Abstract
A particularly challenging problem in chemical biology entails developing systems for modulating the activity of RNA using small molecules. One promising new approach towards this problem exploits the phenomenon of ‘surface borrowing,’ in which the small molecule is presented to the RNA in complex with a protein, thereby expanding the overall surface area available for interaction with RNA. To extend the utility of surface borrowing to include potential applications in synthetic biology, we set out to create an ‘orthogonal’ RNA-targeting system, one in which all components are foreign to the cell. Here we report the identification of small RNA modules selected in vitro to bind a surface-engineered protein, but only when the two macromolecules are bound to a synthetic bifunctional small molecule.
INTRODUCTION
One of the central goals of chemical biology is to discover small molecules that can modulate the function of every gene in the genome. Though such small molecules often produce phenotypes much like those caused by genetic mutations in the target genes, the chemical approach has the often useful advantage of enabling tight control over the timing and dosage of administration. The overwhelming majority of known small molecule modulators do not target genes directly, but instead bind the protein products of genes. Notwithstanding this fact, it now appears that only a modest fraction of proteins are ‘druggable,’ i.e. they possess the distinctive surface features required for high-affinity (Kd <100 nM) interactions with known small (MW <1000 Da) cell-permeable organic molecules (1). This limitation of protein targeting has fueled the search for agents that act upstream on the nucleic acids that encode proteins (2,3). In particular, the globular nature of folded RNA molecules provides some predisposition towards interaction with small molecules; indeed, numerous natural products including the aminoglycoside and macrolide antibiotics are known to form stable interactions with folded RNA (4,5). Whereas nature has been reasonably successful at discovering tight-binding small molecule ligands to RNA, de novo discovery of such ligands in the laboratory has proven to be exceedingly challenging. With few exceptions, synthetic ligands bind with modest affinity (Kd >500 nM) or fail to penetrate cells efficiently, or both. Considerable interest has thus arisen in novel strategies to boost the affinity and specificity of small molecule-RNA interactions.
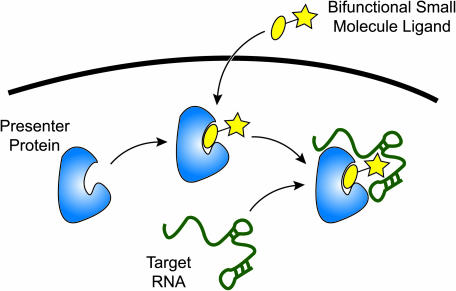
Several years ago we proposed (6) a ‘presenter protein strategy’ (7) to RNA targeting. In this scheme (Figure 1), a small synthetic molecule is designed to possess two binding moieties linked together, one that targets an intracellular protein and another that targets RNA. Upon entry into the cell, the bipartite ligand associates with its protein receptor to form a composite surface which is available for interaction with RNA. Stated otherwise, the small molecule ‘borrows’ the surface of its presenter protein, thus expanding the overall surface area that can come into contact with the RNA target. An obvious attraction of this strategy is that the presenter protein can provide a positively charged binding surface to the RNA, alleviating the impetus to incorporate positive charges in the small molecule, as such charges tend to impede cell permeability. Proof-of-concept for the presenter protein strategy has been reported by Pelletier et al., who found a surprisingly high degree of cooperativity when streptavidin was used to present a biotin–tobramycin conjugate to a tobramycin-binding aptamer (8). The application of this system to regulating gene expression in cells is hampered by the cell-impermeability of the biotin–tobramycin conjugate (8) and its likely interactions with many biotin-utilizing proteins in the cell.
Figure 1.
A presenter protein strategy for targeting RNA. A bipartite small molecule (yellow) composed of a known protein-binding ligand fused to a putative RNA-interacting ligand enters the cell and is captured by its presenter protein partner (blue). Once formed, the complex displays a composite protein-small molecule surface which may bind tightly and specifically to a cellular RNA target. Contacts from both the protein and small molecule are critical for high affinity binding and specific recognition of the RNA.
The goal of the present study was to develop and thoroughly characterize a small molecule RNA-targeting system based on the presenter protein strategy, in which all three components of the system would be foreign to the cell. In principle, this would allow assembly of the ternary protein-small molecule-RNA complex in the cell without interference from other cellular components, and with the ability to control the intracellular concentrations of all three components. By fusing our RNA module to a target RNA of interest, the presenter protein could readily be recruited to the RNA target upon administration of the small molecule. Fusions of the presenter protein with localization domains, RNA modifying enzymes, or other functional units would allow further control over the expression, activity or localization of the targeted RNA. Here we describe the combined use of synthetic chemistry, protein design and RNA selection technology to produce such a cell-orthogonal RNA targeting system.
MATERIALS AND METHODS
Constructs and protein purification
The coding sequences for FKBP* and FKBP*3R were inserted into pTAG2K (9) to generate constructs (tFKBP* and tFKBP*3R, respectively) with an N-terminal 38 amino acid streptavidin-binding peptide (SBP) tag followed by a His6 tag and then the FKBP*- or FKBP*3R-encoding sequence. These protein-coding cassettes were then cloned into a pET24b expression vector. The sequence encoding FKBP*3R with no SBP tag was inserted into pET30b, to give an overproduction vector for His6-FKBP*3R. Proteins were overexpressed in E.coli BL21 (DE3) CodonPlus (Stratagene) and purified with Ni-NTA agarose (Qiagen). His6-FKBP*3R was treated with enterokinase (New England Biolabs) to remove the His6 tag and the untagged FKBP*3R was purified to homogeneity by Mono-S cation exchange chromatography (Amersham Pharmacia Biotech). For tFKBP* and tFKBP*3R, a similar procedure was used, but the enterokinase cleavage was omitted, and for the in-line cleavage assays it was necessary to further purify the proteins using Superdex-200 gel filtration chromatography (Amersham Pharmacia Biotech).
Synthesis of small-molecule ligands
Guanine derivatives 2G, 4G and 8G were prepared by reaction of N9-trityl protected 2-bromohypoxanthine with various lengths of diamine. The resulting amines were then coupled to AP1867 and deprotected. 2A was synthesized similarly from N9-(2-trimethylsilylethoxymethyl)-6-chloropurine. Spectroscopic data for 2G, 4G, 8G and 2A are available online (see Supplementary Data).
In vitro RNA selection
DNA containing a 60 nt region of random sequence (5′-CCCAAGCTTACGTTCAGACCAN60CAATGCGATCCAATGCCCTATAGTGAGTCGTATTAGAATTCCG; N = all 4 nt) was synthesized on an Millipore Expedite DNA synthesizer (1 µmol scale) using a 3:3:2:2 ratio of A:C:G:T and purified by 10% denaturing PAGE. A radiolabeled RNA pool containing ∼6.5 × 1014 unique molecules was obtained from this template annealed to a T7 promoter-containing primer (5′-CGGAATTCTAATACGACTCACTATAGGGCATTGGATCGCATTG) by transcription with T7 RNA polymerase and ∼40 µCi [α-32P]UTP. RNA was treated with RQ1 DNase (Promega), purified by PAGE and refolded by heating at 80°C for 3 min, followed by slow cooling to room temperature.
A negative selection column was generated by incubating Ultralink Immobilized Streptavidin Beads (Pierce) with an equal volume of 20 µM tFKBP*3R in 1× selection buffer [50 mM potassium phosphate (pH 7.4), 5 mM Mg(OAc)2, 150 mM KCl and 1 mM DTT]. This pre-column was washed (beads retain roughly 5 µM protein); 100 µM refolded RNA was applied and the flow through was collected in the selection column [prepared as above with 20 µM tFKBP*3R, 20 µM 2G, 0.2% DMSO and 160 U RNasin (Promega)]. The column was washed with 20 column volumes of 1× selection buffer and bound complexes were eluted with 3 column volumes of elution buffer [2 mM biotin, 50 mM potassium phosphate (pH 7.4), 5 mM Mg(OAc)2, 150 mM KCl and 1 mM DTT]. The elutions were pooled, desalted, reverse transcribed with SuperScript II RT (reverse primer: 5′-CCCAAGCTTACGTTCAGACCA) and amplified by PCR. Six additional rounds of selection were performed similarly except that Immunopure Immobilized Streptavidin Beads (Pierce) were used, as was a specific elution buffer [100 µM FKBP*3R, 100 µM 2G, 50 mM potassium phosphate (pH 7.4), 5 mM Mg(OAc)2, 150 mM KCl, 1 mM DTT, 0.1% DMSO and 8 U RNasin] with an overnight incubation.
Nitrocellulose binding assays
PCR product from round seven was cloned into pCR2.1-TOPO (Invitrogen) and 48 clones were sequenced. The 39 unique sequences were PCR amplified, transcribed, treated with calf intestinal phosphatase (CIP) and end labeled with [γ-32P]ATP. RNA (∼500 pM) was incubated with varying concentrations of a 1:1 mixture of tFKBP*3R and 2G (from 500 pM to 5 µM) in 1× binding buffer [50 mM potassium phosphate (pH 7.4), 5 mM Mg(OAc)2 and 150 mM KCl] for 45 to 90 min at room temperature. Samples (100 µl total volume) were applied to a 0.45 µm nitrocellulose filter (BioRad) in a 96-well dot blot apparatus, washed with 200 µl 1× binding buffer and quantified by phosphorimaging. Specificity assays were performed similarly, using a fixed concentration of 500 nM protein and 500 nM small molecule and data reflect the average of four measurements.
Mutations and truncations were introduced by PCR and cloned into pCR4Blunt-TOPO (Invitrogen). All mutated RNA sequences except M1, M2, M9 and the U75A and A79C mutants also contained a 3′-deletion of five bases that had no effect on binding affinity. Sequence-verified clones were PCR amplified, transcribed, gel purified and labeled. Binding measurements were carried out as described above, with tFKBP*3R-2G varying from 100 pM to 1 µM and ∼100 pM RNA. For M2, tFKBP*3R-2G was assayed from 1 nM to 5 µM. Competition experiments used 500 nM tFKBP*3R-2G, 500 nM to 50 µM competitor (guanine or the N-methyl amide derivative of AP1867) and quantification represents the mean of six assays.
In-line cleavage assays
Refolded 5′ 32P-labeled RNA (∼0.2 pmol and 2 nM) was incubated in in-line cleavage buffer [20 mM MgCl2, 50 mM Tris (pH 8.3) and 100 mM KCl] at room temperature alone or in the presence of 1 µM tFKBP*3R, 1 µM 2G, 1 µM tFKBP*3R-2G complex or 1 µM tFKBP*3R-2A complex. After 40 h, the reactions were extracted with phenol and CHCl3, ethanol precipitated and loaded to an 8% sequencing gel in 10 M urea/1.5 mM EDTA. Gels were run at 40 W for 3.5–4 h, dried and exposed to an imaging plate.
Cell permeability of small molecules
ARIAD Gene Therapeutics, Inc. kindly provided the HT1080 human fibrosarcoma cell line 41–5 and a sample of AP1889. Experimental details have been reported previously (10).
RESULTS
Design and synthesis of small molecule ligands for RNA targeting
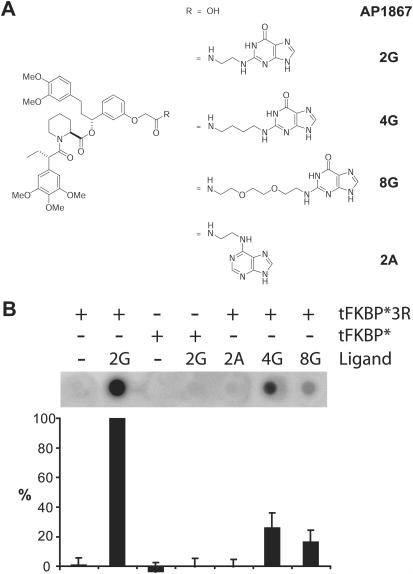
The synthetic ligand AP1867 (Figure 2A) binds with high affinity (0.094 nM) to an engineered version of the FK506-binding protein designated as FKBP* (F36V-FKBP), but has considerably weaker affinity for wild-type FKBP (67 nM) (11). Therefore, under appropriate conditions, AP1867 and its derivatives will preferentially interact with the engineered variant FKBP* over wild-type FKBP in the cell (11). In prior studies, we established that bipartite molecules containing a wide variety of heterocyclic ligands tethered via the carboxyl group of AP1867 bind with high affinity to FKBP* and readily penetrate mammalian cells (10). Importantly, this carboxyl group is surface-exposed in the X-ray structure of the FKBP*-AP1867 complex (11), making it a suitable attachment point for presenting prospective RNA-binding ligands from the protein surface. Given the propensity of RNA to form strong interactions through hydrogen bonding and hydrophobic stacking between nucleobases, we reasoned that nucleobases should be especially effective heterocyclic ligands for use in RNA targeting. This notion drew additional support from reports of RNA aptamers specific for nucleobases (12,13) and nucleotides (14,15). Thus, using a straightforward amide coupling procedure, we conjugated several alkylamine-bearing guanine nucleobases to AP1867, to furnish the bipartite ligands 2G (two-atom tether), 4G (four-atom tether) and 8G (eight-atom tether) (Figure 2A). As a control for nucleobase-specificity, we synthesized the adenine analog of 2G, namely 2A.
Figure 2.
Specificity of the RNA targeting system. (A) Structures of the small molecules used in this study. (B) Characterization of s23. Binding of the s23 aptamer to the 4G-, 8G- or 2A-tFKBP*3R complexes is indicated as a percentage of binding to tFKBP*3R-2G. s23 is highly specific for tFKBP*3R-2G relative to all other protein-small molecule combinations tested. A representative blot is shown above the graph.
Engineering the orthogonal presenter protein FKBP*3R
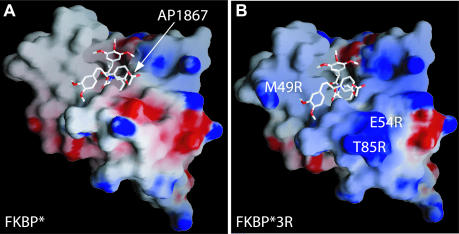
The presenter protein in our studies could contact the target RNA through a combination of electrostatic, hydrogen bonding and non-polar interactions; of these, electrostatic interactions are likely to be especially important, because the surface of RNA has such a high density of negative charge. Inspection of the surface of FKBP* in the vicinity of the ligand-binding site (Figure 3A) reveals the presence of an acidic residue, E54, which might repel RNA. To convert a potentially repulsive interaction into an attractive one, E54 was mutated to Arg. We also mutated two nearby uncharged residues, M49 and T85, to Arg. The resulting triple-mutant protein (M49R, E54R and T85R), designated as FKBP*3R, has abundant positive charge on its ligand-binding surface (Figure 3B). Finally, to provide a defined point of attachment to streptavidin beads, we fused to the N-terminus of the protein a 38 amino acid streptavidin-binding peptide tag (9), to give tFKBP*3R. Although tFKBP*3R expressed at moderately lower levels than tFKBP*, the behavior of the triple-mutant protein on size-exclusion chromatography indicates that it is folded and monomeric; melting temperatures obtained by circular dichroism (CD) measurements also indicate similar thermal stabilities for the two proteins (data not shown).
Figure 3.
Engineering the surface of FKBP*. (A) An electrostatic potential surface (GRASP) representation of FKBP* was generated from the FKBP*-AP1867 co-crystal structure (11). The ligand-binding face of FKBP* shows a surface with sparse patches of positive charge (blue). A region of negative charge (red) is also evident near the ligand-binding pocket. (B) A model of the mutant protein FKBP*3R has a surface lacking this potentially repulsive patch of negative charge while gaining additional sites with positive charge. AP1867 is rendered as a framework model, with carbon atoms shown in white, oxygen in red and nitrogen in blue. The arrow points to the carboxyl group of AP1867 that serves as the site of linkage to a guanine or adenine base.
Aptamer selections
We reasoned that in vitro selection technology (SELEX, Systematic Evolution of Ligands by Exponential Enrichment) (16,17) would enable us to discover an RNA aptamer that binds the composite tFKBP*3R-2G interface and is likely to be distinct in sequence from any RNA native to the cell. This technology has been widely utilized to discover RNA aptamers specific for a variety of small molecule ligands, but to our knowledge has not been used to identify aptamers that cooperatively bind a composite small molecule-protein surface.
Several steps were taken to ensure that only RNAs with high affinity and specificity for the composite protein-small molecule surface were enriched during the selection (see Materials and Methods for additional details). To prevent amplification of RNA sequences that bound to the column matrix or, importantly, to tFKBP*3R alone, a negative selection step was performed by pre-filtering the RNA pool through a column of tFKBP*3R-loaded streptavidin beads. The eluate was then incubated with streptavidin beads containing the small molecule 2G bound to tFKBP*3R. In round 1, all the bound RNA was eluted non-specifically from the column using biotin, while in subsequent rounds, aptamers were recovered by specific elution with non-tagged FKBP*3R-2G. To further favor RNAs that bound the composite FKBP*3R-2G surface, pre-elution with FKBP*3R alone was performed in rounds 4 and 5. Finally, to dislodge weakly bound aptamers, a short pre-elution with FKBP*3R-2G was performed in late rounds (4–6) prior to the overnight elution step.
After 7 rounds of selection and amplification, 48 individual clones were sequenced and the resulting 39 unique RNA sequences were analyzed by nitrocellulose filter binding assays for their ability to bind tFKBP*3R in a small molecule-dependent manner (Supplementary Figure 1). Of these, 16 (41%) clearly showed enhanced binding to tFKBP*3R in the presence of 2G. Quantitative dot blot assays on this panel of 16 RNA molecules revealed 11 that bound tFKBP*3R-2G with an apparent Kd below 50 nM (Supplementary Table 1). These were chosen for further study.
Small molecule- and protein surface-specific recognition
The 11 high-affinity aptamers were assayed for their specificity in binding to the selection target, with respect to both the small molecule and the protein. The ability of each aptamer to discriminate between closely related small molecules was assessed by measuring aptamer-tFKBP*3R binding in the presence of small molecules that varied in linker length (two- versus four- versus eight-atom) or base identity (G versus A) (Figure 2A). Across the panel of 11 aptamers, specificity for a tethered guanine base was exquisite, with 8 of the 11 aptamers showing almost complete (>85%) loss of binding when 2A was substituted for 2G (Supplementary Figure 2). The effects of extending the tether length were less pronounced, on average. Changing the small molecule from 2G to 8G, a six-atom increase in the tether length, resulted in binding levels that ranged between 10 and 60% of the level for 2G; most sequences (7 out of the 11) were grouped between 25 and 50%. Moderate changes were seen for 4G; 9 out of 11 sequences bound nearly as well (>60%) as they bound 2G. However, the aptamer designated s23 exhibited exceptionally high specificity for both the base structure and tether length, as shown in Figure 2B.
Specificity for the surface composition of the presenter protein was assessed by comparing binding of the RNA to tFKBP*3R versus tFKBP* (see Figure 3). All but one of the 11 aptamers, including s23 (Figure 2B) exhibited a dramatic (>85%) loss of binding when tFKBP* was substituted for tFKBP*3R (Supplementary Figure 2). One noteworthy outlier, s36, retained more than 50% binding to tFKBP*. For the more detailed characterization described below, we chose to focus exclusively on s23, owing to the superior specificity of this particular aptamer.
Cooperativity in formation of the ternary tFKBP*3R-2G-RNA complex
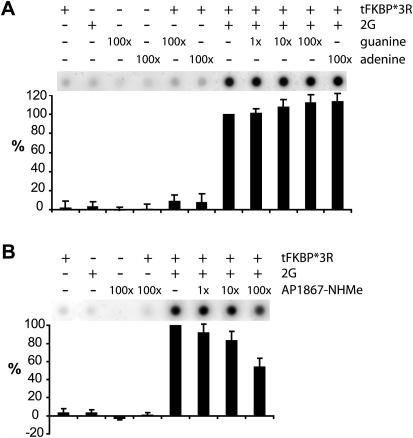
The selection was designed to capture aptamers eluted by the composite protein-small molecule surface while eliminating those that interacted only with tFKBP*3R or 2G individually. We sought to verify that we had successfully obtained aptamers that bound cooperatively to the tFKBP*3R-2G complex. Cooperative binding can be demonstrated by an enhanced affinity of the RNA for the binary tFKBP*3R-2G complex, as opposed to the additive contributions of binding to either tFKBP*3R or 2G alone. We determined that the s23 aptamer binds 2G with a Kd of 4.6 ± 0.8 µM (Supplementary Figure 3). On the other hand, binding of the s23 aptamer to tFKBP*3R at a concentration of 5 µM protein was undetectable (Supplementary Figure 3), indicating a Kd well above this level. In contrast, the affinity of s23 for the tFKBP*3R-2G complex was determined to be 4.3 ± 0.5 nM (Supplementary Figure 3), more than three orders of magnitude below that observed with either the small molecule or protein alone. This large preference clearly establishes that the formation of the ternary complex is cooperative. Further evidence for cooperative assembly of the ternary complex was gained through competition experiments with the separated parts of 2G. The nucleobase guanine, even when present in a 100-fold excess over 2G, failed completely to compete against formation of the ternary complex (Figure 4A). The N-methyl amide derivative of AP1867, when present in 100-fold excess over 2G, did show significant levels of competition (∼45%, Figure 4B). Taken together, these experiments unambiguously establish that the ternary complex is more thermodynamically stable, by at least two orders of magnitude, than either of the constituent binary complexes.
Figure 4.
Competition with the separated components of 2G. (A) Guanine is ineffective at disrupting the tFKBP*3R-2G-s23 ternary complex. Labeled s23 RNA binds only when both tFKBP*3R and 2G are present and binding cannot be competed away by at least a 100-fold excess of free guanine base. (B) The N-methyl amide derivative of AP1867 (AP1867-NHMe) does compete with 2G-s23 for binding to tFKBP*3R. In the presence of 100 equivalents of competitor, only 55% of the ternary tFKBP*3R-2G-s23 complex remains. Values are plotted with tFKBP*3R-2G-s23 binding set to 100 percent.
Ligand binding orders RNA structure
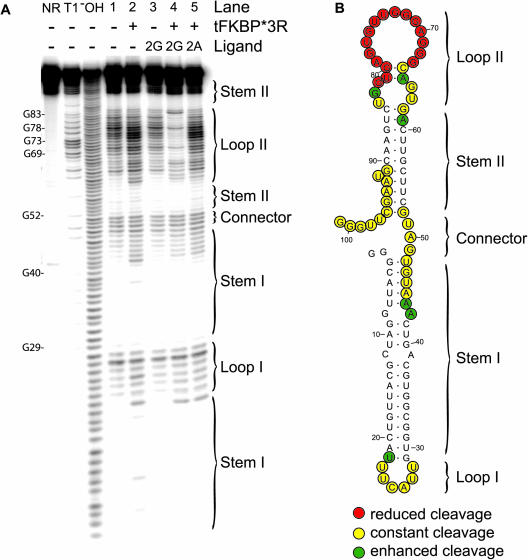
To characterize the binding site on s23 for the tFKBP*3R-2G complex, we used in-line RNA cleavage assays (18) to map sites that undergo conformational reorganization upon ligand binding. Spontaneous cleavage was promoted by incubating s23 RNA at elevated pH and high Mg2+ concentration in the presence or absence of small molecule and/or protein. In-line cleavage data for unbound s23 RNA (Figure 5A, lane 1) correlates well with the computationally predicted secondary structure determined by the RNA fold program (http://rna.tbi.univie.ac.at), which consists of two stem–loops (I and II) separated by a Connector (Figure 5B). Nucleotides belonging to Stem I or to the loop-proximal region of Stem II have substantially retarded cleavage, while Loops I and II, and the Connector readily undergo cleavage.
Figure 5.
Structure probing of s23. (A) s23 RNA subjected to in-line probing conditions shows a distinctly altered pattern of cleavage only in the presence of tFKBP*3R and 2G (lane 4). Protein alone (lane 2) or small molecule alone (lane 3) has little effect on the cleavage pattern that is seen for unbound RNA (lane 1), which is to be expected since both of these components bind s23 RNA with only very weak affinity. NR: unreacted RNA; T1: partial RNase T1 digestion; −OH: alkaline hydrolysis. (B) The predicted secondary structure of the s23 aptamer is shown, along with in-line cleavage data from part (A), lane 4. Yellow: bases that undergo constant levels of cleavage whether tFKBP*3R-2G is bound to s23 or not. Red: bases that have diminished levels of cleavage in the presence of tFKBP*3R-2G. Green: bases at which cleavage is enhanced.
In the presence of the selection target, a pronounced reduction in cleavage was observed at nearly all positions in a 16 nt segment of s23 (Figure 5A). This region corresponds precisely to Loop II, which caps the 11 bp Stem II (Figure 5B). Further corroborating the cooperativity of ternary complex formation, the altered pattern of in-line cleavage at Loop II was observed only with reactions containing both tFKBP*3R and 2G (Figure 5A, lane 4), but not in those containing either the small molecule or protein alone (lanes 2 and 3). When 2A is substituted for 2G (lane 5), cleavage is not reduced, further illustrating the specificity of ternary complex formation. Modest levels of enhanced cleavage in the presence of tFKBP*3R (lanes 2 and 5) are likely due to a residual RNase activity that co-purified with the protein, as extensive purification was critical to avoiding RNA degradation over the course of the in-line cleavage reaction. These results, taken together with those presented above, point to Loop II as the likely site of direct contacts between s23 and tFKBP*3R-2G.
Identification of minimal sequence and critical contacts
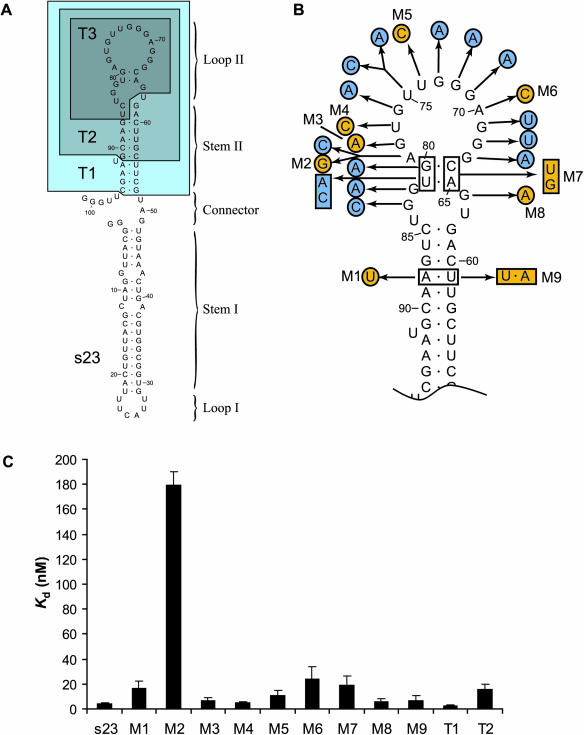
The results of in-line footprinting analysis suggested that Loop II constitutes all or part of the protein-small molecule binding site for the s23 aptamer, and hence that Stem I, Loop I, the Connector, and perhaps even portions of Stem II may be dispensable. In order to evaluate that possibility, we generated truncated versions of s23 in which portions of the sequence corresponding to structural elements were systematically trimmed away (Figure 6A). A truncation variant (T1, nucleotides 52–96) having Stem I, Loop I, the Connector and the 3′-flap deleted bound tFKBP*3R-2G complex with essentially the same affinity as s23 (2.8 ± 0.2 versus 4.3 ± 0.5 nM, respectively), confirming that the protein-small molecule binding site is localized entirely to Stem II/Loop II. Further removal of the 4 bp at the end of Stem II (T2, nucleotides 56–91) resulted in a ∼6-fold loss in binding affinity for the tFKBP*3R-2G complex (16.1 ± 3.6 nM). A 25 nt truncation variant comprising only Loop II had a Kd near 1 µM, thus indicating that Stem II plays an important role in recognition of the tFKBP*3R-2G complex.
Figure 6.
Truncation and mutational analysis of s23. (A) The bases comprising the s23 truncations T1, T2 and T3 are boxed. Truncation variants T1 and T2 bind tFKBP*3R-2G with affinities similar to that of the full-length s23, but complete removal of Stem II in variant T3 results in greatly weakened binding. (B) Point or double mutations that were introduced into s23. Modifications that result in a Kd over 350 nM are indicated in blue; those that bind with tighter affinities are colored orange. (C) Binding affinities of s23 mutations and truncations that are tolerated by the tFKBP*3R-2G complex.
In order to identify nucleobases of the aptamer that make base-specific contacts to the tFKBP*3R-2G complex, and to evaluate the predicted structure, we generated a series of mutations that extensively probed the binding site loop. The majority of loop alterations were not tolerated, showing no detectable levels of binding at a concentration of 1 µM tFKBP*3R-2G (Figure 6B). These sites most likely represent positions that make important contributions to the tertiary fold of s23 RNA or that make base-specific contacts to tFKBP*3R, the guanine base of 2G, or to both. Several mutations in Loop II had only moderate effects on tFKBP*3R-2G binding (Figure 6C), indicating that not all positions are critical for binding.
Cell permeability of the bifunctional small molecules
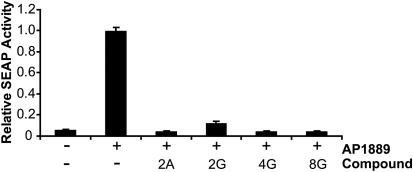
The ability of the bifunctional small molecules to cross the membrane of mammalian cells was assessed using an established secreted alkaline phosphatase (SEAP) assay (10,19,20). In this assay, a small-molecule protein–protein dimerizer recruits an FKBP*-activation domain fusion protein to an FKBP*-DNA-binding domain fusion protein, thus driving transcription of the downstream SEAP cDNA. Our bifunctional molecules are able to interact with either of the FKBP* fusion domains in the assay component proteins, but not both, and hence the molecules act as transcriptional antagonists of SEAP expression if they are able to cross the cell membrane. When added to the growth medium of the HT1080 human fibrosarcoma cell line 41–5 at a concentration of 500 nM, 2G, 4G, 8G and 2A all caused significant suppression of SEAP expression (Figure 7). The nucleobase-AP1867 conjugates therefore exhibit significant cell-permeability at sub-micromolar concentrations.
Figure 7.
Cell permeability of nucleobase compounds. HT1080 41–5 cells were treated for 16 h with 10 nM AP1889 and 500 nM nucleobase compound or blank (DMSO). SEAP activity was determined by the conversion of 4-methylumbelliferyl phosphate (MUP) to fluorescent product. Compounds 2A, 2G, 4G and 8G are able to compete with AP1889 for FKBP* binding and block SEAP expression, implying that they are cell-permeable.
DISCUSSION
A number of RNA-binding proteins and small molecules, such as those that bind to metabolite-sensing riboswitches (21–23), are important cellular regulators of gene expression. In the present work we have developed a completely artificial protein-small molecule-RNA system in which all three components are distinct in structure and function from any known materials that occur naturally. The protein component, tFKBP*3R, contains an active site mutation that substantially weakens binding to the biogenic ligand FK506, in addition to three surface mutations that render the surface chemistry of the protein quite distinct from that of wild-type FKBP. The small molecule in these studies has been tailored to bind specifically to the engineered protein FKBP* in the presence of wild-type FKBP, a property borne out in cellular studies. Finally, the s23 aptamer has no known biologically derived counterpart, as a BLAST search for the s23 aptamer or its truncation variant T1 failed to find any significant matches in the public databases. Although at this stage we cannot rule out the possibility that tFKPB*3R-2G will also bind certain cellular RNA targets, high-affinity binding sites are likely to be rare, as judged by the course of the RNA selection experiments. Thus, if the system were introduced into cells, it is probable that the tFKBP*3R-2G complex will bind tightly only to targeted RNAs to which s23 or T1 had been appended. Since s23 or T1 do not bind tFKBP*3R appreciably in the absence of 2G, we expect to be able to control protein-RNA association in cells simply by adding 2G to cells already expressing the s23 or T1 fusion RNA and tFKBP*3R. Judging by the results of the competition assays in Figure 4, it should be possible to antagonize 2G by adding excess AP1867 or its N-methyl amide derivative.
The results of competition experiments and Kd determinations demonstrate that the ternary tFKBP*3R-2G-s23 complex forms cooperatively, with the small molecule-RNA interaction being stabilized at least 102-fold (2.8 kcal/mol) as the result of presentation by tFKBP*3R. From a practical standpoint, cooperative formation of the complex is important, because it confers resistance to competition by excess small molecule, and favors ternary complex formation at low concentrations of small molecule relative to protein or RNA. The origin of cooperativity in this system is interesting in its own right. In principle, cooperativity could either arise directly, as the result of stabilizing intermolecular contacts between the protein and RNA, or indirectly, from a protein-induced change in the conformation of the ligand that makes the ligand more effective at RNA binding. We are strongly inclined towards the former explanation, because the flexible linker in our ligand is not well suited to transmit conformational effects, and because ternary complex formation is strongly affected by amino acid changes on a well-ordered segment of the protein surface (tFKBP*3R versus tFKBP*). Other aptamers selected in this study may yet display cooperative binding; especially interesting in this regard is s36, which is only modestly affected by the differences in surface chemistry between tFKBP*3R and tFKBP*.
Also of fundamental interest is the nature of the binding interaction between the tethered guanine base in 2G and the RNA aptamers. Recent evidence (24) suggests that guanine- and adenine-binding riboswitches interact with the bound nucleobase through Watson–Crick base pairing. It seems unlikely that this is the operative mechanism for 2G recognition of s23, since the only cytosine present in the putative protein-small molecule-binding loop of the aptamer is non-essential. Wobble-type G/U pairing interactions could possibly play a role at U75, since mutation of this base to either C or A results in greatly diminished binding (>1 µM); wobble interactions with U74 or U77 are ruled out on the basis that mutation to C has almost no effect on binding (Kd = 10.8 ± 3.8 and 5.3 ± 0.8 nM versus 4.3 ± 0.5 nM). Further elucidation of the detailed interactions within the protein-small molecule-RNA complex awaits high resolution structural studies.
In summary, we have created a system by which to induce the binding of a small molecule-protein complex to RNA. Implementation of the system in cells would involve engineering the cells to express tFKBP*3R or perhaps a fusion protein thereof, express the target RNA fused to T1 or s23, and then addition of 2G to the culture medium to trigger formation of the ternary complex. We envisage that this system will serve as a useful tool for a broad range of studies on gene expression and protein function, by enabling temporal control to be exerted over the sub-cellular localization, tissue distribution, splicing and translation of RNA. Efforts along these lines are underway.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Supplementary Material
Acknowledgments
The authors are grateful to Chris Radom for providing a cDNA cassette encoding FKBP*3R, to Ken Wickiser and other members of the Breaker lab for advice on in-line cleavage assays, and to Scott Baskerville for useful advice in the early stages of this project. K.A.P. is the recipient of a National Defense Science and Engineering Graduate Fellowship, M.Y. was supported by the Naito Foundation, and J.M.C. was supported in part by a US National Science Foundation Graduate Fellowship. This work was supported in part by a grant from the US National Institutes of Health (GM53936 to J.W.S). J.W.S. is an investigator of the Howard Hughes Medical Institute. Funding to pay the Open Access publication charges for this article was provided by the Dana Farber/Harvard Program in Cancer Chemical Biology.
Conflict of interest statement. None declared.
REFERENCES
- 1.Hopkins A.L., Groom C.R. The druggable genome. Nature Rev. Drug. Discov. 2002;1:727–730. doi: 10.1038/nrd892. [DOI] [PubMed] [Google Scholar]
- 2.Gottesfeld J.M., Neely L., Trauger J.W., Baird E.E., Dervan P.B. Regulation of gene expression by small molecules. Nature. 1997;387:202–205. doi: 10.1038/387202a0. [DOI] [PubMed] [Google Scholar]
- 3.Werstuck G., Green M.R. Controlling gene expression in living cells through small molecule-RNA interactions. Science. 1998;282:296–298. doi: 10.1126/science.282.5387.296. [DOI] [PubMed] [Google Scholar]
- 4.Tor Y. Targeting RNA with small molecules. Chembiochem. 2003;4:998–1007. doi: 10.1002/cbic.200300680. [DOI] [PubMed] [Google Scholar]
- 5.Vicens Q., Westhof E. RNA as a drug target: the case of aminoglycosides. Chembiochem. 2003;4:1018–1023. doi: 10.1002/cbic.200300684. [DOI] [PubMed] [Google Scholar]
- 6.Stanton J., Vincent P., Basilion J.P., Shen L.X., Verdine G.L. 0052210. EU patent. 2000
- 7.Briesewitz R., Ray G.T., Wandless T.J., Crabtree G.R. Affinity modulation of small-molecule ligands by borrowing endogenous protein surfaces. Proc. Natl Acad. Sci. USA. 1999;96:1953–1958. doi: 10.1073/pnas.96.5.1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harvey I., Garneau P., Pelletier J. Forced engagement of a RNA/protein complex by a chemical inducer of dimerization to modulate gene expression. Proc. Natl Acad. Sci. USA. 2002;99:1882–1887. doi: 10.1073/pnas.042693399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Keefe A.D., Wilson D.S., Seelig B., Szostak J.W. One-step purification of recombinant proteins using a nanomolar-affinity streptavidin-binding peptide, the SBP-Tag. Protein Expr. Purif. 2001;23:440–446. doi: 10.1006/prep.2001.1515. [DOI] [PubMed] [Google Scholar]
- 10.Koide K., Finkelstein J.M., Ball Z., Verdine G.L. A synthetic library of cell-permeable molecules. J. Am. Chem. Soc. 2001;123:398–408. doi: 10.1021/ja0023377. [DOI] [PubMed] [Google Scholar]
- 11.Clackson T., Yang W., Rozamus L.W., Hatada M., Amara J.F., Rollins C.T., Stevenson L.F., Magari S.R., Wood S.A., Courage N.L., et al. Redesigning an FKBP-ligand interface to generate chemical dimerizers with novel specificity. Proc. Natl Acad. Sci. USA. 1998;95:10437–10442. doi: 10.1073/pnas.95.18.10437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kiga D., Futamura Y., Sakamoto K., Yokoyama S. An RNA aptamer to the xanthine/guanine base with a distinctive mode of purine recognition. Nucleic Acids Res. 1998;26:1755–1760. doi: 10.1093/nar/26.7.1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Meli M., Vergne J., Decout J.L., Maurel M.C. Adenine-aptamer complexes: a bipartite RNA site that binds the adenine nucleic base. J. Biol. Chem. 2002;277:2104–2111. doi: 10.1074/jbc.M107130200. [DOI] [PubMed] [Google Scholar]
- 14.Connell G.J., Yarus M. RNAs with dual specificity and dual RNAs with similar specificity. Science. 1994;264:1137–1141. doi: 10.1126/science.7513905. [DOI] [PubMed] [Google Scholar]
- 15.Sassanfar M., Szostak J.W. An RNA motif that binds ATP. Nature. 1993;364:550–553. doi: 10.1038/364550a0. [DOI] [PubMed] [Google Scholar]
- 16.Ellington A.D., Szostak J.W. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 17.Tuerk C., Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 18.Soukup G.A., Breaker R.R. Relationship between internucleotide linkage geometry and the stability of RNA. RNA. 1999;5:1308–1325. doi: 10.1017/s1355838299990891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Amara J.F., Clackson T., Rivera V.M., Guo T., Keenan T., Natesan S., Pollock R., Yang W., Courage N.L., Holt D.A., et al. A versatile synthetic dimerizer for the regulation of protein–protein interactions. Proc. Natl Acad. Sci. USA. 1997;94:10618–10623. doi: 10.1073/pnas.94.20.10618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pollock R., Rivera V.M. Regulation of gene expression with synthetic dimerizers. Methods Enzymol. 1999;306:263–281. doi: 10.1016/s0076-6879(99)06017-6. [DOI] [PubMed] [Google Scholar]
- 21.Nahvi A., Sudarsan N., Ebert M.S., Zou X., Brown K.L., Breaker R.R. Genetic control by a metabolite binding mRNA. Chem. Biol. 2002;9:1043–1049. doi: 10.1016/s1074-5521(02)00224-7. [DOI] [PubMed] [Google Scholar]
- 22.Winkler W., Nahvi A., Breaker R.R. Thiamine derivatives bind messenger RNAs directly to regulate bacterial gene expression. Nature. 2002;419:952–956. doi: 10.1038/nature01145. [DOI] [PubMed] [Google Scholar]
- 23.Winkler W.C., Cohen-Chalamish S., Breaker R.R. An mRNA structure that controls gene expression by binding FMN. Proc. Natl Acad. Sci. USA. 2002;99:15908–15913. doi: 10.1073/pnas.212628899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mandal M., Breaker R.R. Adenine riboswitches and gene activation by disruption of a transcription terminator. Nature Struct. Mol. Biol. 2004;11:29–35. doi: 10.1038/nsmb710. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.