Abstract
Laboratory evolution can be used to address fundamental questions about adaptation to selection pressures and, ultimately, the process of evolution. In this study, we investigated the reproducibility of growth phenotypes and global gene expression states during adaptive evolution. The results from parallel, replicate adaptive evolution experiments of Escherichia coli K-12 MG1655 grown on either lactate or glycerol minimal media showed that (1) growth phenotypes at the endpoint of evolution are convergent and reproducible; (2) endpoints of evolution have different underlying gene expression states; and (3) the evolutionary gene expression response involves a large number of compensatory expression changes and a smaller number of adaptively beneficial expression changes common across evolution strains. Gene expression changes initially showed a large number of differentially expressed genes in response to an environmental change followed by a return of most genes to a baseline expression level, leaving a relatively small set of differentially expressed genes at the endpoint that varied between evolved populations.
One characteristic of biological systems that is both interesting and difficult to describe is the ability of these systems to adapt and to evolve under various environmental conditions. Because of the numerous advantages of using microorganisms as model systems for studying evolution (Elena and Lenski 2003), laboratory evolution recently has grown into a standard tool for studying the evolutionary process in a controlled manner within the microbial community (Helling et al. 1987; Wood and Ingram 1992; Lenski et al. 1998; Nakatsu et al. 1998; Massey et al. 1999; Papadopoulos et al. 1999; Wichman et al. 1999; Cooper et al. 2001; Riehle et al. 2001; Shaver et al. 2002). Despite these efforts, several key questions pertaining to cell biology and evolution remain unanswered (Elena and Lenski 2003).
One foundational concept in evolutionary biology is the notion that organisms traverse a “fitness landscape” during the evolutionary process (Sauer 2001; Kassen 2002; Elena and Lenski 2003; Orr 2005). The fitness landscape depicts an organism's fitness in relation to a specific evolutionary environment where regions of improved fitness are depicted by peaks within the landscape and the shape of the landscape itself is determined by genetic and epigenetic factors (Waddington 1940, 1957; Jablonka and Lamb 2002). A fundamental question associated with this concept pertains to the degree of convergence or reproducibility of the outcome of the evolutionary process. Traditionally, the fitness landscape is depicted as containing multiple peaks of improved fitness, implying the possibility of divergence during evolution. A contrasting perspective proposed by metabolic modeling descriptions suggests that a single global optimal phenotype exists and can be achieved through equivalent usage of the metabolic network (Edwards et al. 2001; Ibarra et al. 2002; Fong et al. 2003; Mahadevan and Schilling 2003).
In addition to phenotype reproducibility at the endpoint of evolution, much interest is given to determining mechanistic changes occurring during the evolutionary process. To investigate mechanistic changes and variability involved in evolution, quantitative metrics are needed that measure cellular phenotypes on a genome scale. Fortunately, a growing number of technologies are now available to provide quantitative, system-wide biological measurements. For example, gene expression microarrays are used to assess genome-wide mRNA transcript levels. Several evolution studies have used gene expression microarrays to study laboratory evolution (Ferea et al. 1999; Cooper et al. 2003; Riehle et al. 2003) but were only able to draw conclusions based on a small subset of genes because of statistical limitations involved in microarray data analysis (Hess et al. 2001; Nadon and Shoemaker 2002). Although these statistical issues may always be present to some degree, recent improvements in gene expression arrays (Venkatasubbarao 2004) and additional statistical methods based on the false-discovery rate (FDR) (Storey and Tibshirani 2003) allow for the study of larger sets of genes with a higher degree of statistical confidence.
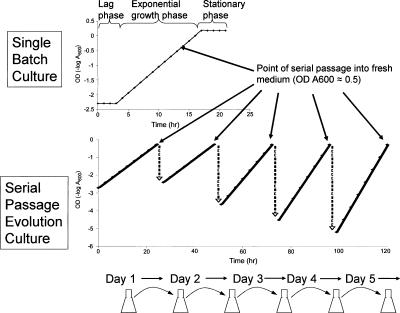
In an effort to study both the phenotypic and the underlying mechanistic changes that occur during evolution, we sought to evaluate the reproducibility of the endpoint of adaptive evolution and to study mechanisms involved in the evolutionary process by conducting parallel, replicate evolution experiments. Evolution cultures were maintained in prolonged exponential growth by daily passage into fresh medium before cultures reached stationary phase (Fig. 1). Evolution experiments were conducted in two independent growth environments, and cellular phenotypes for all evolution populations were determined by measuring growth rates (GRs), substrate uptake rates (SURs), oxygen uptake rates (OURs), GRs on alternative carbon substrates, and genome-wide transcript levels.
Figure 1.
Schematic representation of the experimental evolutionary procedure. Prolonged exponential growth is maintained throughout the course of adaptive evolution by daily passage of cultures into fresh medium prior to entry into stationary phase. Inoculum at time of passage is adjusted to account for increasing growth rates (slope of log plot) over evolutionary time.
Results
In this study, the process of adaptive evolution was investigated using the wild-type K-12 MG1655 strain of Escherichia coli. Seven evolved populations of E. coli were generated through adaptive evolution both on lactate-supplemented M9 minimal medium and on glycerol-supplemented M9 minimal medium. Using these evolved populations, experiments were conducted to investigate evolutionary changes in terms of growth phenotypes and mRNA transcript levels.
Evolving the growth phenotype
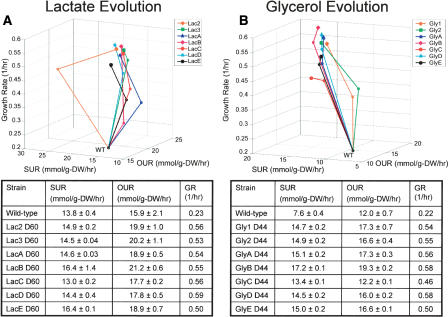
Seven parallel evolution experiments (indexed as Lac2, Lac3, LacA, LacB, LacC, LacD, and LacE) were conducted on L-lactate-supplemented M9 minimal medium at 30°C for 60 days (>1000 generations) and seven parallel evolution experiments (Gly1, Gly2, GlyA, GlyB, GlyC, GlyD, and GlyE) were conducted on glycerol-supplemented M9 minimal medium at 30°C for 44 days (>600 generations). GRs, OURs, SURs, and a biomass correlation were measured for each population at beginning of evolution (wild type), day 20 of evolution, and the endpoint of evolution (day 60 for lactate populations, day 44 for glycerol populations) to track the evolutionary paths of each population. These detailed phenotype measurements allowed the evolutionary paths to be visualized in a three-dimensional fitness landscape where a plane is defined by the SUR and OUR and the GR is graphed on the z-axis as a measure of improved fitness (Fig. 2).
Figure 2.
Fitness landscape depicting phenotype changes during evolution. Measured oxygen uptake rate (OUR, mmol/g-DW/hr) and substrate uptake rate (SUR, mmol/g-DW/hr) values form a plane with improved growth fitness indicated by growth rate (1/hr) on the z-axis. (A) L-lactate evolution populations were characterized at day 1, day 20, and day 60 of evolution. (B) Glycerol evolution populations were characterized at day 1, day 20, and day 44 of evolution. WT denotes the parental wild-type strain and endpoints of evolution are shown with filled symbols. Tables show phenotype measurements for the wild-type (unevolved) and endpoints of evolution with error shown as the standard deviation between replicate measurements.
Results for adaptive evolution on both lactate (Fig. 2A) and glycerol (Fig. 2B) showed convergence of the growth phenotype at the endpoint of evolution in six of the seven evolved populations (with outlier populations LacE and GlyC). In both cases, convergence to a similar growth phenotype was exhibited with GRs, SURs, and OURs all within 12% of each other. The coefficient of variation for each parameter was also calculated to objectively evaluate the degree of variability in these measurements, and this calculation showed decreased variance between evolved populations at the endpoint relative to the day 20 measurements (see Supplemental Table 1). The observed convergence of the growth phenotypes was particularly striking given the differences in evolutionary paths taken by the different populations and the large fitness increase observed (average GR increase: 135% lactate; 145% glycerol). For populations that exhibited lower GRs (LacE and GlyC), evolution was continued for an additional 10 days (∼170 generations) with no observed increase in growth rate (shown in Figures 4 and 5, below).
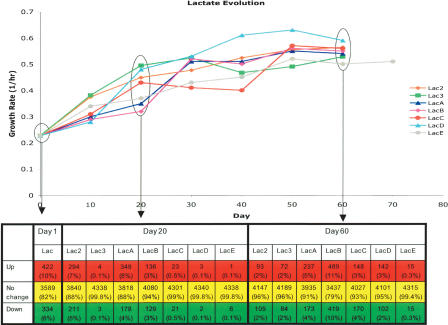
Figure 4.
Gene expression changes in lactate evolution populations throughout the course of adaptive evolution as compared with glucose wild-type expression profile. Growth rate changes throughout evolution are shown as a function of day of evolution with expression profiling performed at days 1, 20, and 60 of evolution. Significant expression changes from the wild-type strain grown on glucose were calculated by t-test using P-value cut-off corresponding to a false-discovery rate of 5%. By this calculation, the number of genes showing statistically significant expression changes tabulated below the plot at either increased expression compared with the wild-type (Up), decreased expression compared with the wild-type (Down), or no change in expression (No change). Percentages given in each cell indicated the percentage of genes falling within the category as a portion of the total genes.
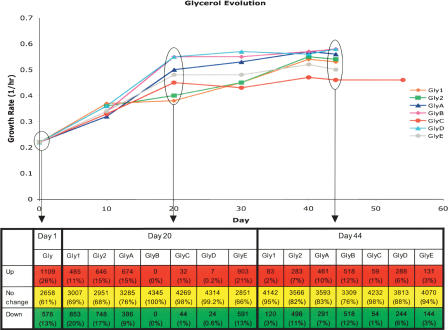
Figure 5.
Gene expression changes in glycerol evolution populations throughout the course of adaptive evolution as compared with glucose wild-type expression profile. Growth rate changes throughout evolution are shown as a function of day of evolution with expression profiling performed at days 1, 20, and 44 of evolution. Significant expression changes from the wild-type strain grown on glucose were calculated by t-test using P-value cut-off corresponding to a false-discovery rate of 5%. By this calculation, the number of genes showing statistically significant expression changes tabulated below the plot at either increased expression compared with the wild-type (Up), decreased expression compared with the wild-type (Down), or no change in expression (No change). Percentages given in each cell indicated the percentage of genes falling within the category as a portion of the total genes.
Growth phenotypes on various carbon sources
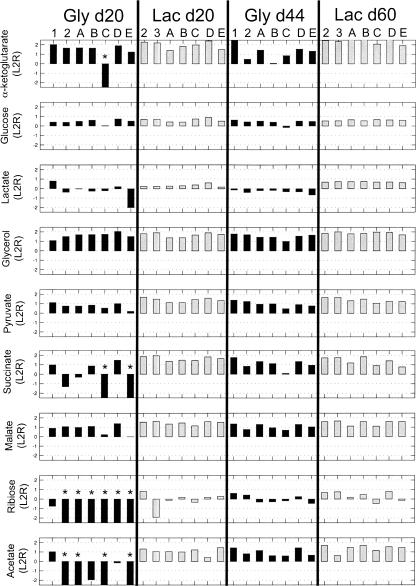
Additional evolved population phenotype characterization was conducted to determine growth properties on nonevolutionary carbon substrates. GRs were measured in replicate for growth on acetate, α-ketoglutarate, glucose, glycerol, lactate, malate, pyruvate, ribose, and succinate for each evolved population at day 20 and the endpoint of evolution. These were compared with the GR of the wild-type strain (Fig. 3). Results were analyzed to screen for large changes in growth characteristics (>50% increase or decrease). Overall, the evolved populations showed improved growth in most conditions tested (77 of 126 conditions at day 20 of evolution and 96 of 126 cases at evolutionary endpoints). Evolved populations showed highly variable growth characteristics on these nonevolutionary carbon substrates at day 20 of evolution when growth phenotypes on the primary evolution substrate were most divergent. In particular, the seven glycerol evolution populations showed differences from each other at day 20 of evolution when grown on acetate, α-ketoglutarate, lactate, malate, ribose, and succinate. The seven lactate evolution populations showed the most differences from each other when grown on ribose at day 20 of evolution. By the endpoint of evolution, growth characteristics were similar between populations evolved on the same substrate.
Figure 3.
Growth rate characterization of evolution populations on nonevolutionary substrates. Columns indicate individual populations grouped by the day of evolution and evolution substrate used. Rows show the substrate used for growth rate testing. Measurements for each population were compared with the growth rate of the wild-type strain and the log2 ratio is reported to compare growth rates. An asterisk (*) indicates cases where the evolved population exhibited slow, almost imperceptible growth relative to the wild-type strain.
mRNA transcriptional profiling
To study mechanisms involved in adaptive evolution and the dynamics of these underlying changes, mRNA transcriptional profiling was performed on day 1, day 20, and at the endpoint of evolution for all evolution populations. For each set of evolution populations (lactate and glycerol), two different baseline expression profiles were used to independently investigate the effects of changing the growth environment and the effects of adaptive evolution. First, all expression profiles for the evolution populations were compared with a wild-type glucose expression profile to determine expression changes that were associated with the shift in growth condition from glucose to lactate or glucose to glycerol (see schematic in Fig. 6, below). An additional set of parallel analyses was focused on determining changes that occurred over the course of evolution by using the day 1 expression profile for each evolution population as the baseline profile. In all analyses, statistically significant gene expression changes were identified by t-test with a P-value cut-off corresponding to a 5% FDR (Benjamini and Hochberg 1995).
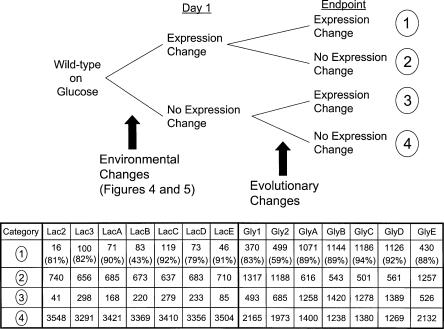
Figure 6.
Evolutionary gene expression changes for lactate- and glycerol-evolved populations. Statistically significant gene expression changes between day 1 expression profiles and endpoint expression profiles (day 60 for lactate, day 44 for glycerol) were tabulated in four categories. Genes in category 1 had changed expression at day 1 in response to the environmental shift and had an additional change in expression during evolution. Genes in category 2 had changed expression at day 1 in response to the environmental shift but no additional expression change during evolution. Genes in category 3 only exhibited an expression change during evolution, and genes in category 4 showed no change in expression at either day 1 or at the endpoint of evolution. It should be noted that genes falling within category 1 can have expression changes during evolution that amplify expression changes observed at day 1 (larger magnitude change in same direction) or that compensate for expression changes at day 1 (similar magnitude change but in opposite direction). The percentage given for numbers in category 1 indicates the number of genes (e.g., 82%: 82 of the 100 genes in Lac3) exhibiting compensatory expression changes during evolution for that strain (i.e., initial environmental change caused increased expression and evolution led to decreased expression back to wild-type expression level).
The expression changes associated with the growth environment shift revealed a large-scale initial gene expression response that was observed at day 1 of evolution for both the lactate-evolved (Fig. 4) and the glycerol-evolved (Fig. 5) populations. This growth shift at day 1 of evolution resulted in 1687 genes (39% of total genes) showing a significant change in gene expression in the glycerol populations and 756 genes (18% of total genes) showing a significant change in the lactate populations. For both sets of evolved populations, this large initial change in gene expression was followed by a dramatic decrease in the number of differentially expressed genes at day 20 and at the endpoint of evolution. On average, 770 genes (18% of total genes) in the glycerol populations showed significant expression changes at day 20 of evolution, and 498 genes (11% of total genes) exhibited significant expression changes at day 44 of evolution. The lactate populations averaged 194 significant, differentially expressed genes at day 20, and 323 genes (7% of total genes) showed significant expression changes at day 60 of evolution. Thus, most genes showing an initial change in transcript level return to the pre-evolution transcriptional state at the endpoint of evolution. In the case of the two phenotypic outlier populations, GlyC and LacE, both populations had very few genes that showed a significant change in expression at day 20 and each had the fewest number of genes (113 and 30 genes, respectively) showing a significant expression change at the end of evolution within each set of evolutions.
Using the day 1 expression profiles for each set of evolution experiments as a baseline, we also studied the gene expression changes that arise during the course of adaptive evolution (Categories 1 and 3 in Fig. 6). Of the evolutionary gene expression changes identified, we sought to distinguish between compensatory gene expression changes and those that could be linked to evolutionary phenotypic improvements and thus were adaptively beneficial. Overall, approximately 87% of the category 1 gene expression changes (involving hundreds of genes) were compensatory expression changes, meaning that the gene expression change during evolution was roughly equal in magnitude but opposite in direction to a gene expression change that occurred at day 1 of evolution in response to the environmental shift. These gene expression changes may have occurred as part of general, nonspecific initial response that was later compensated for by adaptive responses.
Of the remaining evolutionary gene expression changes, genes that exhibited similar significant differential expression in at least six of the seven parallel evolution populations represented consistent gene expression changes that were implicated in general evolutionary mechanisms. Excluding the compensatory gene expression changes, the average number of evolutionary gene expression changes found in the glycerol and lactate evolution populations were 1109 and 203, respectively. Of these changes, only a small percentage was found to be in common across parallel evolution populations. For the glycerol evolution populations, 70 genes (51 annotated) showed changes in common to at least six of the seven populations (Table 1). Evolution on glycerol was found to increase gene expression in seven tRNA genes and decrease gene expression in 23 motility and flagellar genes, as categorized by the MultiFun (Serres and Riley 2000; Serres et al. 2004) gene ontology system. This change in motility was tested and verified using a motility assay in 0.3% semi-solid agar (Kirov et al. 2002) where the endpoint glycerol populations showed decreased motility (see Supplemental Figure 1). The lactate evolution populations had only two genes in common in at least six of the seven lactate-evolved populations (Table 1), one of which was the ppsA gene that encodes phosphoenolpyruvate synthase that would be necessary for conversion of pyruvate to phosphoenolpyruvate after conversion of lactate to pyruvate.
Table 1.
Genes showing statistically significant differential expression during evolution (endpoint vs. day 1) in at least 6 of 7 evolved populations
| Gene category | Lactate | Glycerol | ||
|---|---|---|---|---|
| Hypothetical/putative | None | b0329, | b1045, | b1141, |
| b1266, | b1471, | b1566, | ||
| b1678, | b1784, | b1836, | ||
| b1934, | b2086, | b2263, | ||
| b2302, | b2327, | b2742, | ||
| b3021, | b3293, | b4078, | ||
| b4109, | ||||
| Annotated | ppsA, yihK | proM, | metU, | metT, |
| alaX, | alaW, | mltA, | ||
| murA, | thrV, | trpS, | ||
| prfC, | flgN, | mraY, | ||
| psiF, | wrbA, | msyB, | ||
| rpsV, | sodC, | ptrB, | ||
| motA, | otsB, | fliS, | ||
| fbaB, | yehV, | tktB, | ||
| ptsN, | yjhH, | aceB, | ||
| kdpE, | flgM, | flgA, | ||
| flgC, | flgD, | flgF, | ||
| flgK, | ycfN, | ycgR, | ||
| narX, | cheZ, | cheY, | ||
| cheB, | cheR, | tar, | ||
| cheW, | cheA, | motB, | ||
| fliZ, | fliA, | fliC, | ||
| fliD, | fliL, | menF |
A final analysis was performed on the gene expression data where genes were organized into known regulon (Keseler et al. 2005) associations to identify specific regulatory mechanisms dominantly active during adaptive evolution. Regulons exhibiting changed transcriptional states were selected by tabulating the percentage of genes within the regulon that showed significant expression changes and calculating a P value using the hypergeometric distribution to assess statistical significance. A number of regulatory mechanisms appeared to be active at day 1 of evolution involving global regulators. Both the lactate strains and the glycerol strains exhibited initial changes in regulons associated with phoB, cra, rpoN, and crp (see Supplemental Table 2). These regulatory effects, however, were not generally preserved over the course of evolution in the individual evolution populations and thus likely represent an initial cellular response only during the early stage of evolution.
Discussion
In this study, laboratory evolution of the wild-type E. coli K12 MG1655 strain was used to investigate adaptive evolution and probe the underlying mechanisms driving the evolutionary process. Quantitative phenotype measures of GR, SUR, OUR, GR on alternative carbon sources, and genome-wide transcript levels were conducted to assess the reproducibility of endpoint phenotypes and gain insight into mechanistic processes at work during evolution. It was found that (1) quantitative measurements of the growth phenotype throughout evolution revealed a generally convergent growth phenotype at the end of adaptive evolution despite apparent divergent evolutionary paths; (2) the transcriptional state of each evolution population was very different, despite similarity in endpoint growth phenotypes; and (3) the evolutionary gene expression response involves an initial widespread expression shift followed by a large number of compensatory gene expression changes and a smaller number of adaptively beneficial changes common across parallel evolution strains.
The endpoint growth phenotype convergence shows the generally reproducible phenotypic outcome of adaptive evolution in line with other current findings (Orr 2005). Within the context of the fitness landscape concept, this finding suggests that distinct peaks of increased adaptive fitness exist and that they may be achieved via different evolutionary paths. The divergence of evolutionary paths during evolution and convergence at the endpoint of evolution were supported not only by growth characteristics on the evolutionary substrate but also by growth characteristics on nonevolutionary substrates. Although almost all populations ultimately evolved to a similar improved growth phenotype, they all evolved differently, with some populations (LacE and GlyC) apparently becoming functionally limited. Whether the fitness peaks observed in this study represent global optima and may be addressable in future studies through perturbation of the evolutionary starting point remains an open question.
Global gene expression profiling of the endpoint evolution populations showed wide diversity in evolved transcriptional states despite being evolved in parallel and showing generally convergent growth phenotypes. This observation further provides evidence that the evolution populations used different means of achieving similar growth phenotypes during evolution and hints at the metabolic flexibility and robustness of E. coli. These results also indirectly support computational results that indicate the presence of thousands of metabolic pathway combinations that lead to identical, manifest phenotypes (Mahadevan and Schilling 2003).
Studying the changes in mRNA transcript profiles over the course of adaptive evolution indicated the presence of a dynamic, multiphase transcriptional response. The initial adaptive response to a simple environmental perturbation resulted in a large-scale general change in the transcriptional state of E. coli that appeared to be caused in part by global regulatory effects and may be linked to the dramatic decrease in growth rate (Liu et al. 2005). This initial transcriptional response is followed by compensatory gene expression changes during evolution that act to return most of these mRNA transcripts back to basal levels with only a relatively small subset of the initial gene expression changes remaining at the end of evolution. Just as it is possible to have noncausal genetic mutations (Elena and Lenski 2003), this evolutionary transcriptional process reveals the presence of noncausal changes in gene expression (those that seemingly do not affect the cellular phenotype), particularly at the early stages of adaptive evolution. It is only after many of these noncausal gene expression changes are removed through compensatory changes that adaptively beneficial gene expression changes are found, such as the decreased expression of flagellar and motility genes in the evolved glycerol populations. Interestingly, the conserved intracellular ATP that would result from reduced expression and activity of these gene products could possibly be one factor driving faster adaptation of E. coli to glycerol than to lactate.
Overall, the results of this study provide a novel dynamic view of the adaptive evolution process. The results suggest that growth adaptation and evolution appear to involve compensatory gene expression changes that essentially deselect unnecessary expression changes that occur as part of the general initial adaptive response, as well as positive selection of beneficial gene expression changes. Our analysis indicates that the initial growth adaptation is manifested through widespread gene expression changes that are mediated by global regulators with subsequent growth improvements occurring through an apparently stochastic selection process leading to divergent transcriptional states but convergent growth phenotypes.
Methods
Adaptive evolution
Evolution cultures were propagated from fresh cultures of the wild-type E. coli strain K-12 MG1655 from the American Type Culture Collection (Rockville, MD). Cultures were conducted in 250 mL of M9 minimal medium supplemented with 2 g/L of lactate or glycerol in covered 500-mL Erlenmeyer flasks using magnetic stir bars for aeration (Ibarra et al. 2004) at 30°C. The cells were grown overnight and allowed to reach mid-exponential growth (A600 ≤ 0.5) before being diluted by passage into fresh medium. Passage of each culture into fresh medium was conducted in a biosafety cabinet using standard sterile technique practices. The amount of dilution at each passage was adjusted daily to account for changes in growth rate (typically between 5 × 105 and 5 × 108 cells were passed during each inoculation) shown in Figure 1. The optical density (OD) following dilution typically had an absorbance of A600 ≤ 2.4 × 10-6. Batch growth and serial passage were conducted for 60 days for all lactate cultures (∼1000 generations) and for 44 days for all glycerol cultures (∼600 generations), at which point a stable growth rate was achieved. Evolution populations were generated in identical conditions in four batches: (1) populations Lac2 and Lac3 (Fong et al. 2003); (2) populations LacA, LacB, LacC, LacD, and LacE; (3) populations Gly1 and Gly2 (Ibarra et al. 2002); and (4) populations GlyA, GlyB, GlyC, GlyD, and GlyE. Serial passage also maintained a state of prolonged exponential growth such that each culture never entered stationary phase. Throughout the course of evolution, samples of each evolution population were flash frozen using liquid nitrogen and stored in a freezer at -80°C.
Phenotype testing
Phenotypic characterization of the evolved cultures was conducted at day 0, at day 20, and at the endpoint of evolution to quantitatively determine the phenotypic changes during the adaptive evolutionary process. For each time point examined, pre-cultures were grown overnight and used to inoculate fresh medium for a batch culture. The GR, SUR, and OUR were measured throughout the course of exponential growth (Ibarra et al. 2004) by periodic sampling of each batch culture. The GR was determined by measuring the OD of growing cultures over time using a spectrophotometer (A600 and A420). The SUR was determined by monitoring the depletion of the carbon source in filtered media samples over time with UV detection by HPLC or an enzymatic assay. The OUR was determined by measuring the dissolved oxygen depletion in a respirometer with a polaro-graphic dissolved oxygen probe. Biomass correlations were determined by measuring the optical density of the culture and filtering a set volume of culture onto a preweighed filter that was weighed after being dried to constant weight. All phenotype testing was performed in replicate.
Growth on alternative carbon sources was evaluated using the Bioscreen C plate-reader system (Thermo Labsystems, Franklin, MA). This system measures the optical density of up to 200 cultures (using two 100-well plates) for each experiment in a temperature-controlled environment. For each experiment conducted on the Bioscreen C system, pre-cultures were grown overnight and allowed to reach mid-exponential growth (A600 ≤ 0.5). Approximately 5-7 μL of these cultures were used to inoculate the multi-well Bioscreen C plates containing 300 μL of medium, yielding an initial A600 OD between 0.06 and 0.07. Each experimental run was conducted with an associated negative control sample containing blank medium and a positive control sample with wild-type cells. Growth on nine different carbon sources (acetate, alpha-ketoglutarate, glucose, glycerol, lactate, malate, pyruvate, ribose, and succinate) was tested. The plates were incubated at 30°C and monitored in the Bioscreen C system for a period of 24-48 hours, taking measurements every 15 minutes with continuous shaking between measurements. Growth rates were averaged across a minimum of four replicate cultures with as many as 12 replicates being conducted for some conditions. Replicate growth rate measurements varied by >5%.
Transcriptional analysis
Affymetrix E. coli Antisense Genome Arrays were used for all transcriptional analyses. Each experimental condition was tested in triplicate on the carbon source used for evolution (glycerol or lactate) using independent cultures and processed following the manufacturer-recommended protocols. Although different experimental designs could be used to study the adaptive and time-course responses, analyses in this study used two different types of controls. Three replicates of the wild-type strain grown on glucose were used as a common reference point for all experiments, and replicates of the unevolved wild-type strain grown on either glycerol or lactate were used as a baseline for each evolution experiment. Overall, 96 genome-wide expression profiles were generated in this study (3 wild-type glucose controls, 3 wild-type glycerol controls, 6 wild-type lactate controls, 21 day-20 glycerol profiles, 21 day-44 glycerol profiles, 21 day-20 lactate profiles, and 21 day-60 lactate profiles). All replicate expression profiles were biological replicates. General reproducibility of the expression profiles was assessed using the six replicates of the wild-type strain grown on lactate. No significant differential expression was observed when the profiles were divided into two groups of three and compared with each other via the same procedure used throughout this study (all possible combinations tested, data not shown). Cultures were grown to mid-exponential growth phase (OD A600 ≈ 0.5). Culture (3 mL) was added to 6 mL of RNAprotect (Qiagen), and RNA was isolated using RNeasy kits (Qiagen) following the manufacturer's instructions. Total RNA yields were measured using a spectrophotometer (A260) and quality was checked by visualization on agarose gels and by measuring the sample A260/A280 ratio. cDNA synthesis, fragmentation, and terminal labeling were conducted as recommended by Affymetrix. Raw .CEL files were analyzed using robust multi-array average (Irizarry et al. 2003) for normalization and calculation of probe intensities.
Expression values were then assessed for statistically significant differential expression using t-tests. After conducting pairwise t-test comparisons between evolved and wild-type strains, those genes meeting a 5% FDR-adjusted P-value cut-off were chosen as differentially expressed. Downstream analyses of these differentially expressed genes included examining their associated MultiFun (Serres and Riley 2000; Serres et al. 2004) functional group classifications and studying the percent differential expression of entire regulons (Keseler et al. 2005).
The probability (P-value) of the observed regulon enrichment of differentially expressed genes was calculated using the hypergeometric distribution (Cora et al. 2004):
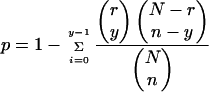 |
where N (=4345) is the total number of E. coli genes listed on the Affymetrix GeneChip, r is the total number of genes that are a part of the regulon, n is the number of differentially expressed genes, and y is the number of genes that are differentially expressed and a member of the regulon.
Supplementary Material
Acknowledgments
We thank T. Durfee and F.R. Blattner for critical input and discussions. This work was supported by NIH grants GM57089 and GM62791. The material in the paper may be related to US patent application US-2002-0142321-A1. The authors thus disclose potential competing interests.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.3832305.
Footnotes
[Supplemental material is available online at www.genome.org.]
References
- Benjamini, Y. and Hochberg, Y. 1995. Controlling the false discovery rate—A practical and powerful approach to multiple testing. J. R. Stat. Soc. B 57: 289-300. [Google Scholar]
- Cooper, V.S., Schneider, D., Blot, M., and Lenski, R.E. 2001. Mechanisms causing rapid and parallel losses of ribose catabolism in evolving populations of Escherichia coli B. J. Bacteriol. 183: 2834-2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper, T.F., Rozen, D.E., and Lenski, R.E. 2003. Parallel changes in gene expression after 20,000 generations of evolution in Escherichia coli. Proc. Natl. Acad. Sci. 100: 1072-1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cora, D., Di Cunto, F., Provero, P., Silengo, L., and Caselle, M. 2004. Computational identification of transcription factor binding sites by functional analysis of sets of genes sharing overrepresented upstream motifs. BMC Bioinformatics 5: 57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards, J.S., Ibarra, R.U., and Palsson, B.O. 2001. In silico predictions of Escherichia coli metabolic capabilities are consistent with experimental data. Nat. Biotechnol. 19: 125-130. [DOI] [PubMed] [Google Scholar]
- Elena, S.F. and Lenski, R.E. 2003. Evolution experiments with microorganisms: The dynamics and genetic bases of adaptation. Nat. Rev. Genet. 4: 457-469. [DOI] [PubMed] [Google Scholar]
- Ferea, T.L., Botstein, D., Brown, P.O., and Rosenzweig, R.F. 1999. Systematic changes in gene expression patterns following adaptive evolution in yeast. Proc. Natl. Acad. Sci. 96: 9721-9726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong, S.S., Marciniak, J.Y., and Palsson, B.O. 2003. Description and interpretation of adaptive evolution of Escherichia coli K-12 MG1655 by using a genome-scale in silico metabolic model. J. Bacteriol. 185: 6400-6408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helling, R.B., Vargas, C.N., and Adams, J. 1987. Evolution of Escherichia coli during growth in a constant environment. Genetics 116: 349-358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess, K.R., Zhang, W., Baggerly, K.A., Stivers, D.N., and Coombes, K.R. 2001. Microarrays: Handling the deluge of data and extracting reliable information. Trends Biotechnol. 19: 463-468. [DOI] [PubMed] [Google Scholar]
- Ibarra, R.U., Edwards, J.S., and Palsson, B.O. 2002. Escherichia coli K-12 undergoes adaptive evolution to achieve in silico predicted optimal growth. Nature 420: 186-189. [DOI] [PubMed] [Google Scholar]
- Ibarra, R.U., Fu, P., Palsson, B.O., DiTonno, J.R., and Edwards, J.S. 2004. Quantitative analysis of Escherichia coli metabolic phenotypes within the context of phenotypic phase planes. J. Mol. Microbiol. Biotechnol. 6: 101-108. [DOI] [PubMed] [Google Scholar]
- Irizarry, R.A., Hobbs, B., Collin, F., Beazer-Barclay, Y.D., Antonellis, K.J., Scherf, U., and Speed, T.P. 2003. Exploration, normalization, and summaries of high-density oligonucleotide array probe level data. Biostatistics 4: 249-264. [DOI] [PubMed] [Google Scholar]
- Jablonka, E. and Lamb, M.J. 2002. The changing concept of epigenetics. Ann. N.Y. Acad. Sci. 981: 82-96. [DOI] [PubMed] [Google Scholar]
- Kassen, R. 2002. The experimental evolution of specialists, generalists, and the maintenance of diversity. J. Evol. Biol. 15: 173-190. [Google Scholar]
- Keseler, I.M., Collado-Vides, J., Gama-Castro, S., Ingraham, J., Paley, S., Paulsen, I.T., Peralta-Gil, M., and Karp, P.D. 2005. EcoCyc: A comprehensive database resource for Escherichia coli. Nucleic Acids Res. 33: D334-D337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirov, S.M., Tassell, B.C., Semmler, A.B., O'Donovan, L.A., Rabaan, A.A., and Shaw, J.G. 2002. Lateral flagella and swarming motility in Aeromonas species. J. Bacteriol. 184: 547-555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenski, R.E., Mongold, J.A., Sniegowski, P.D., Travisano, M., Vasi, F., Gerrish, P.J., and Schmidt, T.M. 1998. Evolution of competitive fitness in experimental population of E. coli: What makes one genotype a better competitor than another? Antonie van Leeuwenhoek 73: 35-47. [DOI] [PubMed] [Google Scholar]
- Liu, M., Durfee, T., Cabrera, J.E., Zhao, K., Jin, D.J., and Blattner, F.R. 2005. Global transcriptional programs reveal a carbon source foraging strategy by Escherichia coli. J. Biol. Chem. 280: 15921-15927. [DOI] [PubMed] [Google Scholar]
- Mahadevan, R. and Schilling, C.H. 2003. The effects of alternate optimal solutions in constraint-based genome-scale metabolic models. Metab. Eng. 5: 264-276. [DOI] [PubMed] [Google Scholar]
- Massey, R.C., Rainey, P.B., Sheehan, B.J., Keane, O.M., and Dorman, C.J. 1999. Environmentally constrained mutation and adaptive evolution in Salmonella. Curr. Biol. 9: 1477-1480. [DOI] [PubMed] [Google Scholar]
- Nadon, R. and Shoemaker, J. 2002. Statistical issues with microarrays: Processing and analysis. Trends Genet. 18: 265-271. [DOI] [PubMed] [Google Scholar]
- Nakatsu, C.H., Korona, R., Lenski, R.E., de Bruijn, F.J., Marsh, T.L., and Forney, L.J. 1998. Parallel and divergent genotypic evolution in experimental populations of Ralstonia sp. J. Bacteriol. 180: 4325-4331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr, H.A. 2005. The genetic theory of adaptation: A brief history. Nat. Rev. Genet. 6: 119-127. [DOI] [PubMed] [Google Scholar]
- Papadopoulos, D., Schneider, D., Meier-Eiss, J., Arber, W., Lenski, R.E., and Blot, M. 1999. Genomic evolution during a 10,000-generation experiment with bacteria. Proc. Natl. Acad. Sci. 96: 3807-3812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riehle, M.M., Bennett, A.F., and Long, A.D. 2001. Genetic architecture of thermal adaptation in Escherichia coli. Proc. Natl. Acad. Sci. 98: 525-530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riehle, M.M., Bennett, A.F., Lenski, R.E., and Long, A.D. 2003. Evolutionary changes in heat-inducible gene expression in lines of Escherichia coli adapted to high temperature. Physiol. Genomics 14: 47-58. [DOI] [PubMed] [Google Scholar]
- Sauer, U. 2001. Evolutionary engineering of industrially important microbial phenotypes. Adv. Biochem. Eng. Biotechnol. 73: 129-169. [DOI] [PubMed] [Google Scholar]
- Serres, M.H. and Riley, M. 2000. MultiFun, a multifunctional classification scheme for Escherichia coli K-12 gene products. Microb. Comp. Genomics 5: 205-222. [DOI] [PubMed] [Google Scholar]
- Serres, M.H., Goswami, S., and Riley, M. 2004. GenProtEC: An updated and improved analysis of functions of Escherichia coli K-12 proteins. Nucleic Acids Res. 32: D300-D302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaver, A.C., Dombrowski, P.G., Sweeney, J.Y., Treis, T., Zappala, R.M., and Sniegowski, P.D. 2002. Fitness evolution and the rise of mutator alleles in experimental Escherichia coli populations. Genetics 162: 557-566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storey, J.D. and Tibshirani, R. 2003. Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. 100: 9440-9445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatasubbarao, S. 2004. Microarrays—Status and prospects. Trends Biotechnol. 22: 630-637. [DOI] [PubMed] [Google Scholar]
- Waddington, C.H. 1940. Organisers and genes. Cambridge University Press, Cambridge, UK.
- ———. 1957. The strategy of genes. Allen & Unwin, London, UK.
- Wichman, H.A., Badgett, M.R., Scott, L.A., Boulianne, C.M., and Bull, J.J. 1999. Different trajectories of parallel evolution during viral adaptation. Science 285: 422-424. [DOI] [PubMed] [Google Scholar]
- Wood, B.E. and Ingram, L.O. 1992. Ethanol production from cellobiose, amorphous cellulose, and crystalline cellulose by recombinant Klebsiella oxytoca containing chromosomally integrated Zymomonas mobilis genes for ethanol production and plasmids expressing thermostable cellulase genes from Clostridium thermocellum. Appl. Environ. Microbiol. 58: 2103-2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.