Abstract
A gram-positive Bacillus sp. that fluoresces yellow under long-wavelength UV light on several common culture media was isolated from soil samples. On the basis of carbon source utilization studies, fatty acid methyl ester analysis, and 16S ribosomal DNA analysis, this bacterium was most similar to Bacillus megaterium. Chemical extraction yielded a yellow-orange fluorescent pigment, which was characterized by X-ray crystallography, mass spectrometry, and nuclear magnetic resonance spectroscopy. The fluorescent compound, chlorxanthomycin, is a pentacyclic, chlorinated molecule with the molecular formula C22H15O6Cl and a molecular weight of 409.7865. Chlorxanthomycin appears to be located in the cytoplasm, does not diffuse out of the cells into the culture medium, and has selective antibiotic activity.
Phenotypic characteristics of microorganisms, such as pigment production on selective media, are important taxonomic tools in the differentiation and classification of bacteria. For example, the pseudomonads are divided into fluorescent and nonfluorescent groups on the basis of diffusible fluorescent pigments or siderophores that are produced during growth on low-iron medium (17) and are visible under long-wavelength UV light (28). Nonfluorescent pigments have also been used in identifying xanthomonads, flavobacters, chromobacters, and acetobacters (16).
While fluorescent pseudomonads are prevalent in soil environments, there are only fragmentary reports of the widely distributed members of the genus Bacillus that produce yellow-green diffusible fluorescent pigments (19, 22, 34, 35). The early reports of Flügge (12, 13), who isolated two fluorescent bacteria from water and soil environments and named them Bacillus fluorescens liquefaciens and Bacillus fluorescens putidus, were erroneous. These bacteria and many other strains of the fluorescent group of Bacillus were reclassified and renamed as fluorescent pseudomonads (1). There is no evidence in the literature of a species belonging to the genus Bacillus that fluoresces yellow under long-wavelength UV light.
Here we describe isolation from soil samples and identification of a member of the genus Bacillus that fluoresces yellow under long-wavelength UV light on several common culture media with and without iron. We purified and characterized the fluorescent compound and showed that it is a new, pentacyclic, chlorinated pyrene with a molecular weight of 409.7865.
MATERIALS AND METHODS
Isolation and growth of bacteria.
Samples collected from the upper 20 cm of soil from agricultural fields over a period of 1 year at the Westside Field Station of the University of California and from agricultural fields from Riverside to Hopland, Calif., were air dried and stored at the ambient temperature. For estimation of the number of yellow fluorescent bacteria, 1 g of soil was suspended in 10 mM phosphate buffer (pH 7.0), serially diluted, and plated onto King's B medium agar (KBM) (17). In addition, the following solid media were used: King's A medium agar (KAM) (17), tryptic soy agar (TSA) (Difco, Detroit, Mich.), yeast dextrose calcium agar with peptone (39), Czapek agar (Difco), nutrient agar (NA) (Difco), Biolog universal glucose medium agar (BUGM) (Biolog Inc., Hayward, Calif.), potato dextrose agar (29), and Luria agar (LA). To determine if iron starvation induced production of the fluorescent compound, the bacteria were grown on iron-rich (BUGM) and iron-limiting (KBM) media (28). To determine if the fluorescent compound was produced during growth in liquid medium, the bacteria were grown on liquid formulations of KBM, KAM, LA, and BUGM (without agar). Fluorescence was detected under UV light (λ = 365 nm) after the plates were incubated for 24 to 30 h at 35°C.
Confocal laser scanning microscopy.
Images of 32- to 120-h-old cells were obtained with a Zeiss 510 confocal laser scanning microscope. Samples were excited with an argon ion laser by using the 488-nm line. A 505-550 band-pass filter was used for scanning emissions. A Plan-Apo 100X with a 1.4 NA objective was used for all scans. The optical slice size was set at 0.8 nm.
Carbon source utilization analysis.
Bacteria were grown on BUGM for 12 h at 31°C. Biolog GP (gram positive) plates were inoculated with bacteria suspended in a sterile 0.85% sodium chloride solution, and the concentration was adjusted to approximately 109 cells/ml. Carbon source utilization patterns were analyzed after 4 and 24 h by using Microlog software (version 4.2; Biolog, Inc.) for gram-positive bacteria (2).
Other bacteria.
Streptomyces resistomycificus DSMZ 40133was obtained from the Deutsche Sammlung fur Mikroorganismen und Zellkulturen.
Fatty acid analysis.
The fatty acid profile was examined by gas chromatographic analysis of the fatty acid methyl esters (GC-FAME) from 0.1 g (wet weight) of cells grown on Trypticase agar plates (10). Two duplicate analyses were performed by Microbe Inotech Laboratories (St. Louis, Mo.). The results were compared with the aerobe GC-FAME databases (TSBA, rev. 3.9, and CLIN, rev. 3.9).
Molecular phylogenetic analysis of SSU rRNA.
PCR amplification of the small-subunit (SSU) rRNA gene was performed by using chromosomal DNA and bacterium-specific SSU rRNA oligonucleotide primers 5′-AGAGTTTGATCCTGGCTCAG-3′ (forward) and 5′-GGTTACCTTGTTACGACTT-3′ (reverse). The 50-μl PCR mixtures contained each deoxynucleoside triphosphate at a concentration of 0.4 mM, 200 ng of the forward PCR primer, 200 ng of the reverse PCR primer, 0.1% NP-40, 1 U of Taq polymerase, and 1× Taq buffer (30 mM Tricine, 50 mM KCl, 1.5 mM MgCl2). The resulting SSU ribosomal DNA PCR product of the novel Bacillus strain was sequenced and aligned with the SSU ribosomal DNA of known Bacillus species. A total of 1,371 homologous nucleotide positions were used for phylogenetic analysis. Phylogenetic relationships were inferred by using the least-squares evolutionary distance of the PAUP 3.0 phylogenetic analysis software (36).
Extraction of pigments.
Bacteria were grown on solid medium (KBM or BUGM) and incubated for 24 h at 35°C before cells were harvested by scraping the bacterial mats from petri plates. In a typical experiment, 100 petri plates yielded 50 g of bacteria, which were placed in an acid-washed glass flask containing 150 ml of 80% cold (4°C) acetone. The mixture was agitated for 1 min with a wrist rotary shaker at full speed and then allowed to settle for 5 min. The pulp was vacuum filtered with Whatman no. 1 filter paper, and the supernatant was saved. The bacterial biomass was washed twice with 100 ml of 80% acetone as described above. The acetone extracts were combined and concentrated to approximately 150 ml under a vacuum in a rotary flask. At this volume, the bright red color of the mixture, which fluoresced yellow, turned brown, and a yellow precipitate formed. The slurry was centrifuged at 5,000 × g for 10 min at 4°C. The nonfluorescent supernatant was discarded, and the pellet was saved and resuspended in 100 ml of 100% acetone, giving it a bright orange color. This mixture was concentrated again under a vacuum until it turned brown and a yellow precipitate formed. This procedure was repeated four times by using 100, 50, 25, and 10 ml of 100% acetone. The final yellow pellet fluoresced orange.
Thin-layer and liquid chromatography.
The purity of the fluorescent compound was monitored at every step of the extraction procedure by thin-layer chromatography by using silica gel sheets without a fluorescent indicator (Eastman Chromogram 13179) or by using 0.5-mm silica gel sheets (Merck 60F-254) and a chloroform-ethyl acetate (3:1) solvent. Compounds were visualized with 5% phosphomolybdic acid reagent in 95% ethanol, iodine vapor, or UV light. The silica gel strips were dried by using a stream of warm air. Fluorescent compounds were detected with a long-wavelength (λ = 365 nm) UV lamp.
High-performance liquid chromatography-mass spectrometry (LC-MS) (model 1100; Agilent, Palo Alto, Calif.) was performed by using a C8 Zorbax column (15 by 4.6 mm; Eclipse) and a methanol-water gradient ranging from 70:30 to 5:95.
Crystal growth and X-ray crystallography.
The purity of the acetone-extracted, vacuum-dried compound was determined by thin-layer chromatography and preparative LC-MS analysis before crystals were grown. Four milligrams of the isolated compound was dissolved in 1 ml of pyridine at 70°C in a small borosilicate glass tube (16 by 100 mm). This tube was secured in a slightly larger glass tube and placed in a glass jar containing 15 ml of pentane. The jar was closed with a screw cap and securely placed in an insulated box containing hot water (90°C). The box was covered and placed at 4°C for 7 days. Slow cooling and the diffusion of pentane into the pyridine induced the formation of rhombohedral crystals. The crystals at the bottom of the glass tube were dried under a constant flow of N2 gas at 25°C for 2 h and then at 40°C for 48 h. Single crystals were removed and used for spectral studies, thin-layer chromatography, LC-MS analysis, and X-ray crystallography. The presence of pyridine is important since the crystals contained one pyridine per fluorescent molecule.
A single crystal of the pigment having approximate dimensions of 0.18 by 0.18 by 0.18 mm was mounted on a glass fiber by using Paratone N hydrocarbon oil. The data were collected at a temperature of −113 ± 1°C. All measurements were made with a SMART charge-coupled device area detector with graphite monochromated Mo-Kα radiation. Frames corresponding to an arbitrary hemisphere of data were collected by using ω scans of 0.3° counted for a total of 10.0 s per frame. Data were integrated by using the SAINT software (version 5.04; Siemens Industrial Automation, Madison, Wis.) to a maximum 2θ value of 49.4°. The data were corrected for Lorentz and polarization effects. Data were analyzed for agreement and possible adsorption by using XPREP software (33). An empirical adsorption correction based on comparison of redundant and equivalent reflections was applied by using SADABS (Tmax = 0.96, Tmin = 0.54) (32). The structure was solved by direct methods and was expanded by using Fourier techniques. The nonhydrogen atoms were refined anisotropically. Hydrogen atoms were included but not refined. Neutral atom scattering factors were obtained from reference 5. Anomalous dispersion effects were included in Fcalc; the values used for Δf′, Δf″, and the mass attenuation coefficients are those of Creagh and McAuley (4). All calculations were performed by using the teXsan crystallographic software package (Molecular Structure Corporation).
Melting points.
Melting points were determined in open-end capillary tubes by using a Thomas-Hoover melting point apparatus and were not corrected.
IR spectroscopy.
The infrared (IR) spectrum was obtained with a Nicolet Magna-IR 550 spectrometer.
NMR spectroscopy.
1H nuclear magnetic resonance (NMR) spectra of the crystallized compound dissolved in pyridine were recorded at 400 MHz with a Bruker AMX spectrometer (Bruker Instruments Inc., Billerica, Mass.).
Mass spectrometry and elemental analysis.
Mass spectra were recorded with a Varian 70-4F or 70 VSE mass spectrometer, and elemental analysis was performed with a Perkin-Elmer series II 2400 analyzer at the Mass Spectrometry Center in the College of Chemistry at the University of California at Berkeley.
UV-visible light and fluorescence spectroscopy.
Single crystals were dissolved in methanol or ethanol in all experiments. UV and visible light spectra were obtained with a Beckman DU-640 spectrophotometer. The fluorescence emission spectrum of the compound in ethanol was measured at wavelengths between 450 and 700 nm with fixed excitation at 519 nm, and the excitation spectrum was measured at wavelengths between 260 and 600 nm with the emission wavelength fixed at 560 nm; both spectra were obtained with a Perkin-Elmer LS 50B luminescence spectrometer. To assure a monochromatic light source, narrow (1.0 mm) excitation slits were used. For spectroscopy of whole cells, cells were centrifuged, resuspended in 100% ethanol, and placed in a quartz cuvette, and the spectrum was determined.
Fluorescence quantum yield measurements.
For fluorescence quantum yield calculations, emission spectra of the fluorescent compound and Rhodamin 6G (φ = 0.90) as a standard in ethanol were acquired at wavelengths from 450 to 700 nm. The absorbance of Rhodamin 6G and the absorbance of the fluorescent compound were matched to within 1% at 529 and 519 nm, respectively. Spectra were measured with an emission bandwidth of 8 nm and a scan speed of 8 nm/s by using a Perkin-Elmer LS 50B luminescence spectrometer. The fluorescence quantum yield was calculated by comparing the integrated intensities of the standard and fluorescent compound emission bands (21).
Antibiotic assays.
One crystal of chlorxanthomycin (40 μg) was dissolved in 1 ml of 100% ethanol or pyridine (final concentration, 97.6 μM) and used as a stock solution. The stock solution was diluted with pyridine to obtain final concentrations of 0.976, 4.88, and 9.76 μM. Petri plates were prepared by using 35 ml of KBM for bacteria and potato dextrose agar for fungi. To test the antimicrobial activity of chlorxanthomycin, five individual spots (50 μl each) of chlorxanthomycin at the concentrations listed above were placed on the petri plates, and the solvent was allowed to evaporate (24). Similar test plates were prepared for control experiments without chlorxanthomycin (solvent only). To test antifungal activity, a loopful of a test organism was placed in the middle of each petri plate, equidistant from each of the five spots; growth was monitored every day for 7 days. To test antibacterial activity, a small amount of a bacterial suspension (1 × 107 cells) in 10 mM phosphate buffer (pH 7.0) was sprayed over the dried spots of chlorxanthomycin; growth was monitored every day for 4 days. The zone of inhibition was measured from the periphery of the spot. The organisms tested were Bacillus subtilis, Bacillus amyloliquefaciens, Erwinia amylovora, Escherichia coli, Fusarium sp., Mucor sp., Phytophthora cinnamoni, Pythium ultimum, Pythium aphanidermatum, Pythium sp., Pseudomonas syringae, Pseudomonas fluorescens, Ralstonia solanacearum, Rhizopus sp., Verticillium sp., and Xanthomonas sp.
Nucleotide sequence accession number.
The sequence of the fluorescent Bacillus sp. described in this paper has been deposited in the GenBank database under accession number AY046591.
RESULTS
Organism isolation and characterization.
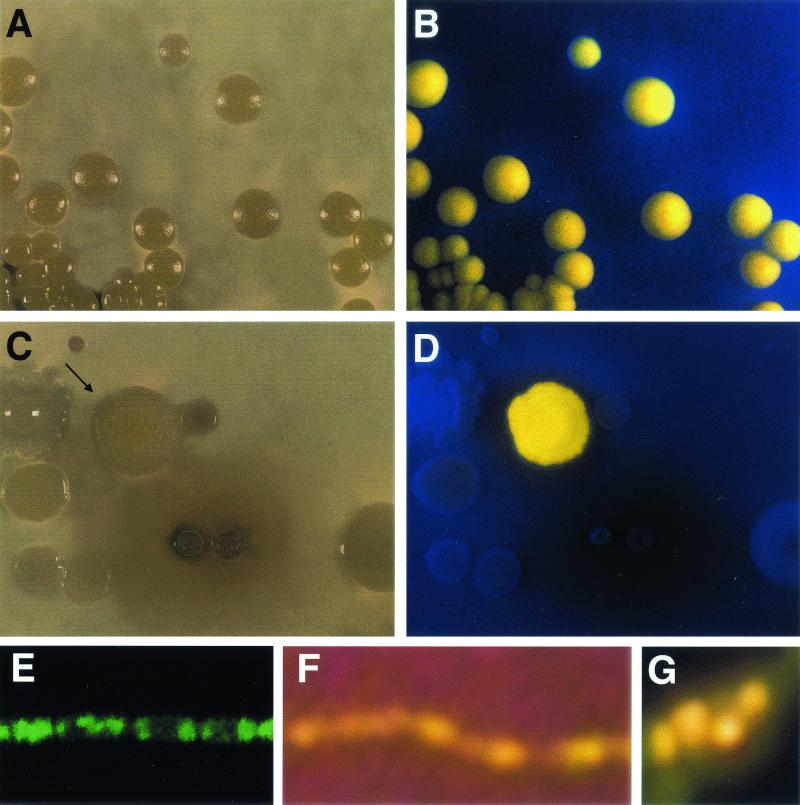
An unusual, fluorescent strain of the genus Bacillus was isolated from a soil sample from the Westside Field Station of the University of California. Subsequently, yellow-pigmented members of the genus Bacillus were found in 4 of 20 soil samples from several other locations in California. The population density was determined by soil dilution and plate count methods and was found to range from 1.8 × 102 to 4.3 × 103 cells per g of soil. (Because bacteria were isolated from air-dried soil, these values may underestimate the total population size of the yellow fluorescent bacilli in natural soils.) The four pigmented isolates fluoresced yellow under long-wavelength UV light on several common culture media, including BUGM, KAM, KBM, LA, TSA, and yeast dextrose calcium agar with peptone (Fig. 1), and had identical spectral characteristics and carbon utilization patterns. Because of the apparent similarities of these isolates, all subsequent studies were performed with the isolate from the Westside Field Station.
FIG. 1.
(A and B) Pure culture of Bacillus sp. on TSA viewed under visible light (A) and long-wavelength UV light (B). (C and D) Mixed culture from soil sample containing Bacillus sp. on TSA viewed under visible light (C) and long-wavelength UV light (D). (E) Confocal microscopy at 0.8-nm thickness of Bacillus sp. Magnification, ×1,000. (F and G) Filament of Bacillus sp. grown on TSA, resuspended in 10 mM phosphate buffer, and visualized by fluorescence microscopy at magnifications of ×400 (F) and ×1,000 (G).
Unlike the fluorescent compounds produced by pseudomonads, the yellow fluorescent compound produced by the Bacillus sp. did not diffuse into the agar, even from old cells. Fluorescent microscopy indicated that the yellow fluorescent compound was contained in the cells (Fig. 1F and G). Confocal microscopy showed clearly that the fluorescent compound was located in nodules in the cytoplasm and was not associated with the cell wall (Fig. 1E).
Production of the fluorescent pigment appears to be regulated by the nutrient source in the medium and by the type of medium (solid or liquid). Cells fluoresced when they were grown on NA containing 20 g of peptone/liter but did not fluoresce when they were grown on NA containing 5 g of peptone/liter. Similarly, the cells fluoresced only when peptone or tryptone was added to Czapek medium. In addition, fluorescence was observed only when glucose or glycerol was added to the medium, suggesting that a rich medium is required for production of the yellow fluorescent pigment. However, iron had no effect on production of the fluorescent pigment, as the pigment was produced on both iron-limiting and iron-rich media. Finally, cells fluoresced only when they were grown on solid medium.
On the basis of carbon source utilization studies performed with Biolog GP plates, the yellow fluorescent bacterium was found to be most closely related to Bacillus megaterium, with a similarity index of 0.52. The next closest relative was Bacillus licheniformis, with a similarity index of 0.42. The results of GC-FAME analysis were similar to the results of the carbon utilization tests; B. megaterium was the first choice, with a similarity index of 0.549, and Bacillus brevis was the second choice, with a similarity index of 0.427.
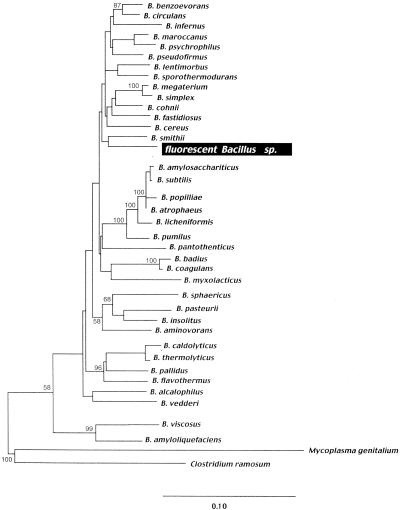
A molecular phylogeny analysis in which maximum likelihood (36) of the 16S rRNA gene was used suggested that the bacterial isolate is a member of the genus Bacillus in the low-G+C-content gram-positive bacteria and does not have a close affiliation with any of the previously identified Bacillus subclasses (Fig. 2) (31). Over 1,394 homologous nucleotide positions, the 16S rRNA gene is equally similar to diverse Bacillus species (94% similar to B. megaterium, B. licheniformis, B. flavis, and B. subtilis).
FIG. 2.
Molecular phylogenetic tree of the SSU rRNA gene of the isolated Bacillus sp. and other Bacillus species, constructed by using the maximum-likelihood tree inference method of PAUP∗ (36). The numbers at the nodes are the numbers of times that the topology was supported in 100 bootstrapped replicates; only values greater than 50 are shown. Scale bar = 10 nucleotide changes per 100 nucleotides.
Pigment characterization.
Extraction and purification of the fluorescent bacterial biomass resulted in a yellow water-insoluble powder that fluoresced orange under long-wavelength UV light. Both the final pellet from the extraction protocol and the crystallized compound from a pyridine-pentane mixture yielded one bright yellow fluorescent spot on a silica gel when it was subjected to thin-layer chromatography. Crystals fluoresced orange without solvent and bright yellow or yellow-orange when they were dissolved in the following solvents: acetone, ammonium hydroxide, dimethyl sulfoxide, formamide, hexyl alcohol, methanol, triethyl amyl alcohol, and pyridine. The fluorescence quantum yield in 100% ethanol was determined to be 0.71 to 0.73. The melting point was >331°C.
LC-MS analysis of the compound revealed only one peak at 8 min with a molecular weight of 409.78. Low-resolution mass spectrometry indicated a molecular weight of 409 [(M+H)+], and high-resolution mass spectrometry indicated a molecular weight of 409.7865 [(C22H15O6Cl+H)+] (26). Elemental analysis indicated that the composition was 62.82% C and 4.06% H; the calculated percentages for C and H in C22H15O6Cl · 0.5 H2O are 62.93 and 3.81%, respectively, indicating that it is highly unsaturated. We propose the name chlorxanthomycin for this compound and refer to it by this name below.
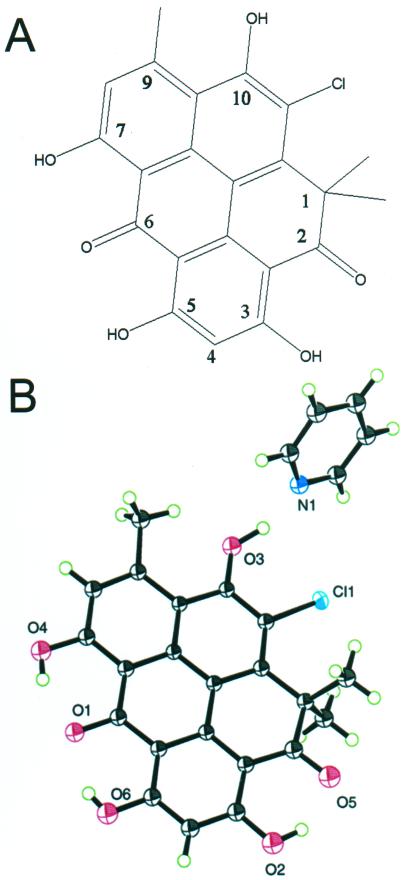
The structure of chlorxanthomycin was determined by X-ray crystallography (Fig. 3). The molecule is a benzopyrene compound consisting of five fused rings in a planar delocalized structure. There are four hydroxyl groups, two carbonyls, one methyl group, a gem dimethyl group, and a chlorine bonded around the edge of the fused ring system. The deviations of the core carbon atoms from the least-squares plane are consistent with a small saddle deformation. The groups bonded around the core followed the same pattern of deviation from planarity as the atoms to which they were bonded.
FIG. 3.
Structure of the fluorescent compound from Bacillus sp. (A) Two-dimensional structure of the compound. (B) Three-dimensional structure deduced from crystallographic data, with the orientation of the pyridine to the fluorescent compound shown. Oxygen atoms are red, chlorine atoms are blue, carbon atoms are black, and hydrogen atoms are white (arbitrary numbering system).
The 1H NMR (pyridine-d5) spectrum of chlorxanthomycin showed the following characteristics: δ 2.14 (s, 6H, 2CH3), 3.22 (s, 3H, CH3), 5.84 (br, s, 4H, OH), 6.71 (s, 1H, Ar-H), 7.21 (s, 1H, Ar-H); 1H NMR δ (CDCl3): δ 1.95 (s, 6H, 2CH3), 3.04 (s, 3H, CH3), 6.55 (br, s, 1H, OH), 7.19 (s, 1H, Ar-H), 7.40 (s, 1H, Ar-H). Interpretation of the 1H NMR spectrum indicated the presence of an isolated methyl group, a gem dimethyl group, and two isolated aromatic protons, each occurring as a singlet.
The IR spectrum of chlorxanthomycin showed absorbance at 3,430, 1,600, 1,575, and 1,495 cm−1. The frequency at 3,430 cm−1 is from the stretching vibration of the OH groups. Absorption at both 1,495 and 1,600 cm−1 clearly indicates that the molecule is aromatic. Although this compound has carbonyl groups, it does not have the IR frequency around 1,700 cm−1 that is typical for stretching of a carbonyl group. Thus, the carbonyl groups must be present in the form of enols or β-hydroxy-α,-β-unsaturated ketones (since the result of 1H NMR ruled out aldehyde, carboxylic acid, and ester), and the carbonyl and hydroxy groups are intramolecularly hydrogen bonded. The IR absorption at both 1,575 and 1,600 cm−1 and the unusually strong fluorescence suggest that chlorxanthomycin may have a large conjugated aromatic system, such as a pentacyclic phenalenone structure.
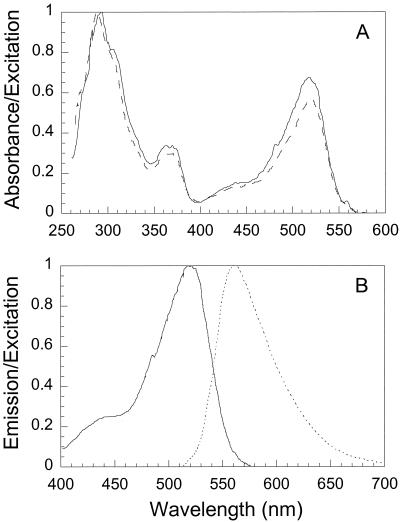
The UV-visible light absorption spectrum of a single crystal dissolved in ethanol had three major peaks, at 519, 368, and 290 nm (Fig. 4). The emission spectrum peaked at 560 nm when the preparation was excited at 290, 368, or 519 nm. The excitation spectrum of the compound followed that of the absorption spectrum when the emission wavelength was fixed at 560 nm. The excitation spectrum of whole cells suspended in 100% ethanol was very similar to that of purified chlorxanthomycin in ethanol, indicating that chlorxanthomycin was the dominant pigment in the cells.
FIG. 4.
Absorption, excitation, and emission spectra of the fluorescent compound dissolved in ethanol. (A) Normalized absorption (dashed line) and excitation (solid line) spectra. (B) Normalized emission (dashed line) and excitation (solid line) spectra. The 400- to 600-nm region of the excitation spectrum in panel A is shown in panel B.
Given the similarity of the structures of chlorxanthomycin and resistomycin, S. resistomycificus, which produces resistomycin, was cultured on media similar to those used to culture the fluorescent bacillus. After 8 days of growth on KBM, S. resistomycificus fluoresced red, but it did not fluoresce on BUGM. In contrast, the Bacillus sp. showed strong yellow fluorescence on BUGM after 24 h of growth, and unlike resistomycin, which diffused into the agar medium, chlorxanthomycin remained inside the cells. This finding agrees with the fluorescence microscopy results (Fig. 1E and F), which showed that fluorescence was found in distinct nodules in the cytoplasm and not in the cell wall.
Chlorxanthomycin at concentrations of 9.76 and 97.6 μM inhibited the growth of B. subtilis and B. amyloliquefaciens, with clear 6-mm zones of inhibition. At lower concentrations, no antibiotic activity was observed. Similarly, no antibiotic activity was observed when gram-negative bacteria were used. Control plates (pyridine without chlorxanthomycin) were overgrown after 1 day.
Chlorxanthomycin at a concentration of 96.7 μM had the most pronounced inhibitory effect on the growth of P. ultimum, P. aphanidermatum, and Pythium sp., with zones of inhibition of 5 mm. At lower concentrations (less than 97.6 μM) fungal growth was observed in the agar around the test compound after 4 days. P. cinnamoni, Mucor sp., and Rhizopus sp. were also inhibited by 96.7 μM chlorxanthomycin, but to a lesser extent (inhibition zones, 3 mm). After 5 days, Mucor sp. and Rhizopus sp. grew over the droplets; no growth was observed on the agar surface in the zones of inhibition. Plates without the test compound (but with the solvent pyridine) were overgrown in 5 days. Chlorxanthomycin had no effect on Fusarium sp. and Verticillium sp.
DISCUSSION
This report is the first report of a fluorescent, chlorinated, pentacyclic, natural compound produced by a member of the genus Bacillus. The bacterium was isolated from several agricultural soils in California with sandy loam textures and showed strong yellow fluorescence on iron-rich and iron-limiting media. Several attempts to isolate the bacterium from sandy nonagricultural soils were unsuccessful.
On the basis of our studies, we believe that the lack of description of this bacterium in the past may have been, in part, because the yellow fluorescence of the organism was masked on culture plates in the vicinity of the strong diffusible green fluorescent pigments of pseudomonads. This was particularly apparent with soils adjacent to the laimosphere, spermosphere, and rhizosphere, where fluorescent pseudomonads are abundant (25). Unlike the diffusible green fluorescent pigments, chlorxanthomycin does not appear to be a siderophore, as it has a different molecular structure and is not regulated by iron.
The yellow fluorescent compound showed strong fluorescence under long-wavelength UV light and had a quantum yield of 0.71 to 0.73 when the yield was compared to that of Rhodamin 6G in 100% ethanol. The other solvents used for quantum yield measurements (namely, acetone, methanol, and pyridine) did not alter the intensity of fluorescence. A compound structurally similar to chlorxanthomycin with flu-orescent properties, 2H-benzo[cd]pyrene-2,6(1H)-dione-3,5,7,10-tetrahydroxy-1,1,9-trimethyl-(8Cl, 9Cl), referred to as resistomycin, resistoflavin, itamycin, or heliomycin, has been isolated from S. resistomycificus (3, 6-9, 20, 38). Like chlorxanthomycin, resistomycin is pentacyclic and highly conjugated and has been reported to be fluorescent (20). However, unlike chlorxanthomycin, resistomycin is not chlorinated and the methyl group is attached at C-9. Resistomycin and its homologues have been reported to inhibit the growth of bacteria (8) and human breast cancer cells (23) and the activity of human immunodeficiency virus protease (30). In particular, resistomycin has been found to be effective against B. subtilis (14, 15), a particularly interesting report since we have found that a species of Bacillus produces a compound that is quite similar. We also found that chlorxanthomycin had antimicrobial effects against B. subtilis. In addition, chlorxanthomycin inhibited the growth of fungi, particularly members of the Zygomycetes and Oomycetes.
The X-ray crystal structure revealed that chlorxanthomycin not only has a flat conjugated ring system, except for gem dimethyl groups at C-1, but also has strong intramolecular hydrogen bonds between the hydroxyl and carbonyl groups. The polycyclic aromatic ring system belongs to the benzo[cd]pyrene structural class and is responsible for the strong fluorescence activity.
Halogenated metabolites are found widely in nature (18, 27, 37). A number of marine organisms produce brominated metabolites, whereas in terrestrial environments most halogenated metabolites are chlorinated (37). Chloramphenicol, the first chlorinated bacterial metabolite to be discovered, is produced by Streptomyces venezuelae (11). Indeed, most of the known bacterially derived halogenated metabolites are diffusible, aromatic compounds, have antibiotic activity, and are produced by actinomycetes. Chlorxanthomycin is also chlorinated and has selective antibiotic properties; however, it is not diffusible and fluoresces on several culture media with and without iron. Additional studies will be necessary to determine why a novel isolate of Bacillus produces chlorxanthomycin and the metabolic pathway responsible for synthesis of this compound.
Acknowledgments
This research was supported by the ERC Program of the National Science Foundation under award EEC-9731725. Equipment was purchased in part by an NSF instrument grant.
The crystal structure analysis was performed at CHEXRAY, the University of California at Berkeley College of Chemistry X-ray Crystallographic Facility. We thank Frederick Hollander and Allen Oliver for their work on the structure, Frank Andel III for help with quantum yield measurements, and Ken Sauer and Javiar Fernandez-Velasco for advice on spectroscopy.
Footnotes
This article is dedicated to the memory of Henry Rapoport.
REFERENCES
- 1.Bergey, D. H., F. C. Harrison, F. C. Breed, B. Hammer, and F. M. Huntoon. 1923. Bergey's manual of determinative bacteriology. Williams & Wilkins, Baltimore, Md.
- 2.Bochner, B. R. 1989. Sleuthing out bacterial identities. Nature 339:157-158. [DOI] [PubMed] [Google Scholar]
- 3.Chugasova, V. A., T. P. Preobrazhenskaya, and N. O. Blinov. 1975. Biologically active pigments of Actinomyces atrovirens. Antibiotiki (Moscow) 20:415-423. [PubMed] [Google Scholar]
- 4.Creagh, D. C., and W. J. McAuley. 1992. International tables for crystallography, vol. C. Kluwer Academic Publishers, Boston, Mass.
- 5.Cromer, D. T., and J. T. Waber. 1974. International tables for X-ray crystallography, vol. IV. The Kynoch Press, Birmingham, England.
- 6.de Mello, J. F., F. Delle Monache, G. B. Marini-Betollo, F. D. D. A. Lyra, M. M. Fernandes de Albuquerque, and O. Goncalves de Lima. 1971. Itamycin, a new antibiotic produced by Streptomyces erythrogriseus. II. Identification with resistomycin. Rev. Inst. Antibiot. Univ. Fed. Pernambuco Recife 11:3-6. [Google Scholar]
- 7.Eckardt, K., G. Bradler, H. Fritzsche, and D. Tresselt. 1970. Resistoflavine, a new antibiotic from an actinomycete. Z. Chem. 10:221. [Google Scholar]
- 8.Eckardt, K., G. Bradler, D. Tresselt, and H. Fritzsche. 1972. Resistoflavin, a new antibiotic of the resistomycin family. Adv. Antimicrob. Antineoplast. Chemother. 1:1025-1027. [Google Scholar]
- 9.Eckardt, K., H. Fritzsche, and D. Tresselt. 1970. Structure of the antibiotic, resistoflavine. Tetrahedron 26:5875-5883. [Google Scholar]
- 10.Eder, K. 1995. Gas chromatographic analysis of fatty acid methyl esters. J. Chromatogr. 671:113-131. [DOI] [PubMed] [Google Scholar]
- 11.Ehrlich, J., Q. R. Bartz, R. M. Smith, D. A. Joslyn, and P. R. Burkholder. 1947. Chloromycetin, a new antibiotic from a soil actinomycete. Science 106:417. [DOI] [PubMed] [Google Scholar]
- 12.Flügge, C. 1886. Die Mikroorganismen. Vogel, Leipzig, Germany.
- 13.Flügge, C. 1896. Die Mikroorganismen, 3rd ed. Vogel, Leipzig, Germany.
- 14.Haupt, I., and K. Eckardt. 1972. Influence of the antibiotics resistomycin and resistoflavin on growth and synthesis of nucleic acid and protein of Bacillus subtilis. Z. Allg. Mikrobiol. 12:573-579. [PubMed] [Google Scholar]
- 15.Haupt, I., U. Waehnert, C. Pitra, G. Loeber, G. Luck, and K. Eckardt. 1975. Effects of the antibiotic resistomycin on the synthesis of macromolecules. Z. Allg. Mikrobiol. 15:411-421. [DOI] [PubMed] [Google Scholar]
- 16.Hayes, P. R., T. A. McMeekin, and J. M. Shewan. 1979. The identification of Gram negative, yellow pigmented rods, p. 177-187. In F. A. Skinner and D. W. Lovelock (ed.), Identification methods for microbiologists, 2nd ed. Academic Press, San Diego, Calif.
- 17.King, E. O., M. K. Ward, and D. E. Raney. 1954. Two simple media for the demonstration of pyocyanin and fluorescin. J. Lab. Clin. Med. 44:301-307. [PubMed] [Google Scholar]
- 18.Kirk, K. L. 1991. Biochemistry of halogenated organic compounds. Plenum Press, New York, N.Y.
- 19.Kolev, D. A., T. N. Antonova, T. A. Nikoevska, and K. P. Petrova. 1991. Synthesis of pigments by the dissociative forms of Bacillus subtilis 76. Acta Microbiol. Bulg. 28:27-33. [Google Scholar]
- 20.Kozhevina, L. S., K. A. Vinogradova, and M. E. Struve. 1982. Morphological and cytochemical differentiation of colonies of actinomycetes—geliomycin producers—in the developmental process on solid media. Biol. Nauki (Moscow) 8:88-91. [PubMed] [Google Scholar]
- 21.Lakowicz, J. R. 1983. Principles fluorescence spectroscopy. Plenum Press, New York, N.Y.
- 22.Laubach, C. A., J. L. Rice, and W. W. Ford. 1916. Studies on aerobic spore-bearing non-pathogenic bacteria. Part II. J. Bacteriol. 1:493-533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee, S.-W., S.-E. Kim, Y.-H. Kim, H.-S. Kim, H.-J. Bang, H.-M. Kim, and J.-J. Lee. 1993. Antitumoral compound, MCS-202, an effector on proliferation and morphology of human breast tumor cell line, MCF-7. Sanop Misaengmul Hakhoechi 21:594-599. [Google Scholar]
- 24.Loo, Y. H., P. S. Skell, H. H. Thornberry, J. Erlich, J. M. McGuire, G. M. Savage, and J. C. Sylvester. 1945. Assay of streptomycin by the paper-disk plate method. J. Bacteriol. 50:701-709. [PMC free article] [PubMed] [Google Scholar]
- 25.Magyarosy, A., and J. G. Hancock. 1972. Microbial population of the laimosphere of squash (Cucurbita maxima). Plant Soil 37:187-190. [Google Scholar]
- 26.McLafferty, F. W., and F. Turecek. 1993. Interpretation of mass spectra, 4th ed. University Science Books, Herndon, Va.
- 27.Neidleman, S. L., and J. Geigert. 1986. Biohalogenation: principles, basic roles and applications. Ellis Horwood Ltd., Chichester, United Kingdom.
- 28.Palleroni, N. J. 1984. Family I. Pseudomonadaceae, p. 140-199. In N. R. Krieg and J. G. Holt (ed.), Bergey's manual of systematic bacteriology, vol. 1. Williams & Wilkins, Baltimore, Md. [Google Scholar]
- 29.Riker, A. J., and R. S. Riker. 1936. Introduction to research on plant diseases. John Swift and Co., St. Louis, Mo.
- 30.Roggo, B. E., F. Petersen, R. Delmendo, H. B. Jenny, H. H. Peter, and J. Roesel. 1994. 3-Alkanoyl-5-hydroxymethyl tetronic acid homologs and resistomycin: new inhibitors of HIV-1 protease. I. Fermentation, isolation and biological activity. J. Antibiot. 47:136-142. [DOI] [PubMed] [Google Scholar]
- 31.Rossler, D., W. Ludwig, K.-H. Schleifer, C. Lin, T. J. McGill, J. D. Wisotzkey, J. P. Jurtshuk, and G. E. Fox. 1991. Phylogenetic diversity in the genus Bacillus as seen by 16S rDNA sequencing studies. Syst. Appl. Microbiol. 14:266-269. [DOI] [PubMed] [Google Scholar]
- 32.Sheldrick, G. M. 1996. SADABS: Siemans area detector absorption correction program. University of Gottingen, Gottingen, Germany.
- 33.Sheldrick, G. M. 1995. SHELXTL crystal structure determination package, 5.03 ed. Siemens Industrial Automation, Inc., Madison, Wis.
- 34.Smith, N. R., R. E. Gordon, and F. E. Clark. 1952. Aerobic spore-forming bacteria. Agric. Monogr. 16:57. [Google Scholar]
- 35.Sneath, P. H. A., N. S. Mair, and M. E. Sharpe. 1989. Endospore-forming Gram-positive rods and cocci, p. 1104-1207. In J. G. Holt (ed.), Bergey's manual of systematic bacteriology, vol. 2. Williams & Wilkins, Baltimore, Md. [Google Scholar]
- 36.Swofford, D. L. 1999. PAUP∗. Phylogenetic analysis using parsimony (∗ and other methods), 4th ed. Sinauer Associates, Sunderland, Mass.
- 37.van Pee, K.-H. 1996. Biosynthesis of halogenated metabolites in bacteria. Annu. Rev. Microbiol. 50:375-399. [DOI] [PubMed] [Google Scholar]
- 38.Vinogradova, K. A., N. P. Kirillova, Z. G. Sokolova, I. V. Yartseva, B. V. Rozynov, and M. N. Preobrazhenskaya. 1991. Structure of heliomycin produced by Streptomyces heliomycini and antibiotic 11-98 produced by Streptomyces olivocinereus. Antibiot. Khimioter. 36:28-29. [PubMed] [Google Scholar]
- 39.Wilson, E. E., F. M. Zeitoun, and D. L. Fredrickson. 1967. Bacterial phloem canker, a disease of Persian walnut trees. Phytopathology 57:618-621. [Google Scholar]