Abstract
We developed the Yeast Gene Order Browser (YGOB; http://wolfe.gen.tcd.ie/ygob) to facilitate visual comparisons and computational analysis of synteny relationships in yeasts. The data presented in YGOB, currently covering seven species, are based on sets of homologous genes that have been intensively manually curated based on both sequence similarity and genomic context (synteny). We reconciled different laboratories' lists of paralogous Saccharomyces cerevisiae gene pairs formed by genome duplication (ohnologs), and present near-exhaustive lists of the ohnolog pairs retained in S. cerevisiae (551, including 22 previously unidentified), Saccharomyces castellii (599), and Candida glabrata (404).
The hemiascomycete yeasts offer great potential for studying many aspects of genome evolution, because the available genome sequence data span a continuum of divergence levels ranging from multiple isolates of the same species (Gu et al. 2005) to genomes that are as different from each other as are those of humans and urochordates (Dujon et al. 2004). Near-complete genome sequences are currently available for 14 species (Goffeau et al. 1996; Cliften et al. 2003; Kellis et al. 2003, 2004; Dietrich et al. 2004; Dujon et al. 2004; Jones et al. 2004). We are interested in particular in the evolutionary consequences of the whole genome duplication (WGD) event that occurred in an ancestor of Saccharomyces cerevisiae and some of its relatives (Wolfe and Shields 1997; Dietrich et al. 2004; Dujon et al. 2004; Kellis et al. 2004), which requires detailed synteny information.
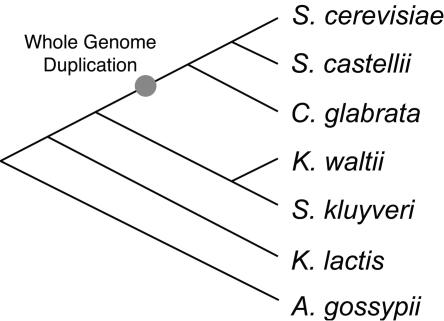
For version 1.0 of the Yeast Gene Order Browser (YGOB), we chose to focus on seven species: three that separated from each other after the WGD (post-WGD species: S. cerevisiae, Saccharomyces castellii, and Candida glabrata), and four outgroups (pre-WGD species: Ashbya gossypii, Kluyveromyces lactis, Kluyveromyces waltii and Saccharomyces kluyveri) (Fig. 1). For the moment we have excluded the four other sequenced Saccharomyces sensu stricto species (Cliften et al. 2003; Kellis et al. 2003) because their gene orders are almost completely collinear with S. cerevisiae. We also excluded more distantly related species (C. albicans, Debaryomyces hansenii, Yarrowia lipolytica) because the level of gene order conservation between them and S. cerevisiae is quite low (Keogh et al. 1998; Llorente et al. 2000; Dujon et al. 2004). However, the YGOB software has been written in a scaleable way, so that other genomes can be added relatively easily.
Figure 1.
Approximate phylogenetic relationship (not drawn to scale) of the yeasts examined in this study, showing the WGD event. The tree is based on the data of Kurtzman and Robnett (2003), but it should be noted that Hittinger et al. (2004) obtained a different branching order among the pre-WGD species.
Although other browsers for comparative genomics have been developed (see Stein et al. 2002; Sherman et al. 2004), they have three limitations that make them inappropriate for our purposes. First, the peculiarity of having a group of genomes of which some are polyploid with respect to others meant that we needed a browser where two chromosomal regions are shown from some species, and only one from others. Second, to maximize what can be fitted onto a computer screen, we wanted to present the gene order rather than a to-scale representation of the chromosomal region. Lastly, and most importantly, we wanted to develop an analytical tool that can score the status of a gene in a species (as either present in its syntenic context or absent from its syntenic context, or uncertain), as opposed to just composing an on-screen image.
YGOB consists of two elements: an engine that can calculate the syntenic context of any gene and thus score it (without making any graphical display), and a “visual browser” that produces a picture of the syntenic context around a gene when a user requests it. The output can be focused on a gene from any genome, not just S. cerevisiae, moving past the exclusively S. cerevisiae worldview of many earlier tools. Underpinning the YGOB software is its curated database of homology assignments across the genomes. We intend YGOB to replace the Yeast Gene Duplications Web site that our laboratory has hosted since 1997 (Wolfe and Shields 1997).
Results and Discussion
Homology assignment and genome editing
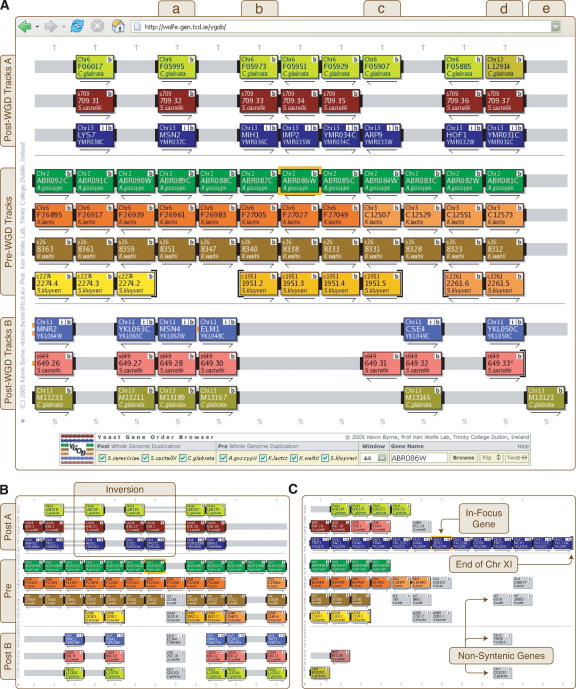
The two key structures behind YGOB are called pillars and tracks. YGOB's visual display (Fig. 2) is a matrix where each column shows a set of homologous genes stored in a pillar, and the horizontal elements (tracks) represent segments of chromosome. There is one track for each pre-WGD species and two tracks (track A and track B) for each post-WGD species. Pillars are the core data structures used to store homology assignments across the species. A pillar has a slot for each gene that can be present, i.e., two slots per post-WGD species and one per pre-WGD species. Each slot either is vacant or contains a gene from that genome homologous to the other genes in the pillar. The pillar data structure takes no account of the syntenic context, which is assigned dynamically by the browser engine's algorithms. Thus, in a pillar, the two slots for genes from a post-WGD species are not preassigned to particular tracks.
Figure 2.
Yeast Gene Order Browser (YGOB) screenshots with a window size of six. Each box represents a gene; each color, a chromosome. The gene in focus is highlighted by an orange border. Connectors join nearby genes: a solid bar for adjacent genes, two bars for loci less than five genes apart, and one bar for loci <20 genes apart. The connectors are extended in gray over intervening space. The end of a chromosome or contig is denoted by a brace. Arrows denote relative transcriptional orientation. The “b” buttons open a window with BLASTP results against YGOB's database, “+” buttons output YGOB data in a tabulated format, “S” buttons click through to a pillar's protein sequences, and “T” buttons draw approximate phylogenetic trees on the fly. Tracks are labeled at left. (A) YGOB focused on the A. gossypii gene ABR086W. The control console at the bottom of the interface allows users to select the window size and the gene to focus on. (B) An inversion in track A of the post-WGD species, marked by orange connectors. (C) The region beside the telomere on the left arm of S. cerevisiae chromosome XI.
The visual browser provides interactive ways to refocus the display on a different gene; to see functional annotation, phylogenetic trees, sequences, and BLASTP results for a gene; and to alter the set of species shown or the number of gene columns (Fig. 2). The browser was also designed with a manual editing interface that allows curators (not all users) to move genes into or out of pillars.
Our assignment of genes into pillars has been built gradually from several sources: the original authors' annotations of genes in other species as orthologs of particular S. cerevisiae genes, automated assignments based on BLASTP searches, and several rounds of manual editing of the entire set of pillars by our laboratory (see Supplemental Methods). The pillars are therefore subjective entities, and what YGOB presents is a curated view of the synteny relations among genomes. We plan to continue to curate YGOB's database and will release updates (with version numbering and access to previous versions) periodically.
Completeness and coverage of the genomes
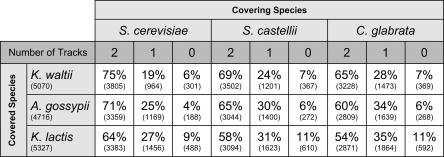
The extent to which the genomes of post-WGD species map onto pre-WGD genomes in a double conserved synteny relationship (Kellis et al. 2004) is summarized in Figure 3, which was calculated by using the engine of YGOB. For example, 75% of the genes in K. waltii are aligned to two syntenic tracks in S. cerevisiae—with the synteny being scored as robust, as described below. This result is identical to that reported by Kellis et al. (2004). A further 19% of K. waltii genes are aligned opposite one track in S. cerevisiae, while 6% can be assigned to none; this class includes K. waltii-specific genes. The S. castellii genome, which is annotated in relatively short contigs, nevertheless provides 69% double syntenic coverage of K. waltii. C. glabrata achieves only 65% double synteny, which seems to be due to a combination of reductive evolution (retention of only one copy of a gene at many loci that remain duplicated in S. cerevisiae) (Dujon et al. 2004) and an increased number of chromosomal rearrangement breakpoints. Similar patterns of coverage are seen when other pre-WGD species are used as the reference instead of K. waltii (Fig. 3). The consistently high level of robustly scored double conserved synteny in all pairwise comparisons between pre-WGD and post-WGD genomes reinforces the conclusion that a polyploidy event occurred in a common ancestor of the post-WGD species.
Figure 3.
Extent of coverage of pre-WGD genomes by two, one, or zero tracks from post-WGD genomes. Numbers in parentheses indicate the numbers of genes in each genome, and numbers covered by tracks from post-WGD species.
Gene content is highly conserved, with 89%-96% of the genes in each genome having a homolog in at least one of the other species (Supplemental Table 1). About 6%-8% of genes remain in singleton pillars in each post-WGD species. The majority of these are located in subtelomeric regions and are often members of multigene families (so we can be fairly sure that they are real genes), but they lack well-defined orthologs in other species. For example, in S. cerevisiae there are 352 genes in singleton pillars, and 213 of them are within 20 genes of the end of a chromosome.
Ohnologs
We have suggested (Wolfe 2000) that paralogs arising from a WGD should be called ohnologs in honor of the late Susumu Ohno (1970). Ohnologs are an important legacy of WGD, representing the genes that did not return to single copy in the genome and that form a pool of genetic material from which new functions (neofunctionalization) or specialization of daughter genes (subfunctionalization) can evolve. The ohnologs that have undergone functional divergence are particularly interesting because they can indicate the adaptation of a species to a certain environment or ecological niche. They are also, given their functional and hence likely sequence divergence, the harder ohnologs to identify, making a synteny tool such as YGOB ideal for finding them.
The number of identified pairs of ohnologs in the S. cerevisiae genome has increased continually since our initial identification of 376 pairs (Wolfe and Shields 1997), as the methods and data available for detecting them have improved. The YGOB data set has 551 likely ohnolog pairs in S. cerevisiae, which means that 19.6% of the genes in S. cerevisiae (1102 of 5516) are members of an ohnolog pair, and we estimate that 11.1% of genes in the immediate pre-WGD genome were retained in duplicate (calculated as 551/[5516 - 551]). The number of ohnologs retained in duplicate in S. castellii is a little higher at 599 (21.4%; note also that the S. castellii genome is not completely sequenced), and in C. glabrata it is significantly lower (404 pairs, 15.6%; P < 0.001 by Fisher's exact test vs. S. cerevisiae). Only 250 ohnolog pairs are present in all three genomes.
Analyses of the pre-WGD species K. waltii (Kellis et al. 2004) and A. gossypii (Dietrich et al. 2004) produced extensive lists of ohnologs in S. cerevisiae. We compared the set of ohnologs identified by YGOB to these two lists and to the list of 450 putative ohnologs previously identified by our laboratory using Génolevures-1 data (Souciet et al. 2000; Wong et al. 2002). The YGOB ohnolog data set was initially produced without consulting any of these previous sets of results, yet 99% of the putative ohnologs that were suggested by any of these studies are present in the YGOB set. A reconciliation of the ohnolog lists in these four studies is given in Supplemental Table 2 along with complete ohnolog lists for S. castellii and C. glabrata. Our initial S. cerevisiae YGOB set overlooked three ohnolog pairs that have particularly long gaps on one of the sister chromosomes, but we subsequently edited some pillars to include these three pairs. The YGOB version 1.0 set also includes 22 new ohnolog pairs that were not detected in previous studies (Table 1) but that have syntenic support.
Table 1.
Previously unidentified ohnolog pairs in S. cerevisiae
| Gene 1 | Gene 2 | Syntenic support | BLASTP E-value |
|---|---|---|---|
| SPO21 (YOL091W) | YSW1 (YBR148W) | Pre-WGD | >1a |
| YJR039W | YEL025C | Pre-WGD | >1a |
| ORC4 (YPR162C) | RIF2 (YLR453C) | Pre-WGD | >1a |
| YNL195C | HBT1 (YDL223C) | Pre-WGD | >1a |
| REG1 (YDR028C) | REG2 (YBR050C) | Pre-WGD | 6.2 |
| STB1 (YNL309W) | YOL131W | Pre-WGD | 0.67 |
| SIP18 (YMR175W) | GRE1 (YPL223C) | Pre-WGD | 0.05 |
| SPO19 (YPL130W) | YOR214C | Post-WGD | 0.027 |
| YFR017C | YOL024W | Pre-WGD | 0.008 |
| YGR035C | YLR346C | Post-WGD | 0.002 |
| ECM13 (YBL043W) | YJR115W | Pre-WGD | 2.00E-06 |
| PEX25 (YPL112C) | PEX27 (YOR193W) | Pre-WGD | 1.00E-08 |
| YAP5 (YIR018W) | YAP7 (YOL028C) | Post-WGD | 6.00E-14 |
| ASI1 (YMR119W) | ASI3 (YNL008C) | Inferredb | 4.00E-24 |
| YJL181W | YJR030C | Inferredb | 2.00E-31 |
| YEL007W | YHR177W | Inferredb | 4.00E-38 |
| HYR1 (YIR037W) | GPX1 (YKL026C) | Post-WGD | 6.00E-55 |
| SNO3 (YFL060C) | SNO2 (YNL334C) | Pre-WGD | 1.00E-117 |
| YFR022W | ROD1 (YOR018W) | Inferredb | 1.00E-118 |
| SAM4 (YPL273W) | MHT1 (YLL062C) | Post-WGD | 1.00E-124 |
| SNZ3 (YFL059W) | SNZ2 (YNL333W) | Pre-WGD | 1.00E-159 |
| MMP1 (YLL061W) | SAM3 (YPL274W) | Post-WGD | 0 |
Sequences do not hit each other in a BLASTP search, but there is indirect BLASTP support for these ohnologs via a mutual homolog
There is no direct syntenic support to suggest that these S. cerevisiae gene pairs are ohnologs, but a combination of some or all of the following convinces us they are ohnologs: synteny in the other post-WGD and pre-WGD genomes at that locus, two copies of the gene in other post-WGD genomes, correct clading in phylogenetic trees, or similar protein lengths
YGOB improves on the previous work because it can identify ohnologs that have extremely weak or only indirect (via a mutual homolog) BLASTP scores, on the basis of synteny established using any of the pre-WGD species (Table 1). Some of the newly identified pairs have high rates of sequence evolution: We calculated the extent of nonsynonymous sequence divergence (KA) between each of the 551 S. cerevisiae ohnolog pairs by using the yn00 method of Yang and Nielsen (2000), and six of the 10 fastest pairs are ones newly identified in this study (Supplemental Table 2). One of these pairs, SPO21/YSW1, is discussed in Wolfe (2004). Another newly identified and rapidly evolving ohnolog pair is ORC4-RIF2. Orc4 is a subunit of the origin recognition complex (ORC), central to the initiation of DNA replication. Rif2 is a protein that interacts with Rap1, which initiates silencing of transcription at telomeres by interacting with Sir3 (Wotton and Shore 1997). This is the second example of an ohnolog pair where one member is one of the six subunits of ORC and the other is involved in transcriptional silencing, the other pair being the more slowly-evolving Orc1 and Sir3 (Wolfe and Shields 1997; Kellis et al. 2004). At the other end of the scale, the 50 slowest-evolving ohnolog pairs are almost all ribosomal protein genes, many of which are being homogenized within species by gene conversion (Gao and Innan 2004; Kellis et al. 2004).
Syntenic configurations and scoring
In each pillar in a post-WGD species, two, one, or zero copies of the gene have been retained since genome duplication. This process of gene loss during evolution can proceed differently in different post-WGD species, a situation referred to as differential gene loss (Lundin 1993; Seoighe and Wolfe 1999; Fischer et al. 2001; Paterson et al. 2004). Figure 2A shows examples of several patterns of gene loss or retention. Column a shows a simple 2:2:2 pattern in the three post-WGD species, meaning that the ohnologs that arose at WGD have survived in two copies in each species. Column b shows a 1:1:1 pattern, where the same syntenic copy of the gene has survived in all species but the other one was lost in all species (or in their shared ancestor). This is the most common pattern seen throughout the genome. Column c shows a case of differential gene loss: Only one gene has been retained in each of the three post-WGD species, but the synteny information shows that the copy retained in S. castellii is a paralog of the one retained in S. cerevisiae and C. glabrata. Column d shows an example of a locus that is single copy in C. glabrata but retained in duplicate in S. cerevisiae and S. castellii. Column e shows a singleton C. glabrata gene without a homolog in its pillar, although there is no information for S. castellii track B in this pillar.
To look at differential gene loss in detail, we ran the YGOB engine along the genomes (i.e., focusing sequentially on every gene) of three pre-WGD species (K. waltii, A. gossypii, and K. lactis), assembling a 20-gene window of genome space around each gene, and scoring the syntenic status of the genes in the three post-WGD species (S. cerevisiae, S. castellii, and C. glabrata) in the same pillar as the gene under focus. Each pillar slot for a post-WGD species was scored by using one of four symbols: 1 means that a gene is present in an unambiguously syntenic context; 0 means that a gene is absent from a clearly syntenic region of the aligned post-WGD genome;! means that a gene is present but has uncertain or no synteny; and? represents an absence without synteny. Thus we used each of three pre-WGD species as an in-focus “scaffold” on which score the synteny of a locus in a post-WGD species, which gave us three sets of scores for the post-WGD genes in each pillar. For most pillars the scores are the same no matter which pre-WGD species is used as a scaffold, but in some places in the genome, it is possible to detect syntenic context by scaffolding on one pre-WGD species, but not another. Accurately assigning and scoring synteny as discussed here was a major challenge in the design of YGOB, and the algorithms used are detailed in Supplemental methods.
For each pairwise comparison of two post-WGD species, and considering only pillars with unambiguously syntenic 1 or 0 scores, on both tracks, in both species, we were able to describe the nature of each pillar (2:2, 2:1, 1:2, 1:1 orthologs or 1:1 paralogs). We did this by using each pre-WGD species as a scoring scaffold and then created a merged (union) data set of all pillars whose scores did not disagree with each other in any of the three choices of scaffold (only 92 pillars had disagreements). This yields reliable information about gene loss or retention patterns for ∼3000 loci in each comparison of a pair of post-WGD species (Table 2).
Table 2.
Distribution of gene loss classes in pairwise comparisons among the post-WGD species
|
S. cerevisiae: C. glabrata
|
S. cerevisiae: S. castellii
|
C. glabrata: S. castellii
|
||||
|---|---|---|---|---|---|---|
| Locus classa | Loci | Percent | Loci | Percent | Loci | Percent |
| 2:2 | 271 | 8.5% | 357 | 10.6% | 239 | 8.5% |
| 2:1 | 168 | 7.5% | 98 | 7.3% | 77 | 10.0% |
| 1:2 | 70 | 146 | 205 | |||
| 1:1 ortholog | 2552 | 80.0% | 2525 | 75.3% | 2102 | 74.3% |
| 1:1 paralog | 127 | 4.0% | 227 | 6.8% | 204 | 7.2% |
| Total | 3188 | 100.0% | 3353 | 100.0% | 2827 | 100.0% |
In the 2:2 class, both species have kept both copies of the gene at the locus. In the 2:1 class, the first species has kept two copies and the second has lost one, vice versa in the 1:2 class. In the 1:1 ortholog class, both species have lost the same syntenic copy of the gene. In the 1:1 paralog class, one species has lost the copy from track A and the other has lost the paralog from track B
In line with the expectation that a polyploidy is followed by a rapid return of most genes to single copy (Nadeau and Sankoff 1997; Kashkush et al. 2002), the vast majority (74%-80%) of traceable loci are 1:1 orthologs, with single orthologous copies of the gene being retained in both species. The loci in all the other categories (20%-26%) were present in two copies at the time of speciation, with much fewer (8%-11%) remaining in two copies now. Two previous studies (Fischer et al. 2001; Kellis et al. 2003) reported a handful of cases of differing fates of ancestrally duplicated genes in two very closely related yeasts, but YGOB permits genome-scale analysis of the phenomenon at an evolutionary depth (and proximity to the WGD) sufficient to capture a significant amount of differential gene loss. Of particular interest is this approach's ability to identify genes that were two copy at speciation but that now have been differentially inactivated in different species. These 1:1 paralogs are loci where the gene was duplicated at the WGD and was still retained in two copies at the time of speciation but has since returned to single copy in both species, with each species losing a different (paralogous) copy of the gene. Between any pair of post-WGD species, 4%-7% of the scorable loci are reciprocal gene losses of this type, confounding the widespread assumption that single-copy homologs between two genomes are always orthologous and revealing an important feature of polyploid genomes. A more detailed analysis of the genes in the categories in Table 2 will be presented elsewhere.
Conclusions
YGOB provides browsing and analytical access to a robust set of ortholog-ohnolog sets (pillars) from the seven yeast genome sequences considered here. These pillars have been extensively curated and manually inspected. The pillar data structure used in YGOB has some limitations, such as its inability to represent homology relationships other than orthology and ohnology (i.e., paralogy resulting from WGD), but for most regions of the genome, YGOB presents accurately assessed and clearly displayed synteny information, via an interface that is intuitive to use. For users interested in genome evolution, the YGOB engine provides a way of systematically harvesting gene order information that takes account of the WGD in some yeast species. The system is scaleable and provides an evolutionary genomics platform into which future genome sequences can be incorporated and additional functionality can be added.
Methods
Details of the genomic data, software, and algorithms used in this study can be found in the Supplemental methods.
Supplementary Material
Acknowledgments
We thank Devin Scannell, Jonathan Gordon, and Simon Wong for discussion, testing, and data editing. This study was supported by Science Foundation Ireland.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.3672305. Article published online before print in September 2005.
Footnotes
[Supplemental material is available online at www.genome.org.]
References
- Cliften, P., Sudarsanam, P., Desikan, A., Fulton, L., Fulton, B., Majors, J., Waterston, R., Cohen, B.A., and Johnston, M. 2003. Finding functional features in Saccharomyces genomes by phylogenetic footprinting. Science 301: 71-76. [DOI] [PubMed] [Google Scholar]
- Dietrich, F.S., Voegeli, S., Brachat, S., Lerch, A., Gates, K., Steiner, S., Mohr, C., Pohlmann, R., Luedi, P., Choi, S., et al. 2004. The Ashbya gossypii genome as a tool for mapping the ancient Saccharomyces cerevisiae genome. Science 304: 304-307. [DOI] [PubMed] [Google Scholar]
- Dujon, B., Sherman, D., Fischer, G., Durrens, P., Casaregola, S., Lafontaine, I., De Montigny, J., Marck, C., Neuveglise, C., Talla, E., et al. 2004. Genome evolution in yeasts. Nature 430: 35-44. [DOI] [PubMed] [Google Scholar]
- Fischer, G., Neuveglise, C., Durrens, P., Gaillardin, C., and Dujon, B. 2001. Evolution of gene order in the genomes of two related yeast species. Genome Res. 11: 2009-2019. [DOI] [PubMed] [Google Scholar]
- Gao, L.Z. and Innan, H. 2004. Very low gene duplication rate in the yeast genome. Science 306: 1367-1370. [DOI] [PubMed] [Google Scholar]
- Goffeau, A., Barrell, B.G., Bussey, H., Davis, R.W., Dujon, B., Feldmann, H., Galibert, F., Hoheisel, J.D., Jacq, C., Johnston, M., et al. 1996. Life with 6000 genes. Science 274: 546, 563-567. [DOI] [PubMed] [Google Scholar]
- Gu, Z., David, L., Petrov, D., Jones, T., Davis, R.W., and Steinmetz, L.M. 2005. Elevated evolutionary rates in the laboratory strain of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. 102: 1092-1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hittinger, C.T., Rokas, A., and Carroll, S.B. 2004. Parallel inactivation of multiple GAL pathway genes and ecological diversification in yeasts. Proc. Natl. Acad. Sci. 101: 14144-14149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, T., Federspiel, N.A., Chibana, H., Dungan, J., Kalman, S., Magee, B.B., Newport, G., Thorstenson, Y.R., Agabian, N., Magee, P.T., et al. 2004. The diploid genome sequence of Candida albicans. Proc. Natl. Acad. Sci. 101: 7329-7334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashkush, K., Feldman, M., and Levy, A.A. 2002. Gene loss, silencing and activation in a newly synthesized wheat allotetraploid. Genetics 160: 1651-1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellis, M., Patterson, N., Endrizzi, M., Birren, B., and Lander, E.S. 2003. Sequencing and comparison of yeast species to identify genes and regulatory elements. Nature 423: 241-254. [DOI] [PubMed] [Google Scholar]
- Kellis, M., Birren, B.W., and Lander, E.S. 2004. Proof and evolutionary analysis of ancient genome duplication in the yeast Saccharomyces cerevisiae. Nature 428: 617-624. [DOI] [PubMed] [Google Scholar]
- Keogh, R.S., Seoighe, C., and Wolfe, K.H. 1998. Evolution of gene order and chromosome number in Saccharomyces, Kluyveromyces and related fungi. Yeast 14: 443-457. [DOI] [PubMed] [Google Scholar]
- Kurtzman, C.P. and Robnett, C.J. 2003. Phylogenetic relationships among yeasts of the “Saccharomyces complex” determined from multigene sequence analyses. FEMS Yeast Res. 3: 417-432. [DOI] [PubMed] [Google Scholar]
- Llorente, B., Malpertuy, A., Neuveglise, C., de Montigny, J., Aigle, M., Artiguenave, F., Blandin, G., Bolotin-Fukuhara, M., Bon, E., Brottier, P., et al. 2000. Genomic exploration of the hemiascomycetous yeasts, 18: Comparative analysis of chromosome maps and synteny with Saccharomyces cerevisiae. FEBS Lett. 487: 101-112. [DOI] [PubMed] [Google Scholar]
- Lundin, L.G. 1993. Evolution of the vertebrate genome as reflected in paralogous chromosomal regions in man and the house mouse. Genomics 16: 1-19. [DOI] [PubMed] [Google Scholar]
- Nadeau, J.H. and Sankoff, D. 1997. Comparable rates of gene loss and functional divergence after genome duplications early in vertebrate evolution. Genetics 147: 1259-1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno, S. 1970. Evolution by gene duplication. George Allen and Unwin, London.
- Paterson, A.H., Bowers, J.E., and Chapman, B.A. 2004. Ancient polyploidization predating divergence of the cereals, and its consequences for comparative genomics. Proc. Natl. Acad. Sci. 101: 9903-9908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seoighe, C. and Wolfe, K.H. 1999. Yeast genome evolution in the post-genome era. Curr. Opin. Microbiol. 2: 548-554. [DOI] [PubMed] [Google Scholar]
- Sherman, D., Durrens, P., Beyne, E., Nikolski, M., and Souciet, J.L. 2004. Genolevures: Comparative genomics and molecular evolution of hemiascomycetous yeasts. Nucleic Acids Res. 32: D315-318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souciet, J., Aigle, M., Artiguenave, F., Blandin, G., Bolotin-Fukuhara, M., Bon, E., Brottier, P., Casaregola, S., de Montigny, J., Dujon, B. et al. 2000. Genomic exploration of the hemiascomycetous yeasts, 1: A set of yeast species for molecular evolution studies. FEBS Lett. 487: 3-12. [DOI] [PubMed] [Google Scholar]
- Stein, L.D., Mungall, C., Shu, S., Caudy, M., Mangone, M., Day, A., Nickerson, E., Stajich, J.E., Harris, T.W., Arva, A., et al. 2002. The generic genome browser: A building block for a model organism system database. Genome Res. 12: 1599-1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe, K. 2000. Robustness: It's not where you think it is. Nat. Genet. 25: 3-4. [DOI] [PubMed] [Google Scholar]
- ———. 2004. Evolutionary genomics: Yeasts accelerate beyond BLAST. Curr. Biol. 14: R392-R394. [DOI] [PubMed] [Google Scholar]
- Wolfe, K.H. and Shields, D.C. 1997. Molecular evidence for an ancient duplication of the entire yeast genome. Nature 387: 708-713.9192896 [Google Scholar]
- Wong, S., Butler, G., and Wolfe, K.H. 2002. Gene order evolution and paleopolyploidy in hemiascomycete yeasts. Proc. Natl. Acad. Sci. 99: 9272-9277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wotton, D. and Shore, D. 1997. A novel Rap1p-interacting factor, Rif2p, cooperates with Rif1p to regulate telomere length in Saccharomyces cerevisiae. Genes & Dev. 11: 748-760. [DOI] [PubMed] [Google Scholar]
- Yang, Z. and Nielsen, R. 2000. Estimating synonymous and nonsynonymous substitution rates under realistic evolutionary models. Mol. Biol. Evol. 17: 32-43. [DOI] [PubMed] [Google Scholar]
WEB SITE REFERENCES
- http://wolfe.gen.tcd.ie/ygob; Yeast Gene Order Browser (YGOB)
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.