanzee chromosome 10 and the homologous chromosome 12 in humans. Cytogenet. Genome Res. 108: 91-97. [DOI] [PubMed] [Google Scholar]
- Kehrer-Sawatzki, H., Szamalek, J.M., Tanzer, S., Platzer, M., and Hameister, H. 2005c. Molecular characterization of the pericentric inversion of chimpanzee chromosome 11 homologous to human chromosome 9. Genomics 85: 542-550. [DOI] [PubMed] [Google Scholar]
- Khaitovich, P., Muetzel, B., She, X., Lachmann, M., Hellmann, I., Dietzsch, J., Steigele, S., Do, H.H., Weiss, G., Enard, W., et al. 2004. Regional patterns of gene expression in human and chimpanzee brains. Genome Res. 14: 1462-1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khaitovich, P., Hellmann, I., Enard, W., Nowick, K., Leinweber, M., Franz, H., Weiss, G., Lachmann, M., and Pääbo, S. 2005. Parallel patterns of evolution in the genomes and transcriptomes of humans and chimpanzees. Science (in press). [DOI] [PubMed]
- Kim, U.J., Shizuya, H., de Jong, P.J., Birren, B., and Simon, M.I. 1992. Stable propagation of cosmid sized human DNA inserts in an F factor based vector. Nucleic Acids Res. 20: 1083-1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar, S. and Hedges, S.B. 1998. A molecular timescale for vertebrate evolution. Nature 392: 917-920. [DOI] [PubMed] [Google Scholar]
- Lahn, B.T. and Page, D.C. 1999. Four evolutionary strata on the human X chromosome. Science 286: 964-967. [DOI] [PubMed] [Google Scholar]
- Lejeune, J., Dutrillaux, B., Rethore, M.O., and Prieur, M. 1973. [Comparison of the structure of chromatids of Homo sapiens and Pan troglodytes (author's transl.).] Chromosoma 43: 423-444. [DOI] [PubMed] [Google Scholar]
- Liu, G., Zhao, S., Bailey, J.A., Sahinalp, S.C., Alkan, C., Tuzun, E., Green, E.D., and Eichler, E.E. 2003. Analysis of primate genomic variation reveals a repeat-driven expansion of the human genome. Genome Res. 13: 358-368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locke, D.P., Archidiacono, N., Misceo, D., Cardone, M.F., Deschamps, S., Roe, B., Rocchi, M., and Eichler, E.E. 2003a. Refinement of a chimpanzee pericentric inversion breakpoint to a segmental duplication cluster. Genome Biol. 4: R50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locke, D.P., Segraves, R., Carbone, L., Archidiacono, N., Albertson, D.G., Pinkel, D., and Eichler, E.E. 2003b. Large-scale variation among human and great ape genomes determined by array comparative genomic hybridization. Genome Res. 13: 347-357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locke, D.P., Segraves, R., Nicholls, R.D., Schwartz, S., Pinkel, D., Albertson, D.G., and Eichler, E.E. 2004. BAC microarray analysis of 15q11-q13 rearrangements and the impact of segmental duplications. J. Med. Genet. 41: 175-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locke, D.P., Jaing, Z., Pertz, L.M., Misceo, D., Archidiacono, N., and Eichler, E.E. 2005. Molecular evolution of the human chromosome 15 pericentromeric region. Cytogenet. Genome Res. 108: 73-82. [DOI] [PubMed] [Google Scholar]
- Lupski, J.R. 2004. Hotspots of homologous recombination in the human genome: Not all homologous sequences are equal. Genome Biol. 5: 242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouse Genome Sequencing Consortium (MGSC). 2002. Initial sequencing and comparative analysis of the mouse genome. Nature 420: 520-562. [DOI] [PubMed] [Google Scholar]
- Needleman, S.B. and Wunsch, C.D. 1970. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J. Mol. Biol. 48: 443-453. [DOI] [PubMed] [Google Scholar]
- Nickerson, E., Gibbs, R.A., and Nelson, D.L. 2005. Breakpoint analysis of a pericentric inversion distinguishing the human and chimpanzee genomes. Genome Res. (in press).
- Ohno, S. 1970. Evolution by gene duplication. Springer Verlag, Berlin/Heidelberg/New York.
- Olson, M.V. 1999. When less is more: Gene loss as an engine of evolutionary change. Am. J. Hum. Genet. 64: 18-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinkel, D., Segraves, R., Sudar, D., Clark, S., Poole, I., Kowbel, D., Collins, C., Kuo, W.L., Chen, C., Zhai, Y., et al. 1998. High resolution analysis of DNA copy number variation using comparative genomic hybridization to microarrays. Nat. Genet. 20: 207-211. [DOI] [PubMed] [Google Scholar]
- Sakaki, Y., Watanabe, H., Taylor, T., Hattori, M., Fujiyama, A., Toyoda, A., Kuroki, Y., Itoh, T., Saitou, N., Oota, S., et al. 2003. Human versus chimpanzee chromosome-wide sequence comparison and its evolutionary implication. Cold Spring Harb. Symp. Quant. Biol. 68: 455-460. [DOI] [PubMed] [Google Scholar]
- Samonte, R.V. and Eichler, E.E. 2002. Segmental duplications and the evolution of the primate genome. Nat. Rev. Genet. 3: 65-72. [DOI] [PubMed] [Google Scholar]
- Sebat, J., Lakshmi, B., Troge, J., Alexander, J., Young, J., Lundin, P., Maner, S., Massa, H., Walker, M., Chi, M., et al. 2004. Large-scale copy number polymorphism in the human genome. Science 305: 525-528. [DOI] [PubMed] [Google Scholar]
- Sharp, A.J., Locke, D.P., McGrath, S.D., Cheng, Z., Bailey, J.A., Vallente, R.U., Pertz, L.M., Clark, R.A., Schwartz, S., Segraves, R., et al. 2005. Segmental duplications and copy-number variation in the human genome. Am. J. Hum. Genet. 77: 78-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snijders, A.M., Nowak, N., Segraves, R., Blackwood, S., Brown, N., Conroy, J., Hamilton, G., Hindle, A.K., Huey, B., Kimura, K., et al. 2001. Assembly of microarrays for genome-wide measurement of DNA copy number. Nat. Genet. 29: 263-264. [DOI] [PubMed] [Google Scholar]
- Stankiewicz, P., Park, S.S., Inoue, K., and Lupski, J.R. 2001. The evolutionary chromosome translocation 4;19 in Gorilla gorilla is associated with microduplication of the chromosome fragment syntenic to sequences surrounding the human proximal CMT1A-REP. Genome Res. 11: 1205-1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stankiewicz, P., Shaw, C.J., Withers, M., Inoue, K., and Lupski, J.R. 2004. Serial segmental duplications during primate evolution result in complex human genome architecture. Genome Res. 14: 2209-2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuzun, E., Sharp, A.J., Bailey, J.A., Kaul, R., Morrison, V.A., Pertz, L.M., Haugen, E., Hayden, H., Albertson, D., Pinkel, D., et al. 2005. Fine-scale structural variation of the human genome. Nat. Genet.. 7: 727-732. [DOI] [PubMed] [Google Scholar]
- Yohn, C.T., Jiang, Z., McGrath, S.D., Hayden, K.E., Khaitovich, P., Johnson, M.E., Eichler, M.Y., McPherson, J.D., Zhao, S., Pääbo, S., et al. 2005. Lineage-specific expansions of retroviral insertions within the genomes of african great apes but not humans and orangutans. PLoS Biol. 3: 1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yunis, J.J. and Prakash, O. 1982. The origin of man: A chromosomal pictorial legacy. Science 215: 1525-1530. [DOI] [PubMed] [Google Scholar]
- Yunis, J.J., Sawyer, J.R., and Dunham, K. 1980. The striking resemblance of high-resolution G-banded chromosomes of man and chimpanzee. Science 208: 1145-1148. [DOI] [PubMed] [Google Scholar]
WEB SITE REFERENCES
- http://genome.ucsc.edu/goldenPath/help/chain.html; UCSC genome Web browser.
- http://www.ncbi.nlm.nih.gov/RefSeq/; NCBI RefSeq Web page.
- http://humanparalogy.gs.washington.edu/CSV; Chimpanzee structural variation database.
- http://humanparalogy.gs.washington.edu/parasight; Parasight software.
- http://www.ncbi.nlm.nih.gov/Traces/trace.cgi?; NCBI trace archive.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.