Abstract
Specific interactions between ribosome recycling factor (RRF) and elongation factor-G (EFG) mediate disassembly of post-termination ribosomal complexes for new rounds of initiation. The interactions between RRF and EFG are also important in peptidyl-tRNA release from stalled pre-termination complexes. Unlike the post-termination complexes (harboring deacylated tRNA), the pre-termination complexes (harboring peptidyl-tRNA) are not recycled by RRF and EFG in vitro, suggesting participation of additional factor(s) in the process. Using a combination of biochemical and genetic approaches, we show that, (i) Inclusion of IF3 with RRF and EFG results in recycling of the pre-termination complexes; (ii) IF3 overexpression in Escherichia coli LJ14 rescues its temperature sensitive phenotype for RRF; (iii) Transduction of infC135 (which encodes a functionally compromised IF3) in E.coli LJ14 generates a ‘synthetic severe’ phenotype; (iv) The infC135 and frr1 (containing an insertion in the RRF gene promoter) alleles synergistically rescue a temperature sensitive mutation in peptidyl-tRNA hydrolase in E.coli; and (v) IF3 facilitates ribosome recycling by Thermus thermophilus RRF and E.coli EFG in vivo and in vitro. These lines of evidence clearly demonstrate the physiological importance of IF3 in the overall mechanism of ribosome recycling in E.coli.
INTRODUCTION
Subsequent to the action of release factors at the step of termination, the deacylated tRNA and the ribosomes remain bound to the mRNA in a post-termination complex. The post-termination complexes are disassembled by the action of ribosome recycling factor (RRF) and elongation factor-G (EFG) in eubacteria and the eukaryotic organelles. Biochemical and genetic studies have shown that specific interactions between RRF and EFG are important for the disassembly of the post-termination complexes. Additionally, it has been revealed that RRF and its specific interactions with EFG are also important in release of peptidyl-tRNAs, at least the ones with shorter chain lengths, in the cell (1,2). Such a phenomenon is generally recognized to be crucial in the rescue of stalled pre-termination complexes (3,4). The peptidyl-tRNAs are, in turn recycled by the esterase activity of peptidyl-tRNA hydrolase (Pth), an essential protein in E.coli (5,6).
A common requirement of specific interactions between RRF and EFG for peptidyl-tRNA release as well as for disassembly of the post-termination complexes suggests that the rescue/recycling of the stalled pre-termination, and the post-termination ribosomal complexes is mechanistically related. Ironically, under in vitro conditions, RRF and EFG fail to recycle the stalled complexes harboring peptidyl-tRNA. However, when these complexes are treated with puromycin to generate model post-termination complexes, they are efficiently recycled by RRF and EFG (7). These observations imply that in vivo, additional factor(s) participate in the overall mechanism of ribosome recycling. Such an argument is further supported by the observation that while T.thermophilus RRF and E.coli EFG carry out low-level ribosome recycling in vivo (8), they do not convert the model post-termination substrates to monosomes in vitro (9).
Three-dimensional structures of RRF-ribosome complexes, and directed hydroxyl radical probing have revealed that binding of RRF to the ribosome results in a remarkable conformational change at helices 69 and 71 in the 23S rRNA of the 50S subunit that form crucial inter-subunit bridges with helix 44 of the 16S rRNA in the 30S subunit. The surface of the 16S rRNA that contacts helix 69 in 23S rRNA also forms the binding site for IF3 (10,11). Based on these observations, it is hypothesized that transient disruption of the inter-subunit bridges may allow access to IF3 for binding to 30S subunit. The anti-association activity of IF3 may thus facilitate RRF and EFG mediated rescue/recycling of the ribosomes. Notably, such a role of IF3 in ribosome recycling is fundamentally different from its previously documented role in the release of the tRNA from the dissociated 30S subunit, mRNA and tRNA complex (12).
Like RRF, EFG and Pth, IF3 is also an essential protein in E.coli. The genes encoding them, frr, fusA, pth and infC are located at 4.2, 74.8, 27.1 and 38.8 min loci, respectively in the E.coli genome. The unlinked nature of these genes lends them for a facile genetic analysis of the ribosome recycling pathway in E.coli. In this study, use of both genetic and the biochemical approaches has allowed us to demonstrate a novel role of IF3 in the recycling of stalled pre-termination and post-termination ribosomal complexes by RRF and EFG in E.coli.
MATERIALS AND METHODS
Bacterial strains, plasmids and growth conditions
Plasmids and strains are listed in Table 1. Unless specified otherwise, Luria-Bertani (LB) liquid or solid (with 1.6% agar) media (Difco, USA) were used for growth (17). The media were supplemented with various antibiotics at the following final concentrations: tetracycline, 7.5 µg/ml; kanamycin, 25 µg/ml; chloramphenicol, 30 µg/ml and ampicillin, 100 µg/ml as required.
Table 1.
List of strains and plasmids
| Strain/Plasmid | Genotype/Details | References |
|---|---|---|
| Strains | ||
| E.coli AA7852 | E.coli harboring a temperature sensitive allele of Pth (pthts) | (6) |
| E.coli AA7852 (frr1) | Derivative of E.coli AA7852 harboring CmR marker at −32 position in the frr gene promoter resulting in down regulation of RRF expression | (13) |
| E.coli STL1670 | +Δ(srlR-recA)304 Tn10-infC135. Contains a R131P mutation in IF3 | (14) |
| E.coli AA7852 (Tn10-infC135) | Derivative of E.coli AA7852 harboring a Tn10 linked infC135 allele encoding R131P mutation in IF3 | This work |
| E.coli AA7852 Tn10-infC | Derivative of E.coli AA7852 harboring Tn10 linked to its native infC | This work |
| E.coli AA7852 Tn10-infC135, frr1 | Derivative of E.coli AA7852 frr1, harboring a Tn10 linked infC135 | This work |
| E.coli AA7852 Tn10-infC, frr1 | Derivative of E.coli AA7852 frr1 harboring a Tn10 linked to its native infC | This work |
| E.coli LJ14 | E.coli MC1061 containing the frrts (frr14) allele | (15) |
| E.coli LJ14 Tn10-infC135 | Derivative of E.coli LJ14 (frrts), harboring infC135 allele linked to Tn10 | This work |
| E.coli LJ14 Tn10-infC | Derivative of E.coli LJ14 (frrts), harboring Tn10 linked to native infC | This work |
| Plasmids | ||
| pET14b-EcoIF3 | pET14b based expression construct of E.coli IF3 (with AUG initiation codon) | This work |
| pTrc-TthRRF | ORF of T.thermophilus RRF cloned into pTrc99C vector | This work |
| pET14b-TthRRF | pET14b based expression construct of T.thermophilus RRF | This work |
| pBR-infC | E.coli IF3 gene with native promoter and initiation codon (AUU) cloned into pBR322 | This work |
| pTrc-EcoRRF | ORF of E.coli RRF cloned into pTrc99C | (2) |
| pACDH | A vector harboring ACYC Ori of replication, compatible with ColE1 Ori of replication | (16) |
| pACDK | A derivative of pACDH with KanR | This work |
| pACDH-TthRRF | ORF of T.thermophilus RRF cloned into pACDH | This work |
| pACDH-EcoRRF | ORF of E.coli RRF cloned into pACDH | This work |
| pACDK-TthRRF | A derivative of pACDH-TthRRF wherein TcR marker has been substituted with KanR | This work |
| pACDK-EcoRRF | A derivative of pACDH-EcoRRF wherein TcR marker has been substituted with KanR | This work |
Purification of E.coli RRF and EFG
E.coli RRF and EFG were purified as described (2).
Cloning of EcoIF3
Cloning of E.coli IF3—E.coli infC with its native promoter and initiation codon (AUU) was amplified from the E.coli genomic DNA with a forward, 5′-CGGGAATTCGTGTTAAAGCAG-3′ and a reverse, 5′-GAAGGATCCACAGACAGT GCT-3′ primers using Pfu DNA polymerase, digested with EcoRI and BamHI, and cloned into similarly digested pUC18R to yield pUC-infC. The EcoRI-BamHI fragment from pUC-infC was subcloned into the same sites of pBR322 to yield pBR-infC and verified by DNA sequencing. To generate a T7 RNA polymerase based expression construct (with AUG initiation codon), E.coli IF3 open reading frame (ORF) was amplified from E.coli HB101 genomic DNA with a forward 5′-AAAGGCGGAAAACGA GTTC-3′ and a reverse 5′-GAAGGATCCACAGA CAGTGCT-3′ primers using Pfu DNA polymerase, digested with BamHI and cloned into the NcoI (end filled) and BglII digested pET14b vector.
Purification of EcoIF3
The pET14b-EcoIF3 was introduced into E.coli Rosetta-pLysS and the transformants were grown in LB broth (2.4 L) to ∼0.6 A595 nm, induced with 0.5 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for 3 h and the cells were harvested by centrifugation at ∼2500 g (SLA-1500, Sorvall) for 10 min, resuspended in 20 ml of TME [25 mM Tris–HCl (pH 8.0), 1 mM Na2EDTA, 2 mM β-mercaptoethanol] and lysed by sonication. The crude cell lysate was centrifuged to obtain S20 lysate, which was subjected to stepwise 0–25%, 25–50% and 50–80% ammonium sulphate cut. The precipitate from the 50–80% cut was dissolved in 5 ml of buffer A [20 mM Tris–HCl (pH 7.5), 10% glycerol, 100 mM NaCl, 1 mM Na2EDTA and 2 mM β-mercaptoethanol] and loaded onto the Sepharose G-100 column, and eluted with buffer A. Fractions enriched for IF3 were pooled, diluted 2-fold using buffer B [20 mM Tris–HCl (pH 7.5), 10% glycerol, 1 mM Na2EDTA and 2 mM β-mercaptoethanol], and loaded onto high-Q and high-S columns (BioRad) connected in series. IF3 was eluted from high-S column with a gradient of 0–1 M NaCl in buffer B. The fractions enriched for IF3 were pooled and subjected to 80% ammonium sulfate cut. The pellet was dissolved in 1 ml of buffer A and subjected to chromatography on Superdex-200 in buffer A. The fractions containing apparently homogenous preparation of IF3 were pooled, dialyzed against 20 mM Tris–HCl (pH 7.5), 50% glycerol, 150 mM NaCl and 2 mM β-mercaptoethanol and stored at −20°C.
Isolation of E.coli polysomes
For the experiment shown in Figure 1A, polysomes were prepared as described (2). The polysomes used in the remaining experiments was prepared by a modified protocol wherein the cells were disrupted by sonication and the treatments with DNase I and Brij-35 were eliminated. Briefly, E.coli MRE600 (2.1 L) was grown to ∼0.6 A595 nm, supplemented with 0.3 mM tetracycline and quick chilled on ice-salt mix. The cells were harvested by centrifugation at ∼2500 g (SLA-1500, Sorvall) for 5 min, suspended in 30 ml of buffer A [25% sucrose in 250 mM Tris–HCl (pH 8.0) and 10 mM MgSO4] and supplemented with 1 ml of lysozyme (40 mg/ml) in 250 mM Tris–HCl (pH 8.0) and 8 mM Na2EDTA. After 10 min of incubation at 4°C, the cells were sonicated thrice in 3 pulses of 2 s each using a microprobe (Heat Systems-Ultrasonics Inc, USA). The lysate was then cleared by centrifugation at ∼17 000 g (AG-508R, Kubota) for 10 min. The supernatant was transferred to a new tube and the pellet was resuspended in 10 ml of buffer B [10 mM Tris–HCl (pH 7.8), 1 M NH4Cl, 40 mM magnesium acetate, 10 mM β-mercaptoethanol and 2 mM Na2EDTA] and centrifuged at ∼17 000 g (AG-508R, Kubota) for 10 min. The two supernatants were pooled and centrifuged at 100 000 g (JA-30.50 Ti, Beckman). The pellet was resuspended in 20 ml of buffer B, layered on 34% sucrose in buffer B and subjected to ultracentrifugation for 7 h at ∼96 000 g (SW28, Beckman). This step was repeated with the pellet obtained after the first round of ultracentrifugation. The pellet thus obtained was resuspended in 2 ml of RRF assay buffer [10 mM Tris–HCl (pH 7.4), 80 mM NH4Cl and 8.2 mM MgSO4], quantified by A260 nm and stored in aliquots at −70°C.
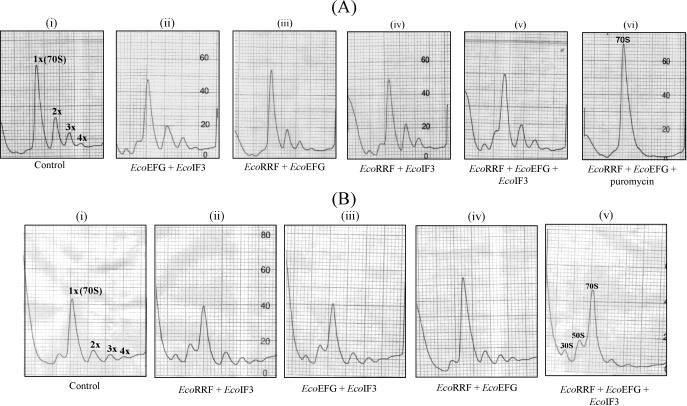
Figure 1.
(A) Ribosome recycling assays. Reactions containing various factors (as indicated in the panels below) were carried out in the absence (panels i–v) or presence (panel vi) of 10 µM puromycin, fractionated on sucrose density gradients and scanned from top to bottom. Amounts of various factors used are EcoEFG (6 µg), EcoIF3 (1.2 µg) and EcoRRF (4 µg). (B) Ribosome recycling assays using the polysomes prepared by sonication method (Materials and Methods). Reactions containing various factors were carried out in the absence of puromycin. Other details of the reactions are same as described for panels in (A).
In vitro ribosome recycling assay
Reactions (250 µl) containing 1 O.D. of polysomes and the specified amounts of factors were incubated at 32°C for 30 min in 10 mM Tris–HCl (pH 7.4), 80 mM NH4Cl, 8.2 mM MgSO4, 1 mM DTT and 160 µM GTP with or without puromycin (7). Reactions were fractionated on 15–30% sucrose density gradients by centrifugation at 190 000 g (SW55Ti, Beckman) for 1 h and scanned from top to bottom at 254 nm using ISCO UA-6 detector.
Allelic exchange of infC with infC135 (R131P)
P1 phage lysate was raised (18) on E.coli STL1670 (kindly provided by Dr Susan Lovett, Brandeis University, Waltham, USA) containing Tn10 (TcR) linked (∼90%) to infC135 allele, and transductional crosses were performed using different E.coli strains. The transductants selected on Tc were screened for the allelic exchange by RFLP analysis of the infC amplicons obtained by using 5′-ATCCTGACCGAAGAGTTC-3′ and 5′-TTTG AAGCGCTTAGCAGC-3′ as forward and reverse primers, respectively using HpaII.
Cloning and purification of T.thermophilus RRF (TthRRF)
TthRRF ORF was amplified from T.thermophilus HB8 DNA (gift from Dr V. Ramakrishnan, MRC, Cambridge, UK) with a forward 5′-GGCGCCATGGCCCTGAAGGAGCT-3′ and a reverse 5′-CGCGGGATCCCGGGGTCA GCCC-3′ primers using Pfu DNA polymerase, digested with NcoI and BamHI, cloned into similarly digested pTrc99C, and verified by DNA sequencing. The NcoI-HindIII fragment from pTrc-TthRRF was subcloned into the same restriction sites of pACDH vector (16) and pET14b vectors to generate pACDH-TthRRF (TcR) and pET14b-TthRRF, respectively. To generate pACDK-TthRRF (KanR), the TcR in pACDH-TthRRF was inactivated by digestion with EcoRV and cloning a KanR cassette (HincII ends) from pUC4K.
TthRRF was purified from E.coli BL21 (DE3) harboring pET14b-TthRRF as described (19). Culture (2.4 L) was grown in LB broth to ∼0.6 A595 nm, induced with 0.25 mM IPTG for 3 h and harvested by centrifugation. The cells were resuspended in 30 ml of buffer A [50 mM Tris–HCl (pH 7.0), 10 mM MgCl2 and 5 mM β-mercaptoethanol] containing 500 mM NH4Cl and lysed by sonication. The crude cell lysate was cleared by centrifugation at ∼15 000 g (AG-508R, Kubota) for 30 min and the supernatant was subjected to centrifugation at 100 000 g for 2 h (JA-30.50 Ti, Beckman). The S100 supernatant thus obtained was heated to 65°C for 15 min and centrifuged at ∼15 000 g (AG-508R, Kubota) for 15 min. The supernatant was subjected to a stepwise 0–30% and 30–80% ammonium sulfate cut. The pellet from the 30–80% cut was dissolved in 3 ml of buffer A, dialyzed against the same buffer, passed through high-Q (BioRad)–heparin Sepharose columns (Amersham Life Sciences) connected in series and pre-equilibrated with buffer A. Proteins from the heparin Sepharose column were eluted with 0–1 M NH4Cl gradient in buffer A. The fractions containing TthRRF were pooled, heated at 65°C for 10 min and centrifuged. The supernatant was loaded onto hydroxyapatite column (BioRad) equilibrated with buffer B [20 mM potassium phosphate buffer (pH 6.8) and 10% glycerol], and eluted with a gradient of 20 mM-1 M potassium phosphate in buffer B. The fractions containing apparently homogenous preparation of TthRRF were pooled, dialyzed against 20 mM Tris–HCl (pH 7.5), 100 mM NaCl, 50% glycerol, 2 mM β-mercaptoethanol and stored at −20°C.
RESULTS
IF3 facilitates recycling of stalled pre-termination complexes
Sucrose density gradient centrifugation of the stalled ribosomal complexes (harboring peptidyl-tRNA) prepared from actively growing E.coli (7), resolves them into fractions corresponding to monosomes (70S ribosomes) and polysomes (disomes, trisomes, tetrasomes etc.) (Figure 1A, panel i). While the monosome peak corresponds to both the free and mRNA bound ribosomes, the polysome peaks correspond to the mRNA bound ribosomes. When these ribosomal preparations are treated with puromycin (to obtain model post-termination complexes harboring deacylated tRNA), RRF and EFG convert them into monosomes (compare panel i with vi) suggesting that the stalled ribosomal preparation was active in the classical assay for recycling of the post-termination complexes (7). However, when the ribosomal preparation not treated with puromycin (thus harboring peptidyl-tRNA) was incubated with RRF and EFG, or RRF, EFG and IF3, conversion of polysomes to monosomes was not detectable (Figure 1A, panels iii and v, respectively). However, as expected of its effect on 70S ribosomes, the reactions with IF3 (panels ii, iv and v) showed an increase in the peaks corresponding to 50S and 30S subunits, indicating that the IF3 preparation used was active.
Interestingly, when we treated a different preparation of stalled ribosomes (wherein cells were disrupted by sonication in a buffer lacking detergent and DNase I, but otherwise processed as before) with RRF and EFG, efficient recycling occurred in the presence of IF3 but not in its absence (Figure 1B, panels iv and v, respectively). This observation not only provides an in vitro evidence for the role of RRF in recycling of stalled pre-termination complexes, but also supports a crucial role of IF3 in this process. Although the exact reasons for why the two preparations of the stalled ribosomes behaved differently have not been investigated, the contribution of non-specific factors (e.g. nucleases) in the detergent untreated preparation is ruled out by the control experiments wherein incubation of this preparation with either RRF and IF3; or EFG and IF3 under the same conditions did not result in conversion of the polysomes to monosomes (panels i–iii). While differences in the biochemical properties of differently prepared ribosomes have been observed in other studies as well (20,21), it was nevertheless important to demonstrate the biological relevance of this observation.
The infC135 allele rescues a Pthts phenotype in E.coli
Haggerty and Lovett (22) identified the infC135 allele, which encodes a functionally compromised IF3 because of an R131P mutation. The E.coli STL1670 wherein this allele is linked (∼90%) to Tn10 (Tn10-infC135) grows normally in rich medium, and serves as an important genetic tool for study of biological functions of IF3 (14,23).
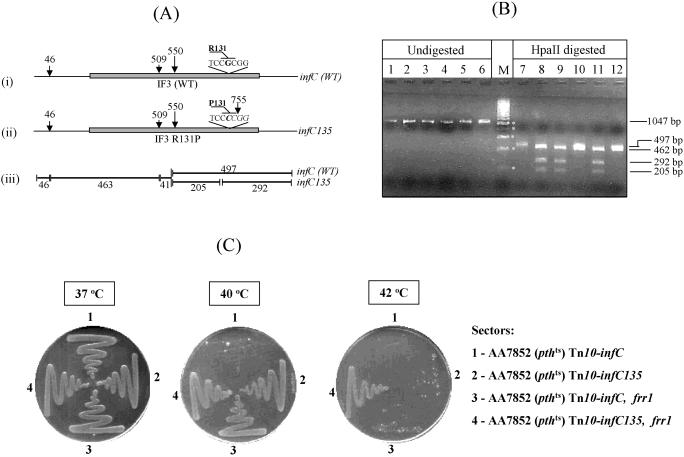
If the biochemically identified role of IF3 in RRF mediated recycling of the stalled ribosomes (Figure 1B) is biologically relevant, an anticipated consequence of a compromised function of IF3 would be a decrease in peptidyl-tRNA release from the ribosomal complexes, a phenomenon known to rescue the growth of E.coli (Pthts) strains at non-permissive temperatures (1,13). To validate this prediction, we introduced by P1 mediated transduction, a Tn10 linked infC135 allele (or as a control, the Tn10 alone) from E.coli STL1670 into E.coli AA7852 (pthts) and its derivative AA7852 (pthts frr1) containing decreased level of cellular RRF [the frr1 allele contains a CmR insertion in the RRF gene promoter, (13)]. The nature of infC allele in the transductants was verified by an associated RFLP for HpaII (Figure 2A and B). A 1.047 kb amplicon from infC produces 497, 463, 46 and 41 bp fragments upon digestion with HpaII. Of these, the 497 and 463 bp migrate as an unresolved doublet, and the 46 and 41 bp fragments are undetectable by agarose gel electrophoresis (Figure 2B lanes 7, 10 and 12). The infC135 allele arises because of a G to C mutation generating a new HpaII site within the 497 bp fragment which splits it into a 292 bp and a 205 bp fragments, separable from each other and the 463 bp fragment (Figure 2B lanes 8, 9 and 11).
Figure 2.
(A) Schematic representation of HpaII cleavage sites in the infC and infC135 amplicons (panels i and ii) and sizes of fragments obtained upon digestion with HpaII (panel iii). (B) Analysis of amplicons from parent strains and the various transductants (lanes 1–6) and their digests with HpaII (lanes 7–12) along with a 200 bp marker ladder (M) in the middle, on 2% agarose gels. Sizes of the bands are indicated on the right. Lanes: 1 and 7, E.coli AA7852 (recipient); 2 and 8, E.coli STL1670 (donor); 3 and 9, E.coli AA7852 (Tn10-infC135); 4 and 10, E.coli AA7852 (Tn10-infC); 5 and 11, E.coli AA7852 (Tn10-infC135, frr1); 6 and 12, E.coli AA7852 (Tn10-infC, frr1). (C) Analysis of complementation of E.coli AA7852 (pthts). Various derivatives of E.coli AA7852 (pthts), as indicated, were streaked on LB agar plates from overnight cultures and incubated for 36 h at the indicated temperatures.
As shown in Figure 2C, all the four pthts strains thus generated, E.coli AA7852 (Tn10-infC; Tn10-infC135; Tn10-infC, frr1 and Tn10-infC135, frr1) grow well at 37°C. As expected (13), unlike the Tn10 alone derivative (Tn10-infC, sectors 1), the frr1 allele in the AA7852 (Tn10-infC frr1) strain allows its growth to a good extent at 40°C and to a detectable level at 42°C (sectors 3). True to the prediction, the infC135 allele in E.coli AA7852 (Tn10-infC135) rescued the growth of the strain (sectors 2) in a manner similar to the frr1 allele (sectors 3). More importantly, simultaneous presence of both the infC135 and frr1 alleles in the AA7852 (Tn10-infC135, frr1) strain conferred a synergistic effect on the rescue (sectors 4, at 42°C). Such a synergistic effect provides a strong genetic evidence for a functional interaction between IF3 and RRF in mediating peptidyl-tRNA release. Taken together with the observations in Figure 1B, and those reported earlier (1,2), it could be concluded that RRF, EFG and IF3 lead to peptidyl-tRNA release from the stalled ribosomes, and that they play a crucial role in their recycling.
Physiological link between RRF and IF3 functions in E.coli
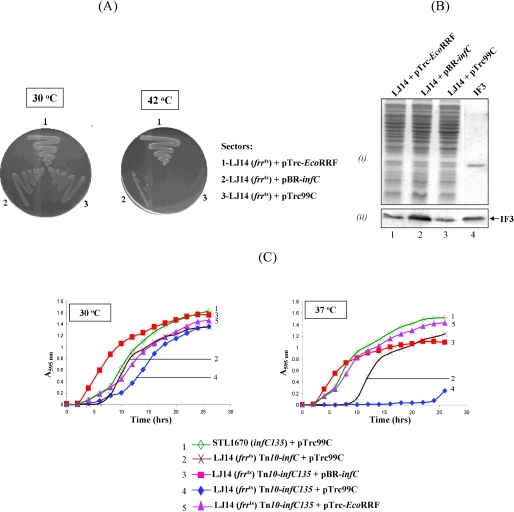
To further our understanding of the functional interaction between RRF and IF3 in ribosome recycling, we introduced either pTrc99C vector, pTrc-EcoRRF or the pBR-infC into E.coli LJ14 (frrts), and monitored growth of the transformants at 30°C, a permissive temperature; and at 42°C, a temperature non-permissive for the growth of E.coli LJ14 (frrts). As shown in Figure 3A, at the permissive temperature, growth of all the three transformants was comparable; and as expected, at the non-permissive temperature, while a transformant harboring the empty vector (pTrc99C) showed no growth, the one harboring pTrc-EcoRRF showed full growth (sectors 3 and 1, respectively). Interestingly, the transformant harboring pBR-infC and producing ∼2- to 3-fold excess of cellular IF3 (Figure 3B), revealed a detectable growth at 42°C (Figure 3A, sectors 2) indicating a functional interaction of IF3 with RRF in ribosome recycling.
Figure 3.
(A) Analysis of complementation of E.coli LJ14 (frrts) harboring pTrc-EcoRRF (sectors 1), pBR-infC (sectors 2) and pTrc99C (sectors 3). Overnight cultures were streaked on LB agar ampicillin plates, and incubated for 36 h at the indicated temperatures. (B) Immunoblot analysis of IF3 levels in total cell extracts (∼15 µg) of E.coli LJ14 harboring pTrc-EcoRRF (lane 1), pBR-infC (lane 2) or vector alone, pTrc99C (lane 3). In lane 4, ∼0.2 µg IF3 was analyzed as marker. A Coomassie blue stained gel is shown in the above panel (i) as a control for equal loading of total proteins. Band corresponding to IF3 is indicated in the immunoblot in the lower panel (ii). (C) Analysis of growth of transformants of various derivatives of E.coli LJ14 (frrts) and E.coli STL1670 in LB broth containing ampicillin, at 30°C (left) and 37°C (right). Saturated cultures grown at 30°C were inoculated (0.1%) into LB broth containing ampicillin and the growth was monitored at 595 nm at regular time intervals. Details of various strains are as shown in the panels.
In yet another approach, we introduced the Tn10 linked infC135 from E.coli STL1670 into the E.coli LJ14 (frrts) transformants by P1 mediated transduction, verified the transductants by the HpaII RFLP analysis, and checked the resulting strains for growth in LB broth at the permissive (30°C) and a semi-nonpermissive (37°C) temperatures (Figure 3C). The parent strain, STL1670 (Tn10-infC135), and a Tn10 derivative of LJ14 (Tn10-infC, frrts) harboring the empty vector (pTrc99C) were included as controls. As expected of their phenotypes, E.coli STL1670 grows well at both the temperatures (curve 1); and E.coli LJ14 (frrts, Tn10 alone derivative) grows well at the permissive temperature (30°C) but shows a weaker growth at the semi-nonpermissive temperature of 37°C (curve 2). Of the three Tn10-infC135 derivatives of E.coli LJ14 (frrts), the one harboring pTrc-EcoRRF (complements the frrts defect) grows well at both the temperatures (curve 5). The one harboring pBR-infC grows well at 30°C and 37°C (curve 3). Notably, the transductant harboring an empty vector pTrc99C grows somewhat weakly at 30°C and becomes hypersensitive to growth at 37°C (curve 4). Such a ‘synthetic severe’ phenotype is another strong genetic evidence to support a role of IF3 in RRF mediated ribosome recycling in E.coli. An argument that the severe phenotype of the double allele (infC135 and frrts) strain is merely a consequence of the simultaneous presence of two unrelated defective alleles, can be ruled out by the fact that combination of infC135 with pthts resulted in a remarkable rescue of E.coli AA7852 (pthts frr1) growth even at 42°C (Figure 2C).
IF3 facilitates ribosome recycling with TthRRF in E.coli
It has been shown that while the TthRRF alone does not complement E.coli (frrts), it sustains a very weak growth upon prolonged incubations at the non-permissive temperature (8). Also, a recent biochemical study (9) showed that TthRRF and EcoEFG lead to partial recycling of E.coli polysomes by effecting efficient release of tRNA but not the mRNA (hence the polysome profile on sucrose density gradients remains largely unchanged).
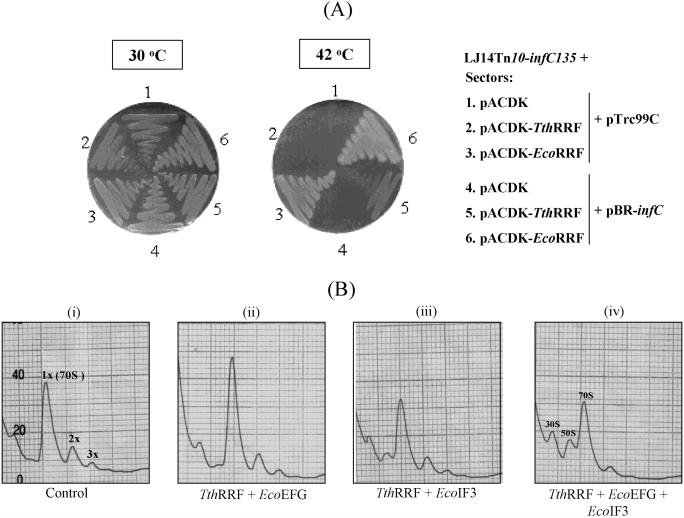
To check the effect of IF3 on ribosome recycling by TthRRF, we introduced into E.coli LJ14 (Tn10-infC135, frrts) harboring pTrc99C or pBR-infC, an empty pACDK (KanR) or pACDK-TthRRF containing TthRRF on a low copy plasmid with a compatible origin of replication (ACYC). And, to enhance the sensitivity of our assay, we monitored growth of the transformants on minimal M9 medium with casamino acids (24). Unlike the reported growth defect of the infC135 strain at 42°C on the 56/2 minimal medium (14), its growth on minimal M9 medium with casamino acids is not temperature sensitive (data not shown). As shown in Figure 4A, in this medium, all the strains grow to similar extent at 30°C in 48 h. At the non-permissive temperature of 42°C, expectedly, the control transformants harboring, pACDK-EcoRRF along with pTrc99C (sectors 3), and pACDK-EcoRRF along with pBR-infC (sectors 6) plasmids show strong growth; and the transformants harboring pTrc99C along with pACDK (sectors 1) or pBR-infC along with pACDK (sectors 4) do not reveal any background growth. Importantly, while the transformant harboring pACDK-TthRRF does not show a growth in the presence of empty vector (pTrc99C), it shows reasonable growth in the presence of pBR-infC (sectors 2 and 5) suggesting that IF3 facilitates ribosome recycling by TthRRF in E.coli. To validate this interpretation, and to further investigate the role of IF3 in recycling of post-termination complexes, we carried out in vitro recycling assays with TthRRF and EcoEFG in the absence and presence of IF3 using the model post-termination complexes. As shown in Figure 4B, while TthRRF along with EcoEFG did not convert polysomes to monosomes (compare panel ii with panel i), inclusion of IF3 in the reaction resulted in such a conversion (compare panel iv with i). As a control, TthRRF with IF3, did not convert polysomes to monosomes (panel iii). Taken together, it can be concluded that TthRRF with EcoEFG, which leads to a partial reaction of tRNA release (9), can be driven to complete recycling by the presence of IF3. Notably, in this heterologous system, inclusion of IF3 with RRF and EFG (panel iv) resulted in a pronounced accumulation of 30S and 50S subunits.
Figure 4.
(A) Analysis of complementation of E.coli LJ14 (frrts) transductants (Tn10-infC135) harboring various constructs (as indicated) incubated at 30°C and 42°C for 48 h using M9 minimal medium containing casamino acids, ampicillin and kanamycin. (B) Ribosome recycling assays using model post-termination complexes. Reactions were carried out in the presence of 10 µM puromycin. Amounts of proteins used are TthRRF (1.7 µg), EcoEFG (6 µg) and EcoIF3 (1.2 µg).
DISCUSSION
In this study, we have presented evidence for a distinct role of IF3 in ribosome recycling, thus resolving a long-standing paradox that existed because of a gap between the genetic studies, which supported a role of RRF in recycling of the stalled ribosomes harboring peptidyl-tRNA, and the biochemical experiments which did not support such a role (25). Just as in a more recent study (26), initially we also did not detect recycling of the stalled pre-termination ribosomal complexes by RRF and EFG despite the presence of IF3 in the reactions (Figure 1A). However, use of a different preparation of stalled ribosomes (Figure 1B), revealed that in the presence of IF3, such complexes were indeed recycled by RRF and EFG. Notably, the subsequent experiments (Figures 2–4) wherein a combination of biochemical and genetic approaches has been employed to resolve the ambiguity arising out of the in vitro experiments, clearly support a physiological role of IF3 in recycling of not only the stalled pre-termination complexes but also of the post-termination complexes by RRF and EFG.
At what step in ribosome recycling does IF3 participate? In the polysome profiles, conversion of polysomes to monosomes results by their release from the mRNA. In our experiments, treatment of stalled pre-termination complexes with EcoRRF and EcoEFG (Figure 1B) or the model post-termination complexes with TthRRF and EcoEFG (Figure 4B), does not cause a detectable change in the polysome profile. However, they do in the presence of IF3 (Figures 1B and 4B). As RRF or EFG alone with IF3 do not result in such a change, these observations clearly suggest that in the presence of RRF and EFG, ribosomes are converted into a state that is compatible to bind IF3, which in turn leads to the release of 50S and 30S subunits from mRNA. Such a role of IF3 in the disassembly process is further supported by the dissociation of empty 70S ribosomes into 50S and 30S subunits by RRF and EFG in the presence of IF3 (27). Thus, our studies reveal yet another cellular role of IF3 at the step of ribosome recycling, and add one more important facet to the coterie of the varied functions of IF3 (28). However, our interpretation of the role of IF3 in antagonizing an association of the 50S and 30S subunits, does not rule out its additional role in dissociation of tRNA from the 30S subunits generated from the post-termination complexes by RRF and EFG, for which the biochemical evidence has been presented earlier (12). It may also be noted that at least under in vitro conditions, recycling of the post-termination complexes with EcoRRF and EcoEFG (a homologous combination which functions efficiently on E.coli ribosomes especially the ones harboring deacylated tRNA) can occur independently in the presence of IF3 [Figure 1A; (29)]. Importantly, our observations espouse for a role of IF3 in the overall mechanism of ribosome recycling in E.coli.
Three-dimensional structures of RRF from several sources have revealed that RRF consists of two domains arranged perpendicular to each other in an L-shaped molecule (19,30–34). Structure of Mycobacterium tuberculosis RRF from five different crystal forms and its comparison with those of the other RRFs (34) highlighted two of the major motions of the RRF-domain II; a rotation in the plane nearly perpendicular to the axis of domain I (the ‘swinging door motion’), and an internal rotation along its own axis (the ‘screw motion’). The structure solution of RRF-ribosome complex by cryo-EM provided evidence that the primary inter-subunit bridge regions B2a and B3 are displaced upon RRF binding. Further, directed hydroxy radical probing (10) and the structures of RRF bound ribosomes (11,35), have shown that RRF binds in the canyon between the 50S and 30S subunits, and while the RRF-domain I establishes an extensive set of interactions with the 50S subunit, the RRF-domain II facing the 30S subunit is highly flexible.
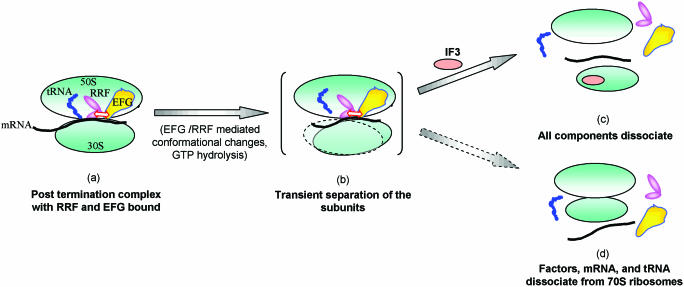
Although, the step at which the energy of GTP hydrolysis is utilized in the overall mechanism of ribosome recycling is not understood, the biochemical and biophysical studies (36,37) have provided important clues to the dynamics of RRF on the ribosome upon EFG binding. These studies show that a high affinity binding of RRF to ribosomes is converted to a low affinity state upon EFG binding. It is not clear whether these affinity differences arise because RRF binds to a different site in the ribosome due to a piston like movement by EFG (36); or due to a distinct mechanism of EFG action leading to a dynamic change in the contact points between RRF and ribosome by a gearwheel action between the RRF-domain II and EFG. While both of these proposals are consistent with the changes in the fluorescence measurements (37), the latter model accommodates both the swinging door and screw motions of RRF-domain II, and is consistent with the genetic studies using heterologous RRF and EFG in E.coli (2,34,38). Further, the ribosome recycling with the heterologous factors suggests that the extent and specificity of interactions between EFG and RRF (Figure 5, intermediate denoted by a) may well determine the efficiency of gearwheel action between the two factors and the degree to which the inter-subunit bridges are distorted to expose the elements for IF3 binding to the 30S subunit (intermediate denoted by b). It can be envisaged that while a stable binding of IF3 to 30S subunit would result in complete dissociation of all the components (as shown in c) occasionally a non-productive approach of IF3 to bind the intermediate (b) may, upon exit from mRNA, lead to re-establishment of the 50S and 30S subunit interactions for release as 70S ribosome (as shown in d), useful for translation of the leaderless mRNAs (39–41). It is interesting to speculate that a critical balance of events occurring at intermediate (b), would also feed the ribosomes to translational re-initiation in polycistronic mRNAs (42).
Figure 5.
Model for ribosome recycling. Post-termination ribosomal complexes harboring deacylated tRNA bind RRF and EFG (intermediate a), and specific interactions between RRF and EFG and GTP hydrolysis lead to a transient state (intermediate b). At this stage, a productive binding of IF3 to 30S subunit leads to dissociation of all components (c), and its occasional failure to bind 30S subunits results in release of 70S ribosomes from mRNA (d). The tRNA release occurs at stage b before IF3 binding (9) or at stage c upon IF3 binding (12).
In the scheme (Figure 5) for recycling of the post-termination complexes, tRNA release has been shown to occur at the intermediate stage b (9) or stage c (12). We believe that release of peptidyl-tRNAs with shorter peptides present in the pre-termination stalled ribosomes such as those arising from minigenes (43–45) could be similar to tRNA release. Whether recycling of the stalled complexes harboring peptidyl-tRNA with longer peptides occurs by a direct release of peptidyl-tRNA or indirectly through generation of a shorter chain peptidyl-tRNA by unknown mechanisms or free tRNA by the action of Pth (46) is not understood (also see below). However, at least in the in vitro experiments, IF3 is more crucial for recycling of stalled pre-termination complexes, than of the post-termination complexes. Hence, some details of these processes are likely to be distinct, which will be a subject of our future research. Importantly, the studies on RRF binding to ribosomes have shown that RRF engages some of the sites on the 50S subunit that are also occupied by the peptidyl-tRNA (11,47). Thus, the observations that RRF recycles pre-termination stalled ribosomes raises an important question as to whether the functionally relevant initial binding of RRF, at least in the pre-termination ribosomes, occurs at a site different from the one seen in the empty ribosomes.
Finally, the use of heterologous systems has remarkably augmented our understanding of important biological processes. Homologous factors, which have co-evolved to function efficiently, make it extremely demanding to trap the intermediates useful for biochemical or genetic studies. Thus a distinct advantage of using heterologous factors is that they serve as ‘naturally optimized mutants’. Earlier studies using heterologous factors have revealed important details of RRF and EFG function (2,9,38,48,49). In this study, we exploited the observation that TthRRF and EcoEFG lead to tRNA but not the mRNA release from the model post-termination complexes (9). As shown in Figure 4, such a system provided an appropriately trapped intermediate, which could be rescued by IF3 both in vivo and in vitro. Importantly, this experiment not only allowed us to identify a step at which IF3 enters the ribosome recycling pathway, but it also provides a suitable intermediate for structural studies. Clearly, further analyses of such systems would reveal more of the crucial mechanistic details of ribosome recycling, and other important biological processes.
Acknowledgments
We thank our laboratory colleagues for their suggestions on the manuscript. This work was supported by grants from Department of Biotechnology, and Department of Science and Technology, New Delhi. N.S.S. and G.D. are supported by a senior research fellowship of Council of Scientific and Industrial Research, New Delhi. A.S. is supported by a senior research fellowship of the University Grants Commission, New Delhi. The Open Access publication charges for this article were waived by Oxford University Press.
Conflict of interest statement. None declared.
REFERENCES
- 1.Heurgue-Hamard V., Karimi R., Mora L., MacDougall J., Leboeuf C., Grentzmann G., Ehrenberg M., Buckingham R.H. Ribosome release factor RF4 and termination factor RF3 are involved in dissociation of peptidyl-tRNA from the ribosome. EMBO J. 1998;17:808–816. doi: 10.1093/emboj/17.3.808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rao A.R., Varshney U. Specific interaction between the ribosome recycling factor and the elongation factor G from Mycobacterium tuberculosis mediates peptidyl-tRNA release and ribosome recycling in Escherichia coli. EMBO J. 2001;20:2977–2986. doi: 10.1093/emboj/20.11.2977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Menninger J.R. The accumulation as peptidyl-transfer RNA of isoaccepting transfer RNA families in Escherichia coli with temperature-sensitive peptidyl-transfer RNA hydrolase. J. Biol. Chem. 1978;253:6808–6813. [PubMed] [Google Scholar]
- 4.Menninger J.R. Accumulation of peptidyl tRNA is lethal to Escherichia coli. J. Bacteriol. 1979;137:694–696. doi: 10.1128/jb.137.1.694-696.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kossel H., RajBhandary U.L. Studies on polynucleotides. LXXXVI. Enzymic hydrolysis of N-acylaminoacyl-transfer RNA. J. Mol. Biol. 1968;35:539–560. doi: 10.1016/s0022-2836(68)80013-0. [DOI] [PubMed] [Google Scholar]
- 6.Atherly A.G., Menninger J.R. Mutant E.coli strain with temperature sensitive peptidyl-transfer RNA hydrolase. Nature New Biol. 1972;240:245–246. doi: 10.1038/newbio240245a0. [DOI] [PubMed] [Google Scholar]
- 7.Hirashima A., Kaji A. Factor-dependent release of ribosomes from messenger RNA. Requirement for two heat-stable factors. J. Mol. Biol. 1972;65:43–58. doi: 10.1016/0022-2836(72)90490-1. [DOI] [PubMed] [Google Scholar]
- 8.Fujiwara T., Ito K., Nakayashiki T., Nakamura Y. Amber mutations in ribosome recycling factors of Escherichia coli and Thermus thermophilus: evidence for C-terminal modulator element. FEBS Lett. 1999;447:297–302. doi: 10.1016/s0014-5793(99)00302-6. [DOI] [PubMed] [Google Scholar]
- 9.Raj V.S., Kaji H., Kaji A. Interaction of RRF and EF-G from E.coli and T.thermophilus with ribosomes from both origins–insight into the mechanism of the ribosome recycling step. RNA. 2005;11:275–284. doi: 10.1261/rna.7201805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lancaster L., Kiel M.C., Kaji A., Noller H.F. Orientation of ribosome recycling factor in the ribosome from directed hydroxyl radical probing. Cell. 2002;111:129–140. doi: 10.1016/s0092-8674(02)00938-8. [DOI] [PubMed] [Google Scholar]
- 11.Agrawal R.K., Sharma M.R., Kiel M.C., Hirokawa G., Booth T.M., Spahn C.M., Grassucci R.A., Kaji A., Frank J. Visualization of ribosome-recycling factor on the Escherichia coli 70S ribosome: functional implications. Proc. Natl Acad. Sci. USA. 2004;101:8900–8905. doi: 10.1073/pnas.0401904101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Karimi R., Pavlov M.Y., Buckingham R.H., Ehrenberg M. Novel roles for classical factors at the interface between translation termination and initiation. Mol. Cell. 1999;3:601–609. doi: 10.1016/s1097-2765(00)80353-6. [DOI] [PubMed] [Google Scholar]
- 13.Singh N.S., Varshney U. A physiological connection between tmRNA and peptidyl-tRNA hydrolase functions in Escherichia coli. Nucleic Acids Res. 2004;32:6028–6037. doi: 10.1093/nar/gkh924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haggerty T.J., Lovett S.T. IF3-mediated suppression of a GUA initiation codon mutation in the recJ gene of Escherichia coli. J. Bacteriol. 1997;179:6705–6713. doi: 10.1128/jb.179.21.6705-6713.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Janosi L., Mottagui-Tabar S., Isaksson L.A., Sekine Y., Ohtsubo E., Zhang S., Goon S., Nelken S., Shuda M., Kaji A. Evidence for in vivo ribosome recycling, the fourth step in protein biosynthesis. EMBO J. 1998;17:1141–1151. doi: 10.1093/emboj/17.4.1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mangroo D., RajBhandary U.L. Mutants of Escherichia coli initiator tRNA defective in initiation. Effects of overproduction of methionyl-tRNA transformylase and the initiation factors IF2 and IF3. J. Biol. Chem. 1995;270:12203–12209. doi: 10.1074/jbc.270.20.12203. [DOI] [PubMed] [Google Scholar]
- 17.Sambrook J.F., Fritsch E.F., Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Edn. Cold Spring Harbor, NY: Cold SpringHarbor Laboratory Press; 1989. [Google Scholar]
- 18.Miller J.H. In Experiments in Molecular Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1972. [Google Scholar]
- 19.Toyoda T., Tin O.F., Ito K., Fujiwara T., Kumasaka T., Yamamoto M., Garber M.B., Nakamura Y. Crystal structure combined with genetic analysis of the Thermus thermophilus ribosome recycling factor shows that a flexible hinge may act as a functional switch. RNA. 2000;6:1432–1444. doi: 10.1017/s1355838200001060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nissen P., Hansen J., Ban N., Moore P.B., Steitz T.A. The structural basis of ribosome activity in peptide bond synthesis. Science. 2000;289:920–930. doi: 10.1126/science.289.5481.920. [DOI] [PubMed] [Google Scholar]
- 21.Thompson J., Kim D.F., O'Connor M., Lieberman K.R., Bayfield M.A., Gregory S.T., Green R., Noller H.F., Dahlberg A.E. Analysis of mutations at residues A2451 and G2447 of 23S rRNA in the peptidyltransferase active site of the 50S ribosomal subunit. Proc. Natl Acad. Sci. USA. 2001;98:9002–9007. doi: 10.1073/pnas.151257098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haggerty T.J., Lovett S.T. Suppression of recJ mutations of Escherichia coli by mutations in translation initiation factor IF3. J. Bacteriol. 1993;175:6118–6125. doi: 10.1128/jb.175.19.6118-6125.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.O'Connor M., Gregory S.T., Rajbhandary U.L., Dahlberg A.E. Altered discrimination of start codons and initiator tRNAs by mutant initiation factor 3. RNA. 2001;7:969–978. doi: 10.1017/s1355838201010184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Croitoru V., Bucheli-Witschel M., Hagg P., Abdulkarim F., Isaksson L.A. Generation and characterization of functional mutants in the translation initiation factor IF1 of Escherichia coli. Eur. J. Biochem. 2004;271:534–544. doi: 10.1046/j.1432-1033.2003.03954.x. [DOI] [PubMed] [Google Scholar]
- 25.Janosi L., Ricker R., Kaji A. Dual functions of ribosome recycling factor in protein biosynthesis: disassembling the termination complex and preventing translational errors. Biochimie. 1996;78:959–969. doi: 10.1016/s0300-9084(97)86718-1. [DOI] [PubMed] [Google Scholar]
- 26.Peske F., Rodnina M.V., Wintermeyer W. Sequence of steps in ribosome recycling as defined by kinetic analysis. Mol. Cell. 2005;18:403–412. doi: 10.1016/j.molcel.2005.04.009. [DOI] [PubMed] [Google Scholar]
- 27.Hirokawa G., Nijman R.M., Raj V.S., Kaji H., Igarashi K., Kaji A. The role of ribosome recycling factor in dissociation of 70S ribosomes into subunits. RNA. 2005;11:1317–1328. doi: 10.1261/rna.2520405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Petrelli D., LaTeana A., Garofalo C., Spurio R., Pon C.L., Gualerzi C.O. Translation initiation factor IF3: two domains, five functions, one mechanism? EMBO J. 2001;20:4560–4569. doi: 10.1093/emboj/20.16.4560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zavialov A.V., Hauryliuk V.V., Ehrenberg M. Splitting of the posttermination ribosome into subunits by the concerted action of RRF and EF-G. Mol. Cell. 2005;18:675–686. doi: 10.1016/j.molcel.2005.05.016. [DOI] [PubMed] [Google Scholar]
- 30.Kim K.K., Min K., Suh S.W. Crystal structure of the ribosome recycling factor from Escherichia coli. EMBO J. 2000;19:2362–2370. doi: 10.1093/emboj/19.10.2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nakano H., Uchiyama S., Yoshida T., Ohkubo T., Kato H., Yamagata Y., Kobayashi Y. Crystallization and preliminary X-ray crystallographic studies of a mutant of ribosome recycling factor from Escherichia coli, Arg132Gly. Acta Crystallogr. D. Biol. Crystallogr. 2002;58:124–126. doi: 10.1107/s0907444901019941. [DOI] [PubMed] [Google Scholar]
- 32.Selmer M., Al-Karadaghi S., Hirokawa G., Kaji A., Liljas A. Crystal structure of Thermotoga maritima ribosome recycling factor: a tRNA mimic. Science. 1999;286:2349–2352. doi: 10.1126/science.286.5448.2349. [DOI] [PubMed] [Google Scholar]
- 33.Yoshida T., Uchiyama S., Nakano H., Kashimori H., Kijima H., Ohshima T., Saihara Y., Ishino T., Shimahara H., Yokose K., et al. Solution structure of the ribosome recycling factor from Aquifex aeolicus. Biochemistry. 2001;40:2387–2396. doi: 10.1021/bi002474g. [DOI] [PubMed] [Google Scholar]
- 34.Saikrishnan K., Kalapala S.K., Varshney U., Vijayan M. X-ray structural studies of Mycobacterium tuberculosis RRF and a comparative study of RRFs of known structure. Molecular plasticity and biological implications. J. Mol. Biol. 2005;345:29–38. doi: 10.1016/j.jmb.2004.10.034. [DOI] [PubMed] [Google Scholar]
- 35.Wilson D.N., Schluenzen F., Harms J.M., Yoshida T., Ohkubo T., Albrecht R., Buerger J., Kobayashi Y., Fucini P. X-ray crystallography study on ribosome recycling: the mechanism of binding and action of RRF on the 50S ribosomal subunit. EMBO J. 2005;24:251–260. doi: 10.1038/sj.emboj.7600525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kiel M.C., Raj V.S., Kaji H., Kaji A. Release of ribosome-bound ribosome recycling factor by elongation factor G. J. Biol. Chem. 2003;278:48041–48050. doi: 10.1074/jbc.M304834200. [DOI] [PubMed] [Google Scholar]
- 37.Seo H.S., Kiel M., Pan D., Raj V.S., Kaji A., Cooperman B.S. Kinetics and thermodynamics of RRF, EF-G, and thiostrepton interaction on the Escherichia coli ribosome. Biochemistry. 2004;43:12728–12740. doi: 10.1021/bi048927p. [DOI] [PubMed] [Google Scholar]
- 38.Ito K., Fujiwara T., Toyoda T., Nakamura Y. Elongation factor G participates in ribosome disassembly by interacting with ribosome recycling factor at their tRNA-mimicry domains. Mol. Cell. 2002;9:1263–1272. doi: 10.1016/s1097-2765(02)00547-6. [DOI] [PubMed] [Google Scholar]
- 39.O'Donnell S.M., Janssen G.R. Leaderless mRNAs bind 70S ribosomes more strongly than 30S ribosomal subunits in Escherichia coli. J. Bacteriol. 2002;184:6730–6733. doi: 10.1128/JB.184.23.6730-6733.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Udagawa T., Shimizu Y., Ueda T. Evidence for the translation initiation of leaderless mRNAs by the intact 70 S ribosome without its dissociation into subunits in eubacteria. J. Biol. Chem. 2004;279:8539–8546. doi: 10.1074/jbc.M308784200. [DOI] [PubMed] [Google Scholar]
- 41.Moll I., Hirokawa G., Kiel M.C., Kaji A., Blasi U. Translation initiation with 70S ribosomes: an alternative pathway for leaderless mRNAs. Nucleic Acids Res. 2004;32:3354–3363. doi: 10.1093/nar/gkh663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Adhin M.R., van Duin J. Scanning model for translational reinitiation in eubacteria. J. Mol. Biol. 1990;213:811–818. doi: 10.1016/S0022-2836(05)80265-7. [DOI] [PubMed] [Google Scholar]
- 43.Dincbas V., Heurgue-Hamard V., Buckingham R.H., Karimi R., Ehrenberg M. Shutdown in protein synthesis due to the expression of mini-genes in bacteria. J. Mol. Biol. 1999;291:745–759. doi: 10.1006/jmbi.1999.3028. [DOI] [PubMed] [Google Scholar]
- 44.Hernandez-Sanchez J., Valadez J.G., Herrera J.V., Ontiveros C., Guarneros G. lambda bar minigene-mediated inhibition of protein synthesis involves accumulation of peptidyl-tRNA and starvation for tRNA. EMBO J. 1998;17:3758–3765. doi: 10.1093/emboj/17.13.3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cruz-Vera L.R., Magos-Castro M.A., Zamora-Romo E., Guarneros G. Ribosome stalling and peptidyl-tRNA drop-off during translational delay at AGA codons. Nucleic Acids Res. 2004;32:4462–4468. doi: 10.1093/nar/gkh784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brun G., Paulin D., Yot P., Chapeville F. Peptidyl-tRNA hydrolase: demonstration in various organisms. Enzymatic activity in the presence of ribosomes. Biochimie. 1971;53:225–231. doi: 10.1016/s0300-9084(71)80054-8. [DOI] [PubMed] [Google Scholar]
- 47.Gao N., Zavialov A.V., Li W., Sengupta J., Valle M., Gursky R.P., Ehrenberg M., Frank J. Mechanism for the disassembly of the posttermination complex inferred from cryo-EM studies. Mol. Cell. 2005;18:663–674. doi: 10.1016/j.molcel.2005.05.005. [DOI] [PubMed] [Google Scholar]
- 48.Yamami T., Ito K., Fujiwara T., Nakamura Y. Heterologous expression of Aquifex aeolicus ribosome recycling factor in Escherichia coli is dominant lethal by forming a complex that lacks functional co-ordination for ribosome disassembly. Mol. Microbiol. 2005;55:150–161. doi: 10.1111/j.1365-2958.2004.04387.x. [DOI] [PubMed] [Google Scholar]
- 49.Atarashi K., Kaji A. Inhibitory effect of heterologous ribosome recycling factor on growth of Escherichia coli. J. Bacteriol. 2000;182:6154–6160. doi: 10.1128/jb.182.21.6154-6160.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]