Abstract
The transcriptional machinery in individual cells is controlled by a relatively small number of molecules, which may result in stochastic behavior in gene activity. Because of technical limitations in current collection and recording methods, most gene expression measurements are carried out on populations of cells and therefore reflect average mRNA levels. The variability of the transcript levels between different cells remains undefined, although it may have profound effects on cellular activities. Here we have measured gene expression levels of the five genes ActB, Ins1, Ins2, Abcc8, and Kcnj11 in individual cells from mouse pancreatic islets. Whereas Ins1 and Ins2 expression show a strong cell–cell correlation, this is not the case for the other genes. We further found that the transcript levels of the different genes are lognormally distributed. Hence, the geometric mean of expression levels provides a better estimate of gene activity of the typical cell than does the arithmetic mean measured on a cell population.
A typical eukaryotic cell contains ∼20 pg of RNA, of which less than 5% is mRNA. This corresponds to a few hundred thousands of transcripts of the ∼10,000 genes that are expressed in each cell at any given time (Kuznetsov et al. 2002), and the composition of this expression palette, or transcriptome, influences cell function and is a record of its recent history. When a small number of molecules determine the outcome of a reaction, a certain degree of stochastic variation is expected. As the number of mRNA molecules increases, so does the predictability of the number of proteins synthesized per cell. Likewise, if the number of enhancer and transcription activator molecules in a cell is low, large variation in mRNA copy number is expected (Walters et al. 1995; McAdams and Arkin 1997; Fiering et al. 2000; Elowitz et al. 2002; Raser and O'Shea 2004). These effects may account for the large cell-to-cell variations observed in isogenic cell populations (Levsky and Singer 2003; Peixoto et al. 2004). Furthermore, such variability can be anticipated to play important roles in biological processes and even establish asymmetries that determine cell differentiation (Fiering et al. 2000). These fundamental questions can be addressed by quantitative single-cell mRNA expression studies. Some insight about the gene expression profile in single cells can be obtained by microarray analysis following amplification of the mRNA by T7 polymerase (Luo et al. 1999; Kamme et al. 2003) or RT-PCR (Chiang and Melton 2003; Tietjen et al. 2003). These studies have demonstrated cellular heterogeneity of seemingly homogenous, morphologically defined cell types (Kamme et al. 2003; Levsky and Singer 2003). Other methods for single-cell gene expression analysis employ either the luciferase reporter gene (Castano et al. 1996) or green fluorescent protein (GFP) constructs to monitor variations at the protein level in yeast (Blake et al. 2003; Raser and O'Shea 2004) and bacteria (Elowitz et al. 2002; Ozbudak et al. 2002). By single-cell high-resolution multiplex fluorescence in situ hybridization, mRNA levels of genes expressed under the same promoter have been found to correlate, while high variation was found among unrelated genes (Levsky et al. 2002).
Here we have studied the expression of five genes in individual mouse pancreatic islet cells and MIN6 insulinoma cells (Miyazaki et al. 1990) by reverse transcriptase quantitative real-time PCR (RT-QPCR). This technique affords superior sensitivity, accuracy, and dynamic range compared with that of alternative methods for gene expression analysis (Bustin 2000; Peixoto et al. 2004). The pancreatic islets of Langerhans play a central role in the regulation of plasma glucose level. Each islet contains on average a total of 1000 cells of at least four to five different endocrine cell types (Bishop and Polak 1997). This diversity complicates the study cell-type–specific effects on gene transcription. By combining the patch-clamp technique with real-time PCR (Sucher et al. 2000; Liss et al. 2001; Gehwolf et al. 2002), we have determined the expression levels of several genes in individual pancreatic cells exposed to either low or high glucose concentrations. These include β-actin (ActB), insulin I (Ins1), insulin II (Ins2), as well as the KATP-channel subunits sulfonylurea receptor 1 (SUR1, a member of ATP-binding cassette family; Abcc8) and the inwardly rectifying channel Kir6.2 (subfamily J, member 11; Kcnj11).
Results and Discussion
Out of a total of 169 mouse islet cells, 84 were incubated in the presence of 5 mM glucose (low) and 85 in 20 mM glucose (high). In Table 1, the arithmetic means of the expression level of the five genes indicate that whereas the number of insulin transcripts per cell is in the order of several thousand copies, ActB and Abcc8 transcripts are present in a few hundred copies. The number of transcripts of the KATP-channel subunit Kcnj11 is <30 copies per cell. Based on the presence of Ins1 or Ins2 transcripts, it was concluded that at least 123 cells (73%) were β-cells. The fraction of β-cells in islet preparations is known to show large variations, and the average is 70%–80% (Barg et al. 2000).
Table 1.
Statistical parameters describing gene expression in single (insulin-expressing) β-cells at 5 and 20 mM glucose
| Glucose | Gene | Na | Arithmetic meanb | Geometric meanc | log10 Geometric mean (SD)d | Shapiro-Wilk P-valuee | Skewnessf |
|---|---|---|---|---|---|---|---|
| 5 mM | ActB | 51 | 130 | 61 | 1.79 (0.51) | 0.56 | -0.02 |
| Ins1 | 70 | 1700 | 190 | 2.29 (0.94) | 0.32 | 0.06 | |
| Ins2 | 44 | 5200 | 1100 | 3.03 (1.03) | 0.005 | -0.12 | |
| Abcc8 | 28 | 130 | 98 | 1.99 (0.34) | 0.97 | -0.05 | |
| Kcnj11 | 15 | 30 | 24 | 1.38 (0.22) | 0.77 | 0.31 | |
| 20 mM | ActB | 45 | 420 | 300 | 2.47 (0.37) | 0.53 | 0.01 |
| Ins1 | 49 | 7700 | 3200 | 3.51 (0.67) | 0.09 | -0.70 | |
| Ins2 | 47 | 16000 | 10000 | 4.01 (0.53) | 0.05 | -0.70 | |
| Abcc8 | 23 | 180 | 150 | 2.16 (0.33) | 0.21 | -0.01 | |
| Kcnj11 | 18 | 34 | 31 | 1.49 (0.23) | 0.48 | -0.19 |
N is the number of cells expressing the tested gene. A total of 60 (84 for Ins1 and Ins2) cells were collected in 5 mM glucose, and 61 (85 for Ins1 and Ins2) cells in 20 mM glucose
The arithmetic mean is calculated as μA = (Σ Xn)/N
The geometric mean is derived from the formula μG = (Π Xn)1/N
Logarithm of the geometric mean and corresponding standard deviations
For the Shapiro-Wilk test, a high P value reflects a good fit. If P ≥ 0.05, then the null hypothesis that data are lognormal cannot be rejected
Skewness is calculated as γ = Σ(Xn - μ)3/(N - 1)σ3
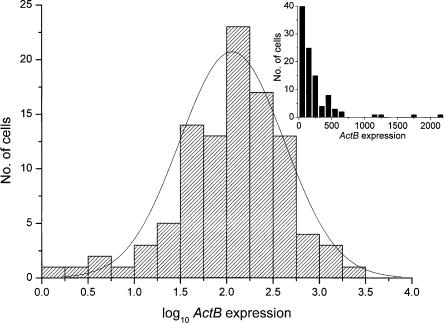
To visualize the gene expression profile in a population of cells, distribution plots were used. Figure 1 shows histograms of ActB expression levels in both logarithmic and linear scale. As confirmed by the Shapiro-Wilk normality test, the transcript distribution is lognormal at 95% significance level (P = 0.05) (Table 1). The transcript distribution of the other genes is also lognormal at the same significance level. Corresponding data for the MIN6 cells are in Supplemental Table 1.
Figure 1.
Histograms showing the expression levels of 96 cells expressing ActB in logarithmic and linear scale (inset). Logarithms of transcript levels are mean-centered for the two glucose concentrations. Solid line describes lognormal distribution centered on the geometric mean (2.06) of the ActB expression levels. Inset shows histogram of the expression levels in linear scale.
Lognormal distributions are common in nature. They are occasionally mistaken for normal (Gaussian) distributions, although the difference is fundamental (Limpert et al. 2001). In lognormal distribution, random, independent effects are multiplicative, while they are additive in a normal distribution. Bacteria in exponential growth are a classic example of a biological system where the numbers of organisms in the different colonies show lognormal distribution. Many more examples are known (Koch 1966; Limpert et al. 2001). The parameters describing the lognormal distributions of the expression levels of the studied genes at low and high glucose concentrations are given in Table 1. Whereas the arithmetic mean is representative for a normally distributed population, the geometric mean is a better indicator for a lognormal population. The differences between the geometric and arithmetic means of cellular gene expression levels for the Ins1 and Ins2 genes in the non–glucose-stimulated cells were substantial and amounted to nine- and fivefold, respectively. This is consistent with lognormal distribution of expression levels.
We ascertained that the observed lognormal distribution of expression levels reflects true biological variability and is not an artifact of the technology or the approach used (see Supplemental information; Methods; Supplemental Fig. 1). The finding that cellular transcript levels are lognormally distributed has implications on the interpretation of gene expression data in general. If mRNA expression levels among cells are lognormally rather than normally distributed, then the average expression measured on a cell population does not reflect the expression in the typical cell in the population. The average value is strongly biased by a small population of cells with very active transcription of the particular gene. Accordingly, it may not be valid to extrapolate results of gene expression measurements on cell populations to the single-cell level. We analyzed this aspect by measuring the distribution of the ratios between the expression levels at high and low glucose concentration for Ins1 and Ins2 (see Table 3). Glucose stimulation has been reported to increase Ins1 and Ins2 expression two- to fivefold (Nielsen et al. 1985), and this is in the range of the arithmetic means of our single cell expression data. By contrast, the ratios of the geometric means of the single cell expression levels are 17 and 9.5 for Ins1 and Ins2, respectively. Thus, glucose may have a much stronger effect on insulin gene transcription in some β-cells than is indicated by measurements on populations of cells.
Table 3.
Ratios of mean expression levels in cell populations incubated in 20 mM and 5 mM glucose
| Arithmetic | Geometric | |
|---|---|---|
| ActB | 3.3 | 4.9 |
| Ins1 | 4.6 | 17 |
| Ins2 | 3.0 | 9.5 |
| Abcc8 | 1.4 | 1.5 |
| Kcnj11 | 1.1 | 1.3 |
Arithmetic and geometric mean value of the expression level at high glucose concentration divided with the corresponding mean value at low glucose concentration. The mean values are calculated according to legends in Table 1.
The expression levels of Ins1 and Ins2 are affected similarly by glucose, suggesting that the two genes are regulated by similar or perhaps even a common mechanism (Wicksteed et al. 2001). By comparing at single-cell level, we find the expressions of the insulin genes highly correlated (Pearson coefficient of 0.90) (Table 2). In contrast, their expression does not correlate with that of the other genes studied (Table 2). Thus, there are not two populations with overall high and low transcriptional activity, but some cells have low and others have high Ins1 and Ins2 expression independently of the expression of the other genes. This conclusion provides additional support of the notion that the observed variation of gene expression reflects true biological variation in cellular mRNA levels. Our analyses also provide the novel observation that the expression of ActB is also stimulated by glucose (Table 3), which casts doubt on the use of ActB as expression standard (Vandesompele et al. 2002). Interestingly, we find no correlation between ActB and Ins1/Ins2 transcription, suggesting that the molecular mechanisms behind the stimulations of ActB and Ins1/Ins2 are different.
Table 2.
Pearson correlation coefficients based on logarithms of expression levels
| ActB | Ins1 | Ins2 | Abcc8 | Kcnj11 | |
|---|---|---|---|---|---|
| ActB | 1 | ||||
| Ins1 | 0.15 | 1 | |||
| Ins2 | 0.12 | 0.90 | 1 | ||
| Abcc8 | -0.02 | -0.01 | 0.06 | 1 | |
| Kcnj11 | 0.11 | -0.02 | 0.24 | -0.15 | 1 |
Pearson coefficient is calculated as 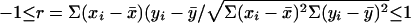 , where x and y are the logarithms of expression levels of the two genes, i indexes the cell, and the bar indicates average. r close to 1 signifies strong positive correlation, while a value close to zero means no correlation. Negative r value would be anti-correlation.
, where x and y are the logarithms of expression levels of the two genes, i indexes the cell, and the bar indicates average. r close to 1 signifies strong positive correlation, while a value close to zero means no correlation. Negative r value would be anti-correlation.
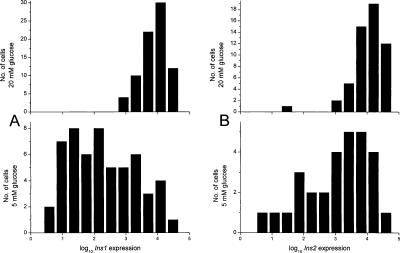
Figure 2 shows the logarithm of Ins1 and Ins2 expression at low and high glucose levels in histograms. The large shift toward higher expression levels at elevated glucose concentration reflects 100-fold increase in insulin expression triggered by the sugar. Asymmetry, or skewness, of the distribution constitutes evidence of deviation from lognormal behavior. The skewness values for the genes analyzed, quantified as Σ(X – μ)3/(N – 1)σ3, are presented in Table 1. A positive value suggests that the distribution is skewed toward higher values, and vice versa. Ins1 distributions exhibited high skewness of opposite signs at low and high glucose concentrations. This may suggest the existence of two subpopulations of cells: one active to secrete insulin and one dormant, exhibiting a bimodal distribution. The bimodal gene induction model (Ko 1992; Fiering et al. 2000) suggests that a subpopulation of the β-cells has enhanced transcription levels characterized by a high mean value, while another subpopulation has low transcriptional activity characterized by a much lower mean value. Elevated glucose level thus increases the probability that individual cells are activated. The currently prevailing model of the action of enhancers is consistent with the binary model for gene activation (Fiering et al. 2000; Paldi 2003).
Figure 2.
(A) Histograms of Ins1 expression levels in cells incubated in 5 mM (bottom) and 20 mM (top) glucose. Horizontal axis has logarithmic scale, which is identical in the two histograms. Scale on the vertical axis indicates the cell count in each histogram bin. (B) Corresponding histograms for Ins2 expression levels.
An alternative interpretation of the data shown in Figure 2 is that high glucose “locks” the insulin gene in a high-expression state, whereas its expression at low glucose concentrations is more random. It should be noted that a fairly large portion of the cells at low glucose concentration exhibit as high insulin gene expression as that observed for most cells during stimulation with 20 mM glucose.
When the expression levels of two genes are correlated, a common regulatory mechanism is often assumed. It may be through a mechanism that actually affects the two genes the same way, such as a common transcription factor, resulting in correlation at the single cell level, but it can also be a general increase in transcriptional activity due to, for example, environmental factors. The latter would also give rise to correlation in expression between genes, but not necessarily at the level of the individual cell. In our system, Ins1 and Ins2 are correlated on the cell level, while Ins1/Ins2 and ActB are correlated only on the population level. Our technology offers means to distinguish between these two cases and is expected to become especially useful for studies of molecular mechanisms underlying complex biological processes as well as disease.
Methods
Preparation and culture of cells
Animals used in this study were healthy female National Maritime Research Institute (NMRI) mice aged 3–4 mo that were obtained from a commercial breeder (Bomholtgaard, Ry, Denmark) and fed a normal diet ad libitum. Care and use of animals were approved by the ethical committee of Lund University. The mice were sacrificed by cervical dislocation, and pancreatic islets were isolated by collagenase P digestion (Roche) (Olofsson et al. 2002). Single islet cells were then prepared by gently shaking the collected islets at low extracellular [Ca2+] as described (Eliasson et al. 1997). Dispersed cells were plated on plastic Petri dishes (Nunc) in RPMI 1640 (SVA) supplemented with 10% fetal calf serum, 100 U/mL penicillin, and 10 μg/mL streptomycin (all from Invitrogen) in the presence of either 5 or 20 mM glucose (Sigma-Aldrich). The cells were maintained in culture for 20–24 h. The data presented are from four batches of cells from different animals that were exposed to either 5 (two animals) or 20 (one animal) mM glucose. To account for individual variation between the animals, a fourth batch of cells were prepared from one animal and incubated in both 5 and 20 mM glucose. The individual variation was found to be negligible compared with the effect of glucose.
Mouse insulinoma MIN6-cells (passage 30 and above) were cultured in DMEM medium (10 mM glucose, Invitrogen) supplemented with 10% fetal calf serum, 100 U/mL penicillin, and 10 μg/mL streptomycin (all from Invitrogen) to ∼50% confluence by using standard culture techniques.
Single cell isolation and cDNA synthesis
Cells that adhered to the dish were thoroughly washed with extracellular solution containing 138 mM NaCl, 5.6 mM KCl, 1.2 mM MgCl2, 2.6 mM CaCl2, and 5 mM HEPES (pH 7.4 with NaOH). Cell content was collected with a borosilicate glass patch-clamp pipette with an average diameter of 5 μm mounted on a hydraulic micromanipulator. By controlling the pressure inside the pipette, it was possible to collect intact or nearly intact cells with minimum volume of extracellular solution. Pipettes were emptied in 0.2-mL tubes initially containing 2 μL of nonchaotropic lysis solution. Tubes were then vortexed for 10 sec. The lysis buffer contained 0.5% Igepal CA-630, 50 mM Tris-Cl (pH 8.0), 140 mM NaCl, 1.5 mM MgCl2 (all Sigma), and 1 U/μL Prime RNase inhibitor (Eppendorf). Samples were immediately frozen in ethanol cooled with dry ice (temperature: –78°C), and stored at –25°C for subsequent reverse transcription. A reverse transcription master mix contained dNTP (Sigma-Aldrich) and oligo(dT) primers (TAGC Copenhagen) at final concentrations of 0.5 mM and 10 μM, respectively. It was also supplemented with 150 ng of random hexamer primers (TAGC Copenhagen) and 15 U of Prime RNase inhibitor (Eppendorf); 150 ng Linear Polyamide (GenElute LPA, Sigma-Aldrich) was added to some samples, but this did not have an effect on the cDNA synthesis efficiency. It was also found that the cell could be emptied directly into the RT-base, without the lysis buffer, with maintained reaction efficiency. After incubation for 5 min at 80°C and subsequent cooling on ice, the cDNA synthesis was initiated by addition of reverse transcription enzyme (SuperScript III, Invitrogen) and 2-h incubation at 50°C. The reaction was terminated by heating for 15 min at 70°C. Final volume for all reverse transcription reactions was 10 μL. The cDNA was either immediately quantified by real-time PCR or stored at –20°C pending later analysis. A total of 169 cells were analyzed, of which 84 were incubated in 5 mM glucose and 85 cells in 20 mM glucose. Twenty-four of the cells in each group were analyzed with primers for Ins2 only; the remaining cells, with primers for ActB, Ins1, Ins2, Abcc8, and Kcnj11.
Quantitative real-time PCR
Two instruments were used for real-time PCR measurements: the LightCycler (Roche) and the ABI PRISM 7900HT Sequence Detection System (Applied Biosystems). Ten-microliter reactions were used on the LightCycler containing 3 mM MgCl2, 0.3 mM dNTP, 0.1 mg/mL BSA (all Sigma-Aldrich), 400 μM primer (MWG and TAGC Copenhagen), 0.5× SYBR Green I (Molecular Probes/Invitrogen), and 1 U JumpStart Taq polymerase (Sigma-Aldrich). Primer sequences are available upon request. A similar master mix was used with the ABI PRISM 7900HT system, with ROX as passive reference dye but without BSA and a final volume of 20 μL. Single-cell cDNA (1 μL) was added to the reaction mixture when using the LightCycler, while 2 μL cDNA was analyzed in the 7900. Absolute quantification of each cDNA species was performed by dilution series of purified PCR products (QIAquick PCR purification reagent set, Qiagen), and concentrations were measured with a NanoDrop ND-1000 spectrophotometer (Nanodrop Technologies). Real-time PCR data analysis was performed according to established procedures (Bustin 2000; Stahlberg et al. 2005).
Some cells appeared to lack at least one transcript; 96% of all collected samples contained detectable transcripts from at least one gene, and 83% had detectable levels of ActB transcript. For the analysis of primary cells in this article, only the insulin-producing β-cells were used. The detection limit of the method is 10–20 copies, which is below the range of expected levels of ActB, Ins1, Ins2, and Abcc8. It is possible that some of the cells that lack insulin gene transcripts are members of an inert subpopulation of β-cells, but the extent would be limited to a maximum 10% of the cells.
Supplementary Material
Acknowledgments
We thank Kristina Borglid, Britt-Marie Nilsson, and Lena Thiman for technical assistance with cell preparations. Research was supported by the Crafoord Foundation, the Royal and Hvitfeldtska Foundation, Swedish Research Council (8647), the Swedish Diabetes Association, the NovoNordisk Foundation, the Swedish Strategic Foundation, the Goran Gustafsson Foundation for Research in the Natural Sciences and Medicine, and the Medical Faculty Lund University. We also thank Prof. Miyazaki for providing us with the MIN6 cell line. P.R. is a Royal Society-Wolfson Research Fellow.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.3820805.
Footnotes
[Supplemental material is available online at www.genome.org.]
References
- Barg, S., Galvanovskis, J., Gopel, S.O., Rorsman, P., and Eliasson, L. 2000. Tight coupling between electrical activity and exocytosis in mouse glucagon-secreting α-cells. Diabetes 49: 1500-1510. [DOI] [PubMed] [Google Scholar]
- Bishop, A.E. and Polak, J.M. 1997. The anatomy, organisation and ultrastructure of the islets of Langerhans. In Textbook of diabetes (eds. J. Pickup and G. Williams), pp. 6.1-6.16. Blackwell Science, Oxford, UK.
- Blake, W.J., Kaern, M., Cantor, C.R., and Collins, J.J. 2003. Noise in eukaryotic gene expression. Nature 422: 633-637. [DOI] [PubMed] [Google Scholar]
- Bustin, S.A. 2000. Absolute quantification of mRNA using real-time reverse transcription polymerase chain reaction assays. J. Mol. Endocrinol. 25: 169-193. [DOI] [PubMed] [Google Scholar]
- Castano, J.P., Kineman, R.D., and Frawley, L.S. 1996. Dynamic monitoring and quantification of gene expression in single, living cells: A molecular basis for secretory cell heterogeneity. Mol. Endocrinol. 10: 599-605. [DOI] [PubMed] [Google Scholar]
- Chiang, M.K. and Melton, D.A. 2003. Single-cell transcript analysis of pancreas development. Dev. Cell 4: 383-393. [DOI] [PubMed] [Google Scholar]
- Eliasson, L., Renstrom, E., Ding, W.G., Proks, P., and Rorsman, P. 1997. Rapid ATP-dependent priming of secretory granules precedes Ca(2+)-induced exocytosis in mouse pancreatic B-cells. J. Physiol. 503: 399-412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elowitz, M.B., Levine, A.J., Siggia, E.D., and Swain, P.S. 2002. Stochastic gene expression in a single cell. Science 297: 1183-1186. [DOI] [PubMed] [Google Scholar]
- Fiering, S., Whitelaw, E., and Martin, D.I. 2000. To be or not to be active: The stochastic nature of enhancer action. Bioessays 22: 381-387. [DOI] [PubMed] [Google Scholar]
- Gehwolf, R., Griessner, M., Pertl, H., and Obermeyer, G. 2002. First patch, then catch: Measuring the activity and the mRNA transcripts of a proton pump in individual Lilium pollen protoplasts. FEBS Lett. 512: 152-156. [DOI] [PubMed] [Google Scholar]
- Kamme, F., Salunga, R., Yu, J., Tran, D.T., Zhu, J., Luo, L., Bittner, A., Guo, H.Q., Miller, N., Wan, J., et al. 2003. Single-cell microarray analysis in hippocampus CA1: Demonstration and validation of cellular heterogeneity. J. Neurosci. 23: 3607-3615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko, M.S. 1992. Induction mechanism of a single gene molecule: Stochastic or deterministic? Bioessays 14: 341-346. [DOI] [PubMed] [Google Scholar]
- Koch, A.L. 1966. The logarithm in biology, 1: Mechanisms generating the log-normal distribution exactly. J. Theor. Biol. 12: 276-290. [DOI] [PubMed] [Google Scholar]
- Kuznetsov, V.A., Knott, G.D., and Bonner, R.F. 2002. General statistics of stochastic process of gene expression in eukaryotic cells. Genetics 161: 1321-1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levsky, J.M. and Singer, R.H. 2003. Gene expression and the myth of the average cell. Trends Cell Biol. 13: 4-6. [DOI] [PubMed] [Google Scholar]
- Levsky, J.M., Shenoy, S.M., Pezo, R.C., and Singer, R.H. 2002. Single-cell gene expression profiling. Science 297: 836-840. [DOI] [PubMed] [Google Scholar]
- Limpert, E., Stahel, W.A., and Abbt, M. 2001. Log-normal distributions across the sciences: Keys and clues. Bioscience 51: 341-352. [Google Scholar]
- Liss, B., Franz, O., Sewing, S., Bruns, R., Neuhoff, H., and Roeper, J. 2001. Tuning pacemaker frequency of individual dopaminergic neurons by Kv4.3L and KChip3.1 transcription. EMBO J. 20: 5715-5724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo, L., Salunga, R.C., Guo, H., Bittner, A., Joy, K.C., Galindo, J.E., Xiao, H., Rogers, K.E., Wan, J.S., Jackson, M.R., et al. 1999. Gene expression profiles of laser-captured adjacent neuronal subtypes. Nat. Med. 5: 117-122. [DOI] [PubMed] [Google Scholar]
- McAdams, H.H. and Arkin, A. 1997. Stochastic mechanisms in gene expression. Proc. Natl. Acad. Sci. 94: 814-819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazaki, J., Araki, K., Yamato, E., Ikegami, H., Asano, T., Shibasaki, Y., Oka, Y., and Yamamura, K. 1990. Establishment of a pancreatic β cell line that retains glucose-inducible insulin secretion: Special reference to expression of glucose transporter isoforms. Endocrinology 127: 126-132. [DOI] [PubMed] [Google Scholar]
- Nielsen, D.A., Welsh, M., Casadaban, M.J., and Steiner, D.F. 1985. Control of insulin gene expression in pancreatic β-cells and in an insulin-producing cell line, RIN-5F cells, I: Effects of glucose and cyclic AMP on the transcription of insulin mRNA. J. Biol. Chem. 260: 13585-13589. [PubMed] [Google Scholar]
- Olofsson, C.S., Gopel, S.O., Barg, S., Galvanovskis, J., Ma, X., Salehi, A., Rorsman, P., and Eliasson, L. 2002. Fast insulin secretion reflects exocytosis of docked granules in mouse pancreatic B-cells. Pflugers Arch. 444: 43-51. [DOI] [PubMed] [Google Scholar]
- Ozbudak, E.M., Thattai, M., Kurtser, I., Grossman, A.D., and van Oudenaarden, A. 2002. Regulation of noise in the expression of a single gene. Nat. Genet. 31: 69-73. [DOI] [PubMed] [Google Scholar]
- Paldi, A. 2003. Stochastic gene expression during cell differentiation: Order from disorder? Cell. Mol. Life Sci. 60: 1775-1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peixoto, A., Monteiro, M., Rocha, B., and Veiga-Fernandes, H. 2004. Quantification of multiple gene expression in individual cells. Genome Res. 14: 1938-1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raser, J.M. and O'Shea, E.K. 2004. Control of stochasticity in eukaryotic gene expression. Science 304: 1811-1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahlberg, A., Zoric, N., Aman, P., and Kubista, M. 2005. Quantitative real-time PCR for cancer detection: The lymphoma case. Expert Rev. Mol. Diagn. 5: 221-230. [DOI] [PubMed] [Google Scholar]
- Sucher, N.J., Deitcher, D.L., Baro, D.J., Warrick, R.M., and Guenther, E. 2000. Genes and channels: Patch/voltage-clamp analysis and single-cell RT-PCR. Cell Tissue Res. 302: 295-307. [DOI] [PubMed] [Google Scholar]
- Tietjen, I., Rihel, J.M., Cao, Y., Koentges, G., Zakhary, L., and Dulac, C. 2003. Single-cell transcriptional analysis of neuronal progenitors. Neuron 24: 161-175. [DOI] [PubMed] [Google Scholar]
- Vandesompele, J., De Preter, K., Pattyn, F., Poppe, B., Van Roy, N., De Paepe, A., and Speleman, F. 2002. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 3: research0034. [DOI] [PMC free article] [PubMed]
- Walters, M.C., Fiering, S., Eidemiller, J., Magis, W., Groudine, M., and Martin, D.I. 1995. Enhancers increase the probability but not the level of gene expression. Proc. Natl. Acad. Sci. 92: 7125-7129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicksteed, B., Herbert, T.P., Alarcon, C., Lingohr, M.K., Moss, L.G., and Rhodes, C.J. 2001. Cooperativity between the preproinsulin mRNA untranslated regions is necessary for glucose-stimulated translation. J. Biol. Chem. 276: 22553-22558. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.