Abstract
Accessing and analyzing the exponentially expanding genomic sequence and functional data pose a challenge for biomedical researchers. Here we describe an interactive system, Galaxy, that combines the power of existing genome annotation databases with a simple Web portal to enable users to search remote resources, combine data from independent queries, and visualize the results. The heart of Galaxy is a flexible history system that stores the queries from each user; performs operations such as intersections, unions, and subtractions; and links to other computational tools. Galaxy can be accessed at http://g2.bx.psu.edu.
Currently available genome browsers (UCSC Genome Browser [Kent et al. 2002, http://genome.ucsc.edu], NCBI MapViewer [Wheeler et al. 2005], and Ensembl [Birney et al. 2004, http://www.ensembl.org]) allow experimental biologists with no programming experience to locate and visualize genomic regions using intuitive graphical interfaces. However, more sophisticated analyses (e.g., “find all DNase I hypersensitive sites within introns of RefSeq genes on human chromosome 22 that are also conserved in the mouse and rat genomes but not in the dog genome”) still rely on programming and database skills. To solve this problem we designed Galaxy, a system for the integration of genomic sequences, their alignments, and functional annotation. Galaxy is not a browser. Instead, it allows users to gather and manipulate data from existing resources in a variety of ways. Every action of the user is recorded and stored in the history system, a key element of Galaxy. This allows users to conduct independent queries on genomic data from different sources and then use Galaxy to combine or refine them, perform calculations, or extract and visualize corresponding sequences or alignments. Operations such as join, union, intersection, and subtraction can be accomplished using a simple interface.
Galaxy differs from existing systems in its specificity for access to, and comparative analysis of, genomic sequences and alignments. For example, the premier metaserver for the retrieval, analysis, and display of protein and DNA sequences, SRS (Etzold and Argos 1993; Zdobnov et al. 2002), does not provide access to precomputed genome sequence alignments, scores derived from those alignments, expression data, or other genomic data types that are central to Galaxy. Other examples of efforts integrating various data sources and analysis tools include ISYS (Siepel et al. 2001) and the Biology Workbench (Subramaniam 1998). ISYS requires programming experience and serves as a development framework rather than a ready-to-use tool. Biology Workbench is one of the most comprehensive Web-based collections of sequence analysis software. However, it is unsuitable for the analysis of genomic data as it cannot handle large sequence data sets. Here we describe the presently implemented functionality of the Galaxy system, show examples of usage, and discuss some aspects of its design.
Results and Discussion
Data retrieval and manipulation
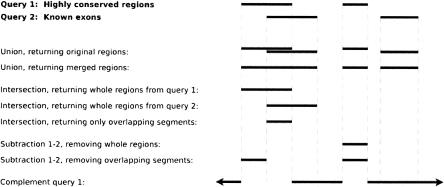
Presently, Galaxy contains three major classes of data manipulation: query operations, sequence analysis tools, and output displays. The first class includes standard set operations such as union, intersection, subtraction, and complement as well as filters based on region size, proximity to regions from another query, and clustering by distance of regions within a single query (Fig. 1). Sequence analysis tools are stand-alone modules designed to perform biologically oriented calculations such as finding orthologous regions in another species, extracting genomic alignments, computing Ka/Ks ratios, and retrieving GC content or conservation in the regions of interest. Finally, displays allow retrieving/viewing of the results generated by the user in a variety of formats. Current options include displaying the query results as a custom track at the UCSC or Ensembl Genome Browsers and downloading a text file in various formats (standard BED, Ensembl upload, or raw); additional formats are provided by individual tools (e.g., score distribution plot, Ka/Ks sliding window profile). Alignment viewers such as Laj (Wilson et al. 2001) and zPicture (Ovcharenko et al. 2004) are planned for the near future.
Figure 1.
Galaxy supports several variations of the basic set operations, to accommodate the fact that our elements are coordinate-based regions rather than simple atomic objects.
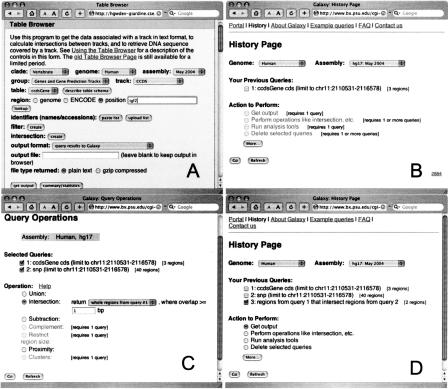
The basic functionality of Galaxy is best illustrated with an example. Here we use the Galaxy history system to combine independent queries to find single nucleotide polymorphisms (SNPs) within coding exons of the human insulin-like growth factor II (IGF-II) gene. At the Galaxy portal page the user first chooses a genomic region of interest (the IGF-II locus) from the UCSC Table Browser (Karolchik et al. 2004) (Fig. 2A), which sends its results (genomic coordinates of coding exons) directly to Galaxy (for this purpose the Table Browser interface features a “Send Results to Galaxy” option). The Galaxy history page then displays one query, which contains the genomic coordinates for each protein-coding exon of the IGF-II gene (Fig. 2B). Because our goal is to find all SNPs associated with coding exons, we go back to the Table Browser and repeat the process, this time requesting all SNPs that fall in the genomic region of the IGF-II gene. Now the requested SNPs will appear as the second query on the history page (Fig. 2C). However, we are only interested in SNPs that fall within coding exons, so to identify these we apply the intersection operation to the two queries (Fig. 2C). The result (six SNPs are found within protein-coding exons of IGF-II) is displayed as a new item on the history page (Fig. 2D). At this point, the user can download the results or display them as a custom track at the UCSC Genome Browser (generating an image similar to Fig. 3).
Figure 2.
Galaxy history system for querying UCSC Table Browser. (A) UCSC Table Browser page sending results to Galaxy. (B) Galaxy's history page with a single query. (C) History page showing how Galaxy can be used to find intersection between two queries. (D) History page displaying intersection results.
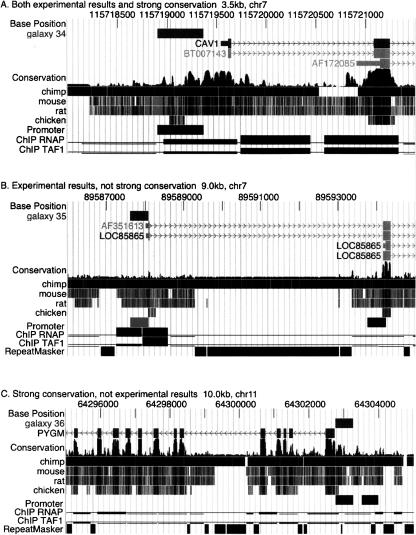
Figure 3.
Examples of promoters characterized by binding of the transcription initiation complex and/or high conservation. Images from the UCSC Genome Browser generated via Galaxy illustrate (A) a promoter that has strong conservation (indicative of purifying selection) and biochemical evidence of binding by RNA polymerase II and TAF1, (B) a promoter that is poorly conserved but is strongly bound by RNA polymerase II and TAF1, and (C) a strongly conserved promoter that is not bound by the transcription initiation machinery in the cells tested. The track labeled “galaxy” is the custom track automatically generated by Galaxy for each query number (34, 35, and 36). Genes are labeled and have exons as boxes and introns as lines with arrowheads pointing in the direction of transcription. “Conservation” is the phastCons track followed by positions of aligning DNA in homologous regions of other species. The positions of promoters are shown as rectangles. The results of chromatin immunoprecipitations (ChIP data) are plotted as the negative log of the p-value, ranging in the vertical direction from 0 to 10.0 and with a continuous thin line placed at the threshold of 2.0. Positions of repeats identified by RepeatMasker (A.F.A. Smit and P. Green, unpubl., http://ftp.genome.washington.edu/RM/RepeatMasker.html) are shown as black rectangles in panels B and C.
Combining and comparing ENCODE data to find promoters
Locating promoters is one of the aims of the ENCODE consortium (ENCODE Project Consortium 2004). Six data tracks relevant to this goal are already deposited at the UCSC ENCODE portal. These include empirical results (experimentally validated promoters [Trinklein et al. 2003], DNase I hypersensitive sites, and regions bound by RNA polymerase II or TAF1 [Kim et al. 2005]) and computational predictions (multi-spe-cies conserved sequences [Margulies et al. 2003], phastConsElements [Siepel et al. 2005], and regions with high regulatory potential [Kolbe et al. 2004]). The ability to combine and compare these diverse data is critical for their biological interpretation. The following example shows that Galaxy is ideally suited for this purpose. Starting at the Galaxy portal, the UCSC Table Browser was used to retrieve genomic intervals that passed reasonable thresholds for each of the six data types (see online supplement). Galaxy operations (intersection and subtraction) were then applied to compare the data sets, determining what fraction of experimentally verified promoters had the other properties investigated (Table 1). Of the 289 promoters, 95 (33%) are both highly conserved (phastConsElement) and have significant binding by TAF1 in HeLa cells. Thus, these promoters can be identified by either strong conservation or by experimental results (such as TAF1 binding). One example is the CAV1 promoter (Fig. 3A). Of the remaining 194 promoters, 52 intersect with a segment having significant binding by TAF1, and 66 intersect with a phastConsElement. Thus, some promoters are characterized by TAF1 binding but not strong conservation, exemplified by LOC85865 (Fig. 3B). These will be more difficult to identify by comparative genomics approaches. Others are characterized by strong conservation but do not show evidence of TAF1 binding. The example of PYGM (Fig. 3C) could be explained by the fact that the glycogen phosphorylase encoded by the gene is made primarily in muscle cells, whereas the binding data are from HeLa cells, which were derived from a cervical carcinoma.
Table 1.
Number of regions within ENCODE targets with properties associated with gene promoters
| Type of region | Number of regions exceeding threshold | Number of promoters that overlap regions | Percentage of promoters that overlap regions |
|---|---|---|---|
| Promoters | 289 | 289 | 100 |
| DNase HSs | 230 | 71 | 25 |
| Bound by RNA polymerase | 2121 | 175 | 61 |
| Bound by TAF1 | 573 | 153 | 53 |
| MCS | 23,148 | 179 | 62 |
| phastConsElements | 7479 | 139 | 48 |
| RP | 16,170 | 161 | 56 |
(DNase I HSs) DNase I hypersensitive sites; (MCS) multispecies conserved sequence; (phastConsElements) DNA sequence whose multispecies alignment falls within the 5% most highly conserved genomic intervals in human; (RP) regulatory potential.
Evolutionary analyses with Galaxy
Our system will allow users to apply existing molecular evolution algorithms directly to sequences and alignments retrieved through Galaxy queries. The current release of Galaxy features a tool for calculation of synonymous (Ks) and non-synonymous (Ka) substitution rates using the Yang-Neilsen algorithm (Yang and Nielsen 2000). The tool allows traditional estimation across the entire length of a selected sequence as well as estimation using a sliding window approach. The estimates obtained with the tool can be used to perform the Ka/Ks ratio test, the most widely used predictor of selection acting on a protein coding region (Li 1997). The sliding window Ka/Ks test is a simple analysis that can provide a wealth of information about the selection regime of a gene of interest (Endo et al. 1996; Presgraves et al. 2003). This test provides significantly greater resolution compared with the conventional Ka/Ks test, which is overly conservative for detecting deviations from a negative selection scenario as it averages Ka and Ks estimates over the entire sequence (Li 1997). Galaxy users are now able to apply this analysis to any coding sequence available from the UCSC Table Browser (e.g., as shown in Fig. S1).
Conclusions
The Galaxy system pioneers a new generation of interactive tools for large-scale genome analysis. It allows large-scale analyses that previously required users to have substantial programming experience and database management skills. The Galaxy history page is simple to use, yet quite powerful, and is able to handle large genome annotation data sets. Users have the ability to perform multiple types of analyses (e.g., query intersections, subtractions, and proximity searches) and then display the results using existing browsers (e.g., the UCSC Genome Browser or Ensembl). In the future we plan to add a powerful toolbox that will include the most popular sequence and genome analysis algorithms. Galaxy's permanent Web site address is http://www.g2.bx.psu.edu.
Methods
Modularity
Galaxy is designed as a set of separate software components that work together to perform tasks. The central “core” component orchestrates the action, executes queries, and keeps track of user histories, while the user interface(s) (UIs) and operation/tool/output libraries are implemented separately. All communication with other sites (UCSC Table Browser, etc.) is handled by the core component. Benefits of this arrangement include extensibility (ease of adding new tools and interfaces) and convenient division of labor and expertise among programmers. Also, the operation libraries are available for use by other projects, such as ENCODEdb.
The UIs communicate with the core component via HTTP (Web) requests, using the GET or POST methods. The core provides an API (application program interface) consisting of the requests it is prepared to handle, such as using a tool, retrieving a user's query history for a particular assembly of a genome, etc. When the user runs a query at another source site (e.g., the Table Browser), the core passes its connection with the user's Web browser on to the Galaxy UI via HTTP redirection. Using an HTTP API makes it easy to support a variety of UIs, which do not have to be running on the same server. In fact, any site on the Web could set up its own UI for Galaxy by crafting the appropriate HTTP requests, and individual researchers can use the API directly for programmatic access to Galaxy's features.
Language
The Galaxy core component and operation libraries are written in C and are built to the standards of the Bioinformatics group at UCSC. Thus, if it turns out to be more effective to run some Galaxy functions from UCSC instead of PSU, the programs are ready to be run there. Also, this code makes use of UCSC utility libraries to avoid duplication of effort.
Our initial UI (called HUI for History User Interface) is written in Perl for convenient text manipulation and CGI access, but one could use any language that can generate an HTTP request.
Local storage
Although Galaxy primarily processes source data obtained from other sites, it does have a local database for storing user histories (implemented in MySQL for compatibility with UCSC). It also stores some precomputed query results. We originally implemented this as a way to avoid recomputing popular and/or time-consuming queries again and again, but now we also view this “featured data sets” facility as a way to provide public access to newly obtained research results before they are available on the primary data sites, and to data sets that are too large for uploading from the Table Browser.
Additional local storage is used for reference data and temporary workspace needed by some of the tools, and for caching query results, output files, and custom data sets (uploaded by users) for further manipulation and/or subsequent retrieval.
Data format
The primary format that Galaxy uses to store query results is the BED (Browser Extensible Data) format that is used at UCSC for Genome Browser tracks and also is accepted at several other sites. This is a tab-separated text format readable by both humans and computer programs. The BED format is convenient for interoperability with UCSC's Table Browser, Genome Browser, and other tools, but it has fairly strict limitations on the associated fields since it is primarily geared toward displaying the regions rather than conducting further analysis. Currently we are working on extensions to the BED format by adding extra columns that can be readily truncated back to true BED for the tools that require it, but ultimately we will probably need to use a more generic format, or several formats, to handle a broader range of data types. In particular, there are well-established file formats for alignment data (such as AXT and MAF) that are used directly. But regardless of the data formats we end up using internally, it will be important for Galaxy to provide a suitable complement of conversion tools so users can easily obtain output in whatever format they need.
Response time
Galaxy's history scheme enables the UI to return a response page quickly, even for queries that may take minutes to run. HUI's history page displays the status of each query as “in queue,” “running,” “N regions” (N = the number of regions selected by the completed query), “error” (which is a link to a more detailed error message). This status currently is updated by clicking the “Refresh” button, though we intend to explore ways to make this happen automatically. Currently, for queries using the UCSC Table Browser the average response time is only 44 sec. Genome-wide queries on large tables can take up to 25 min, but genome-wide queries on small data sets (e.g., UCSC Known Genes), or queries on large tables that are limited to a relatively small genomic range can be done in close to the average time (<1 min). The Galaxy operations are more uniform with an average execution time of 53 sec and a typical maximum of 8 min.
User identity
Galaxy needs to keep track of individual users in order to maintain their personalized query histories. Currently this is accomplished by assigning a sequential ID number and storing it as a cookie in the user's Web browser. This is distinct from the IDs that are assigned by the Table Browser and other data sources, and indeed Galaxy records these as well so it can add new queries from those sources to the proper history. Cookies can also keep track of user preferences; currently HUI stores the most recently used genome and assembly so it can open to the same page next time. We are also considering adding a login facility so users can easily carry their histories from one computer to another, and so multiple users can have different identities on the same computer.
Web browser requirements
Our initial HUI uses simple HTML markup that should work with most or all modern Web browsers. It endeavors to adhere to W3C standards for HTML 4.01 Transitional. The only “fancy” features it uses are cookies (discussed above), style sheets (CSS), and a small amount of JavaScript, which is used only to refresh the page as the user makes choices. Users who prefer to turn off scripting can still use the interface; they simply need to click the Refresh button manually.
Supplementary Material
Acknowledgments
We thank David Haussler for his support of the project and members of the Center for Comparative Genomics and Bioinformatics at Penn State for their input. This work is supported by funds provided by the Eberly College of Science, Huck Institutes of the Life Sciences at Penn State University, and NIH grants DK65806 (R.H.) and HG02238 (W.M.).
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.4086505. Article published online before print in September 2005.
Footnotes
[Supplemental material is available online at www.genome.org.]
References
- Birney, E., Andrews, D., Bevan, P., Caccamo, M., Cameron, G., Chen, Y., Clarke, L., Coates, G., Cox, T., Cuff, J. et al. 2004. Ensembl 2004. Nucleic Acids Res. 32: D468-D470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ENCODE Project Consortium. 2004. The ENCODE (ENCyclopedia Of DNA Elements) Project. Science 306: 636-640. [DOI] [PubMed] [Google Scholar]
- Endo, T., Ikeo, K., and Gojobori, T. 1996. Large-scale search for genes on which positive selection may operate. Mol. Biol. Evol. 13: 685-690. [DOI] [PubMed] [Google Scholar]
- Etzold, T. and Argos, P. 1993. SRS—An indexing and retrieval tool for flat file data libraries. Comput. Appl. Biosci. 9: 49-57. [DOI] [PubMed] [Google Scholar]
- Karolchik, D., Hinrichs, A.S., Furey, T.S., Roskin, K.M., Sugnet, C.W., Haussler, D., and Kent, W.J. 2004. The UCSC Table Browser data retrieval tool. Nucleic Acids Res. 32: D493-D496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent, W.J., Sugnet, C.W., Furey, T.S., Roskin, K.M., Pringle, T.H., Zahler, A.M., and Haussler, D. 2002. The human genome browser at UCSC. Genome Res. 12: 996-1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, T.H., Barrera, L.O., Zheng, M., Qu, C., Singer, M.A., Richmond, T.A., Wu, Y., Green, R.D., and Ren, B. 2005. A high-resolution map of active promoters in the human genome. Nature 436: 876-880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King, D.C., Taylor, J., Elnitski, L., Chiaromonte, F., Miller, W., and Hardison, R.C. 2005. Evaluation of regulatory potential and conservation scores for detecting cis-regulatory modules in aligned mammalian genome sequences. Genome Res. 15: 1051-1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolbe, D., Taylor, J., Elnitski, L., Eswara, P., Li, J., Miller, W., Hardison, R., and Chiaromonte, F. 2004. Regulatory potential scores from genome-wide three-way alignments of human, mouse and rat. Genome Res. 14: 700-707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, W.H. 1997. Molecular evolution. Sinauer, Sunderland, MA.
- Margulies, E.H., Blanchette, M., NISC Comparative Sequencing Program, Haussler, D., and Green, E.D. 2003. Identification and characterization of multi-species conserved sequences. Genome Res. 13: 2507-2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ovcharenko, I., Loots, G.G., Hardison, R.C., Miller, W., and Stubbs, L. 2004. zPicture: Dynamic alignment and visualization tool for analyzing conservation profiles. Genome Res. 14: 472-477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presgraves, D.C., Balagopalan, L., Abmayr, S.M., and Orr, H.A. 2003. Adaptive evolution drives divergence of a hybrid inviability gene between two species of Drosophila. Nature 423: 715-719. [DOI] [PubMed] [Google Scholar]
- Ren, B. and Dynlacht, B.D. 2004. Use of chromatin immunoprecipitation assays in genome-wide location analysis of mammalian transcription factors. Methods Enzymol. 376: 304-315. [DOI] [PubMed] [Google Scholar]
- Siepel, A., Farmer, A., Tolopko, A., Zhuang, M., Mendes, P., Beavis, W., and Sobral, B. 2001. ISYS: A decentralized, component-based approach to the integration of heterogeneous bioinformatics resources. Bioinformatics 17: 83-94. [DOI] [PubMed] [Google Scholar]
- Siepel, A., Bejerano, G., Pedersen, J.S., Hinrichs, A.S., Hou, M., Rosenbloom, K., Clawson, H., Spieth, J., Hillier, L.W., Richards, S., et al. 2005. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 15: 1034-1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramaniam, S. 1998. The Biology Workbench—A seamless database and analysis environment for the biologist. Proteins 32: 1-2. [PubMed] [Google Scholar]
- Trinklein, N.D., Aldred, S.J., Saldanha, A.J., and Myers, R.M. 2003. Identification and functional analysis of human transcriptional promoters. Genome Res. 13: 308-312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler, D.L., Barrett, T., Benson, D.A., Bryant, S.H., Canese, K., Church, D.M., DiCuccio, M., Edgar, R., Federhen, S., Helmberg, W., et al. 2005. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 33: D39-D45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson, M.D., Riemer, C., Martindale, D.W., Schnupf, P., Boright, A.P., Cheung, T.L., Hardy, D.M., Schwartz, S., Scherer, S.W., Tsui, L.C., et al. 2001. Comparative analysis of the gene-dense ACHE/TFR2 region on human chromosome 7q22 with the orthologous region on mouse chromosome 5. Nucleic Acids Res. 29: 1352-1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Z. and Nielsen, R. 2000. Estimating synonymous and nonsynonymous substitution rates under realistic evolutionary models. Mol. Biol. Evol. 17: 32-43. [DOI] [PubMed] [Google Scholar]
- Zdobnov, E.M., Lopez, R., Apweiler, R., and Etzold, T. 2002. The EBI SRS server—New features. Bioinformatics 18: 1149-1150. [DOI] [PubMed] [Google Scholar]
WEB SITE REFERENCES
- http://ftp.genome.washington.edu/RM/RepeatMasker.html; RepeatMasker Web site
- http://www.g2.bx.psu.edu; Galaxy home page
- http://genome.ucsc.edu; UCSC Genome Browser and Table Browser Web sites.
- http://www.ensembl.org; Ensembl Web site.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.