Abstract
Diversity of cultured ammonia-oxidizing bacteria in the γ-subdivision of the Proteobacteria was investigated by using strains isolated from various parts of the world ocean. All the strains were very similar to each other on the basis of the sequences of both the 16S rRNA and ammonia monooxygenase genes and could be characterized as a single species. Sequences were also cloned directly from environmental DNA from coastal Pacific and Atlantic sites, and these sequences represented the first Nitrosococcus oceani-like sequences obtained directly from the ocean. Most of the environmental sequences clustered tightly with those of the cultivated strains, but some sequences could represent new species of Nitrosococcus. These findings imply that organisms similar to the cultivated N. oceani strains have a worldwide distribution.
The first marine ammonia-oxidizing strain to be isolated from seawater was described in 1965 by Watson (22) and was called Nitrosocystis oceanus. This species is now known as Nitrosococcus oceani and, along with Nitrosococcus halophilus, comprises the known species of ammonia-oxidizing bacteria (AOB) in the γ-subdivision of the Proteobacteria. All other cultivated aerobic AOB are in the β-subdivision, and while some β-subdivision AOB have been isolated from and detected in both freshwater and marine environments, γ-subdivision AOB have only been found in marine environments. A possible exception is the detection of N. oceani by immunofluorescence and fluorescent in situ hybridization (FISH) in the saline waters of Lake Bonney, a permanently ice-covered lake in Antarctica (14), but this is not really an exception to the obligately halophilic nature of this group. Probably because N. oceani was the first AOB isolated from seawater, this species and the γ-subdivision have long been regarded as the major AOB group in seawater, but much less is known about their diversity and distribution. The recent explosion of interest and research into the ecology and phylogeny of AOB has been almost entirely focused on the β-subdivision, because this group is monophyletic and relatively accessible to molecular ecological analysis (8).
On the basis of nitrite production rates in enrichment cultures, Watson (22) originally estimated the abundance of N. oceani to be 1 to 103 cells liter−1 (depending on the lag time in culture). After the original isolation of N. oceani (which was isolated from samples collected in 1957), additional isolates from distant parts of the ocean were described by Carlucci and Strickland (2), and similar low in situ concentrations were estimated. Both groups of workers estimated that the in situ nitrification rates were on the order of 0.02 to 0.7 μmol liter−1 year−1 and concluded that these rates were too low to account for the observed distributions of nitrite and nitrate in the ocean. They suggested that either higher numbers of AOB must be present in the ocean or some mechanism for ammonia oxidation other than the known biologically mediated reaction might exist.
N. oceani has since been detected in many marine environments by immunofluorescence at concentrations on the order of 103 to 104 cells ml−1 (17, 18, 19, 24). The β-subdivision AOB Nitrosomonas marina has also been detected by immunofluorescence at similar concentrations (the antisera employed for this study would have detected most known strains of β-subdivision AOB [18]), and both types of organisms were several orders of magnitude more abundant in the Chesapeake Bay (17). Subsequent direct measurements of nitrification rates by 15N tracer methods detected rates of up to 50 μmol liter−1 year−1 in open ocean surface waters and rates of less than 0.02 μmol liter−1 year−1 in the deep sea (15, 16, 20). The depth distribution of ammonia oxidation rates showed that most nitrification occurs in the near-surface waters, and early estimates of activity in deep water were surprisingly accurate.
Because immunofluorescence enumeration depends on antibodies derived from cultured microbes, it is expected that abundance estimates obtained in this way should be vast underestimates of the true nitrifier population (because immunofluorescence might not be able to detect uncultivated strains). However, if AOB detected by immunofluorescence are responsible for the total ammonia oxidation rates, per-cell rates of 1 to 1,000 × 10−10 μmol cell−1 day−1 are estimated. These rates are much lower than the rate observed by Watson (22) in pure cultures of N. oceani (2 × 10−6 μmol cell−1 day−1), implying that the immunofluorescence abundance estimates might not be serious underestimates.
The congruence of rates and concentrations suggests that the known AOB assemblages may represent the diversity of AOB in the ocean rather well. Nevertheless, we still do not know what level of diversity might be contained in the assemblages detected by polyclonal antibodies. The strains characterized by Watson (22), Watson and Mandel (23), and Carlucci and Strickland (2) varied in their responses to different temperatures, their responses to different ammonium concentrations, and their maximum growth rates. Thus, although the strains were all isolated with similar enrichment media, there does appear to be significant physiological diversity represented in the culture collection. In the present study, we investigated the molecular phylogeny of several N. oceani isolates in terms of both the 16S rRNA and amoA genes and compared the sequences of these isolates to cloned 16S rRNA sequences obtained from marine environments. amoA encodes ammonia monooxygenase, the first essential and unique enzyme in the ammonia oxidation pathway, and has been found to contain phylogenetically useful information in cultivated and environmental AOB sequences (10).
Bacterial strains and field sampling.
The sites of origin and dates of isolation of the strains whose genes were sequenced in this study are listed in Table 1. Strains whose designations begin with SW C- originated in the culture collection of Stanley Watson at the Woods Hole Oceanographic Institution. They were kindly provided by Stanley Watson and Frederica Valois to one of us (B.B.W.) at various times over the last 25 years and have been maintained in liquid N2 in her laboratory. Strains whose designations begin with AFC originated in the culture collection of Angelo Carlucci at Scripps Institution of Oceanography (SIO). They were given to one of us (B.B.W.) between 1978 and 1982 and have been maintained in liquid culture and liquid N2 in her laboratory. N. oceani strains (Table 1) were grown in the seawater medium (W medium) of Watson (22) after retrieval from long-term storage in liquid nitrogen. Nitrifier cultures were grown at room temperature (18°C) in the dark with no agitation in 1- or 2-liter flasks. Cells of the cultivated strains were harvested by filtration onto polycarbonate filters and then washed, resuspended in Tris-EDTA buffer, and stored frozen until DNA extraction.
TABLE 1.
Strain and sequence identification information
| Identification | Site of origin | Date of isolation | Accession no.
|
|
|---|---|---|---|---|
| amoA/pmmo | 16S rDNA | |||
| New N. oceani strains included in this study | ||||
| N. oceani (AFC) | Unknown | 1964 | AF50899 | AF508993 |
| N. oceani (SW) | Unknown | Unknown | AF509003 | AF508994 |
| AFC 3 | SIO pier (33°N, 117°W) | 1964 | AF508998 | AF508989 |
| AFC 5 | SIO pier (33°N, 117°W) | 1964 | AF509000 | AF508990 |
| AFC 24P | North Pacific (47°N, 155°W) | 1964 | AF509002 | AF508987 |
| AFC 27 | North Pacific (43°N, 155°W) | 1964 | AF509001 | AF508988 |
| AFC 36 | Central North Pacific (31°N, 155°W) | 1964 | AF508995 | AF508989 |
| AFC 132 | South Pacific (13°S, 76°W) | 1966 | AF508996 | AF508986 |
| SW C-19 | Continental shelf, Peru | 1966 | AF508997 | AF508991 |
| Additional database sequences used in the analysis | ||||
| SW C-113 | Red Sea | 1966 | AF153344 | AF153343 |
| N. oceani C-27 | Barbados Harbor | 1964 | NDa | M96398 |
| N. oceani C-107 | North Atlantic | 1957 | AF047705 | M96395 |
| Methylomonas methanica | U31653 | L20842 | ||
| Methylococcus capsulatus | L40804 | AF150806 | ||
| Nitrosomonas europaea | AF058691 | |||
| Nitrosococcus halophilus | AF272521 | AF2872998 | ||
ND, not determined.
Seawater was collected in March 2001 from Cape Cod by submerging a carboy from a jetty off the beach near Salt Pond in Woods Hole, Mass. The water was taken to Princeton, N.J., within 1 day and was stored at 12°C until filtration the next day. Cells from 1.2 liters of seawater were vacuum filtered onto 0.2-μm-pore-size polycarbonate filters; then they were washed off the filters by vortexing, concentrated into a pellet by centrifugation, and frozen in Tris-EDTA until DNA extraction. Monterey Bay seawater was collected from 36 m at the midbay station (total depth, approximately 1,000 m) in October 1999 by using Niskin bottles on a CTD rosette and was concentrated by tangential flow filtration from a volume of 4 liters. The concentrate was filtered onto Supor cellulose acetate filters and frozen in 0.8 ml of 0.5 M EDTA until extraction.
DNA extraction, amplification, and sequence analysis.
Standard procedures for DNA extraction were followed (1). DNA extracts were tested for PCR inhibition by amplification with universal eubacterial 16S ribosomal DNA (rDNA) primers EUB1 and EUB2 (9). PCR primers NOC1 and NOC2 for N. oceani 16S rRNA genes described by Voytek (12) were used with the amplification protocols described previously (21). When necessary, the EUB fragment was used as a template for amplification with the NOC1 and NOC2 primers as reported previously (13). Amplification of a fragment of the ammonia monooxygenase (amoA) gene was performed by using primers AMO189 and AMO682 described by Holmes et al. (6).
EUB (∼1,400-bp), NOC (∼1,100-bp), and AMO (∼550-bp) amplification products were extracted from 1% agarose gels with a Qiaquick gel extraction kit (Qiagen) or were purified directly from the reaction mixture by using Qiagen columns and cloned with a TOPO-TA cloning kit (Invitrogen). White transformant colonies were screened for inserts of the correct size by PCR by using vector primers T7 and M13 as previously described (3). The PCR cycling protocol recommended by the manufacturer (Invitrogen) was used for T7-M13 amplification. T7-M13 PCR products of the expected size were used as templates for cycle sequencing of both strands with the T7 and M13 primers and a BigDye terminator kit (Perkin-Elmer). Additional internal primers (NOC1 plus NOC2 and 773f plus 795r [12]) were used for sequencing to obtain the sequences of the internal regions of the 16S rRNA genes. Cycle sequencing products were precipitated by using the manufacturer's instructions and were sequenced with an ABI310 genetic analyzer (Perkin-Elmer). amoA sequences were obtained from nine strains of N. oceani. 16S rDNA sequences were obtained from the same nine strains, from six clones from Cape Cod samples, and from six clones from Monterey Bay samples.
Sequences were assembled by using AutoAssembler V 1.4.0 (Perkin-Elmer) or Sequencher 4.1 (Gene Codes Corp.) into a single consensus sequence. Sequence Navigator V 1.0.1 (Perkin-Elmer) or Clustal W (www.genebee.msu.su/clustal/basic/html) was used to align homologous regions of 16S rRNA and amoA gene sequences from different organisms. The aligned sequences were analyzed by distance matrix methods by using DNADIST in the phylogenetic inference package PHYLIP 3.572 (4). Distances were calculated by using the Kimura two-parameter model for nucleic acid sequences (7). The input files were each bootstrapped 1,000 times by using the SEQBOOT program of PHYLIP prior to distance matrix analyses. Neighbor-joining trees were produced for each pseudoreplicate analysis. The CONSENSE program was used to compute the majority rule consensus tree for each set of molecular sequences, and trees were drawn by using the TreeViewPPC program of Roderic D. M. Page.
16S rRNA phylogeny.
When the nine new sequences from cultivated strains of N. oceani were analyzed together with two other N. oceani sequences (C-107 and C-27) from the database by using about 1,400 bp of the 16S rRNA sequence, bootstrap values of >75% were obtained for several branch points (data not shown). At least three of the strains, N. oceani (SW), N. oceani (AFC), and AFC 132, were essentially identical as determined by this analysis (100% similarity). The remaining sequences all differed by 1.8% or less when pairwise comparisons were performed, indicating that all of the strains are representatives of the species N. oceani.
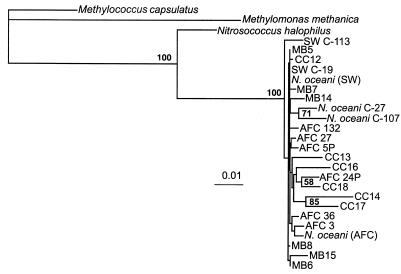
Six N. oceani-like sequences were obtained from Monterey Bay, and six N. oceani-like sequences were obtained from Cape Cod. These sequences were compared to sequences obtained from the N. oceani strains described above, as well as the sequences of Nitrosococcus. sp. strain C-113 (probably an N. oceani strain), N. halophilus NC-4, and two methanotrophs, Methylococcus capsulatus and Methylomonas methanica (Fig. 1). The NOC primers generate an approximately 1,100-bp fragment, but a shorter region (964 bp) was used in the analysis in order to include shorter sequences from the database. All the cloned sequences from marine samples were very similar to the sequences of cultivated N. oceani strains, as opposed to the sequences of the closely related N. halophilus or methanotroph lineages (Fig. 1). The N. halophilus sequence branched distinctly from all the N. oceani sequences; the levels of sequence difference between N. halophilus and the N. oceani strains ranged from 4.5 to 5.6%. Nitrosococcus sp. strain C-113 branched independently from the other cultivated N. oceani strains, which formed a tight cluster with all of the marine clones, indicating that there was great similarity among cultivated strains and environmental clones. Clones from Monterey Bay and Cape Cod did not cluster separately by region (bootstrap values, <50%), although some clones (e.g., CC14 and CC17) formed distinct branches and were slightly more than 2.5% different from some of the cultivated N. oceani strains.
FIG. 1.
Distance tree showing phylogenetic relationships, determined on the basis of 964 bp of the 16S rDNA sequence, among cultivated strains of N. oceani, its cultivated relatives, and cloned sequences derived from seawater. CC, Cape Cod; MB, Monterey Bay. The accession numbers of sequences of cultivated strains are shown in Table 1. Bootstrap values greater than 50% are indicated at branch points. Scale bar = 0.01 substitution per site.
amoA phylogeny.
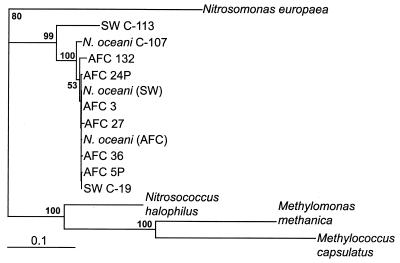
Approximately 520 bp of the amoA sequence was obtained from the N. oceani culture collection, but only 369 bp was used in the phylogenetic analysis (Fig. 2) in order to include sequences from the database that did not overlap entirely (e.g., the sequence of N. halophilus). An analysis of all 520 bp produced a nearly identical tree topology in an analysis that included only the N. oceani strains, and it detected significant branch points for most sequences (data not shown). The genetic distances between amoA sequences were small but generally were larger than those detected in the 16S rDNA sequences for the same cultivated strains. When compared in a phylogenetic tree by using pmoA genes (pmoA encodes the analogous enzyme, methane monooxygenase, in methanotrophs) from methanotrophs and amoA genes from related ammonia oxidizers (Fig. 2), the cultivated N. oceani sequences clustered tightly with few significant branch points. Interestingly, however, AFC 132, which was virtually identical to N. oceani (SW) and N. oceani (AFC) on the basis of rDNA sequences, is noticeably different on the basis of amoA sequences, although its placement in the tree is not supported by a very high bootstrap value (53%). amoA sequences were not retrieved from the seawater samples, but an investigation of amoA sequences representing the γ-subdivision from marine environments is ongoing.
FIG. 2.
Distance tree showing phylogenetic relationships among the amoA sequences (369 bp) of cultivated strains of N. oceani and its relatives. The accession numbers of sequences are shown in Table 1. Bootstrap values greater than 50% are indicated at branch points. Scale bar = 0.1 substitution per site.
Worldwide distribution of N. oceani.
Denaturing gradient gel electrophoresis analysis of several of the cultivated N. oceani strains had implied that there is great similarity among these organisms at the 16S rDNA level (Ward, unpublished data), and the more sophisticated analysis described here verified that they are indeed very closely related. The cultivated strains included in this study were obtained in the 1960s and 1970s by Watson and Carlucci from a variety of marine locations, including both the Pacific and Atlantic oceans, and from the open ocean as well as coastal sites (Table 1). The history of some of these strains is somewhat unclear because specific strain designations were not always retained when the two initial owners of the culture collections sent strains to each other (Angelo Carlucci, personal communication) and to one of us (B.B.W.). The strains designated N. oceani (AFC) and N. oceani (SW) were given to B.B.W. in the late 1970s by the initial owners. It seems clear from this analysis that they originated from the same strain prior to that time.
Despite their diverse origins, all the cultivated N. oceani strains appear to form one species and are very closely related to each other. The levels of sequence dissimilarity ranged from 0% [among N. oceani (AFC), N. oceani (SW), and AFC 132] to 1.8% (between AFC 24P and N. oceani C-107) for the 1,400-bp region of the 16S rRNA gene, values which were within the 2.5% similarity convention used for the definition of a species (11). The presence of significant clusters (bootstrap values, >75%) (data not shown) implies, however, that distinct strains are represented in the collection. Shorter sequences from the database (most of which were less than 92% similar) were not included in this analysis in order to maximize the resolution of the analysis. The most obvious implication of the great similarity among these strains is that the cultivation methods used to isolate N. oceani have all tended to enrich for the same species, even though Carlucci and Watson used slightly different enrichment media, referred to as A medium (2) and W medium (22), respectively. The main difference between the media was the inclusion of solid CaCO3 in A medium, while W medium required periodic addition of dissolved K2CO3 during growth. In both cases, the carbonate served to buffer the acidification caused by growth of the ammonia oxidizers, but the CaCO3 also provided a solid surface around which cells usually clustered preferentially. In addition, subsequent isolation attempts with W medium also yielded Nitrosomonas marina-like strains, which have been described and characterized on the basis of immunofluorescence reactivity (2) and 16S rDNA sequences (12, 3). This demonstrates that nitrifiers not closely related to N. oceani can be isolated with W medium. Thus, although isolation bias cannot be ruled out, it is clear that N. oceani and related strains are found throughout the world ocean.
As expected for functional genes, the diversity of amoA genes in the cultivated strains exceeded the 16S rDNA sequence diversity only slightly (only the nucleic acid phylogeny is shown here, because it has higher resolution than the parallel amino acid analysis [10]). The greatest sequence dissimilarity observed among the amoA sequences of the newly sequenced cultivated Nitrosococcus strains was the 1.93% dissimilarity between AFC 27 and N. oceani C-107. The amoA genes are expected to show greater diversity than the 16S rRNA genes, but this is an imperfect comparison due to the much shorter fragment used for analysis of the functional gene. The Nitrosococcus sp. strain C-113 amoA sequence was significantly more different from the AFC 27 sequence (11% dissimilarity). The sequences of all the cultivated N. oceani strains were more than 40% different from those of Nitrosomonas europaea and the methanotrophs and between 27 and 30% different from those of N. halophilus in terms of the amoA sequence.
The NOC PCR primers were developed initially (12) on the basis of two N. oceani sequences, which were produced by Head et al. (5) from N. oceani C-107 and C-27. Voytek (12) screened the primers for PCR specificity against a large culture collection of nitrifiers and unrelated heterotrophic and autotrophic bacteria and found them to be completely specific for N. oceani. These primers were used to amplify the DNA fragment of the correct size from DNA extracted from seawater, but no sequences were obtained from these environmental samples. Detection of the fragment of the correct size usually required a two-stage amplification (amplification with a set of eubacterial primers, followed by reamplification of the template with the NOC primers). The same primers were used as fluorescent probes in FISH analysis of samples from permanently ice-covered Lake Bonney in the Taylor Valley of Antarctica, and N. oceani was detected in that environment by both semiquantitative PCR and FISH (14). The primers were also used in an attempt to amplify DNA from Mono Lake in California, but no NOC fragments were recovered from this environment, even with a two-stage amplification method (21). N. halophilus is reported to have a higher salt requirement and salt tolerance than N. oceani; it would be worthwhile to use a similar approach to search for it in Mono Lake.
The NOC1 and NOC2 primers differ only slightly from the homologous region of the N. halophilus 16S rRNA sequence; there is a 1-bp mismatch in the NOC1 fragment, and there is a 4-bp mismatch at the 3′ end of the NOC2 primer. The N. halophilus sequence was not available at the time that the NOC1 and NOC2 primers were designed, but Voytek has subsequently shown that N. halophilus DNA is amplified only with a modified version of the NOC primers under the amplification conditions used here (M. A. Voytek, personal communication). Therefore, it is not surprising that no N. halophilus-like sequences were retrieved by PCR from the seawater samples investigated here.
Because the NOC primers were developed on the basis of only two sequences, it is possible and indeed likely that they do not amplify the sequences of even other strains of N. oceani. In fact, however, the sequences of all cultured putative N. oceani strains were amplified satisfactorily, and they all fall within the 2.5% similarity convention for designation of a species. Most of the cloned sequences derived from the two seawater sites were fairly similar to the sequences in the culture collection and to each other, but some of them clearly fall outside the 2.5% convention. The greatest difference detected among clones was the 4.0% difference between CC14 and CC13, and these two clones were more than 2.5% different from most of the cultivated strains. Thus, although the cultivated strains could be characterized as members of one species, this analysis detected greater diversity among environmental clones. The use of nondegenerate primers probably minimized the diversity of clones retrieved from the environment, and these sequences may not reflect the greater diversity of N. oceani-like sequences that may be present in the ocean. Nevertheless, the environmental data indicate the presence of strains that are essentially the same as those in culture and also the existence of closely related but distinct species. The cloned sequences could be used to design slightly more general primers that might amplify related sequences from the environment in order to investigate the diversity of additional species of Nitrosococcus that may be present in the marine environment.
Nucleotide sequence accession numbers.
The partial amoA sequences and 16S rRNA gene sequences of the N. oceani strains have been deposited in the GenBank database under accession numbers AF508986 to AF509003. The partial 16S rRNA gene sequences of the environmental clones have been deposited under accession numbers AF509004 to AF509015.
Acknowledgments
We thank B. Song for help with the initial sequencing. We appreciate comments on the manuscript from C. Francis and K. Casciotti.
Funding was provided by the National Science Foundation (grant OCE-9896240).
REFERENCES
- 1.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, and K. Struhl (ed.). 1987. Current protocols in molecular biology. John Wiley and Sons, New York, N.Y.
- 2.Carlucci, A. F., and J. D. H. Strickland. 1968. The isolation, purification and some kinetic studies of marine nitrifying bacteria. J. Exp. Mar. Biol. Ecol. 2:156-166. [Google Scholar]
- 3.Casciotti, K. L., and B. B. Ward. 2001. Nitrite reductase genes in ammonia-oxidizing bacteria. Appl. Environ. Microbiol. 67:2213-2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Felsenstein, J. 1989. Phylip—phylogeny inference package. Cladistics 5:164-166. [Google Scholar]
- 5.Head, I. M., W. D. Hiorns, T. M. Embley, A. J. McCarthy, and J. R. Saunders. 1993. The phylogeny of autotrophic ammonia-oxidizing bacteria as determined by analysis of 16S ribosomal-RNA gene-sequences. J. Gen. Microbiol. 139:1147-1153. [DOI] [PubMed] [Google Scholar]
- 6.Holmes, A. J., A. Costello, M. E. Lidstrom, and J. C. Murrell. 1995. Evidence that particulate methane monooxygenase and ammonia monooxygenase may be evolutionarily related. FEMS Microbiol. Lett. 132:203-208. [DOI] [PubMed] [Google Scholar]
- 7.Kimura, M. 1980. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16:111-120. [DOI] [PubMed] [Google Scholar]
- 8.Kowalchuk, G. A., and J. R. Stephen. 2001. Ammonia-oxidizing bacteria: a model for molecular microbial ecology. Annu. Rev. Microbiol. 55:485-529. [DOI] [PubMed] [Google Scholar]
- 9.Liesack, W., H. Weyland, and E. Stackebrandt. 1991. Potential risks of gene amplification by PCR as determined by 16S rDNA analysis of a mixed culture of strict barophilic bacteria. Microb. Ecol. 21:191-198. [DOI] [PubMed] [Google Scholar]
- 10.Rotthauwe, J.-H., K.-P. Witzel, and W. Liesack. 1997. The ammonia monooxygenase structural gene amoA as a functional marker: molecular fine-scale analysis of natural ammonia-oxidizing populations. Appl. Environ. Microbiol. 63:4704-4712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stackebrandt, E., and B. M. Goebel. 1994. Taxonomic note: a place for DNA-RNA reassociation and 16S rRNA sequence analysis in the present species definition of bacteriology. Int. J. Syst. Bacteriol. 44:846-849. [Google Scholar]
- 12.Voytek, M. A. 1996. Relative abundance and species diversity of autotrophic ammonia-oxidizing bacteria in aquatic systems. University of California, Santa Cruz, Calif.
- 13.Voytek, M. A., and B. B. Ward. 1995. Detection of ammonium-oxidizing bacteria of the beta-subclass of the class proteobacteria in aquatic samples with the PCR. Appl. Environ. Microbiol. 61:1444-1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Voytek, M. A., B. B. Ward, and J. C. Priscu. 1998. The abundance of ammonium-oxidizing bacteria in Lake Bonney, Antarctica, determined by immunofluorescence, PCR and in situ hybridization, p. 217-228. In J. C. Priscu (ed.), Ecosystem dynamics in a polar desert: the McMurdo Dry Valleys, Antarctica, vol. 72. American Geophysical Union, Washington, D.C. [Google Scholar]
- 15.Ward, B. B. 2002. Nitrification in aquatic systems, p. 2144-2167. In D. G. Capone (ed.), Encyclopedia of environmental microbiology. John Wiley & Sons, New York, N.Y.
- 16.Ward, B. B. 1987. Nitrogen transformations in the Southern California Bight. Deep-Sea Res. 34:785-805. [Google Scholar]
- 17.Ward, B. B. 1982. Oceanic distribution of ammonium-oxidizing bacteria determined by immunofluorescent assay. J. Mar. Res. 40:1155-1172. [Google Scholar]
- 18.Ward, B. B., and A. F. Carlucci. 1985. Marine ammonium- and nitrite-oxidizing bacteria: serological diversity determined by immunofluorescence in culture and in the environment. Appl. Environ. Microbiol. 50:194-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ward, B. B., H. E. Glover, and F. Lipschultz. 1989. Chemoautotrophic activity and nitrification in the oxygen minimum zone off Peru. Deep-Sea Res. 36:1031-1051. [Google Scholar]
- 20.Ward, B. B., K. A. Kilpatrick, E. Renger, and R. W. Eppley. 1989. Biological nitrogen cycling in the nitracline. Limnol. Oceanogr. 34:493-513. [Google Scholar]
- 21.Ward, B. B., D. P. Martino, C. M. Diaz, and S. B. Joye. 2000. Analysis of ammonia-oxidizing bacteria from hypersaline Mono Lake, California, on the basis of 16S rRNA sequences. Appl. Environ. Microbiol. 66:2873-2881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Watson, S. W. 1965. Characteristics of a marine nitrifying bacterium, Nitrosocystis oceanus sp. n. Limnol. Oceanogr. 10(Suppl.):R274-R289. [Google Scholar]
- 23.Watson, S. W., and M. Mandel. 1971. Comparison of the morphology and deoxyribonucleic acid composition of 27 strains of nitrifying bacteria. J. Bacteriol. 107:563-569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zaccone, R., G. Caruso, and M. Azzaro. 1996. Detection of Nitrosococcus oceanus in a Mediterranean lagoon by immunofluorescence. J. Appl. Bacteriol. 80:611-616. [DOI] [PubMed] [Google Scholar]