Abstract
Gram-negative bacteria can communicate with each other by N-acyl homoserine lactones (AHLs), which are quorum-sensing autoinducers. Recently, the aiiA gene (encoding an enzyme catalyzing the degradation of AHL) has been cloned from Bacillus sp. strain 240B1. During investigations in the course of the ongoing Bacillus thuringiensis subsp. morrisoni genome project, an aiiA homologue gene in the genome sequence was found. These results led to consideration of the possibility of the widespread existence of the gene in B. thuringiensis. aiiA homologue genes were found in 16 subspecies of B. thuringiensis, and their sequences were determined. Comparison of the Bacillus sp. strain 240B1 aiiA gene with the B. thuringiensis aiiA homologue genes showed high homologies of 89 to 95% and 90 to 96% in the nucleotide sequence and deduced amino acid sequence, respectively. Among the subspecies of B. thuringiensis having an aiiA gene, the subspecies aizawai, galleriae, kurstaki, kyushuensis, ostriniae, and subtoxicus were shown to degrade AHL. It was observed that recombinant Escherichia coli producing AiiA proteins also had AHL-degrading activity and could also attenuate the plant pathogenicity of Erwinia carotovora. These results indicate that insecticidal B. thuringiensis strains might have potential to compete with gram-negative bacteria in natural ecosystems by autoinducer-degrading activity.
Quorum sensing is a signaling mechanism used by bacteria for monitoring their population density and coordinating gene expression in response to changes in cell density (7). Many gram-negative bacteria secrete and accumulate signaling molecules, N-acyl homoserine lactones (AHLs), as the cells grow, and members of a bacterial strain can communicate with each other by these diffusible quorum-sensing autoinducers. AHLs consist of a lactone ring covalently linked to 4- to 14-carbon acyl side chains through an amide bond (18, 19, 28). These AHL molecules have critical roles in controlling a number of phenotypic traits, such as antibiotic production, biofilm formation, and hydrolytic enzyme production and, especially, the generation of virulence factors in some pathogenic bacteria (10, 18). It has been reported that pathogens use a quorum-sensing system to escape premature detection by host defenses and to overwhelm the host efficiently at the appropriate time (6, 13). In the case of Erwinia carotovora, which causes soft rot in a variety of plants, pathogenicity depends on the production of several exoenzymes which are involved in the maceration of plant tissue. The production of these exoenzymes is regulated by the accumulation of N-3-oxohexanoyl-l-homoserine lactone (10, 22). Erwinia carotovora does not produce plant tissue-degrading enzymes until sufficient bacterial density has been achieved for successful evasion of plant defenses.
Because many pathogenic bacteria use quorum sensing for the regulation of virulence, it is suggested that interfering with the bacterial communication system by disrupting quorum sensing is a way of treating or preventing infection. As one of the anti-quorum-sensing strategies, degradation of AHL-signaling molecules could have potential applications in attenuating plant disease. Signal-degrading enzymes from one bacterium interrupt the signaling among other pathogenic bacteria and can prevent plant disease such as soft rot caused by E. carotovora (5). Recently, autoinducer-degrading microbes or enzymes have been reported: Leadbetter and Greenberg (12) reported that some strains of Variovorax paradoxus could grow in a minimal medium containing an AHL as the sole energy and nitrogen source. Variovorax paradoxus cleaves the acyl-amide bond of AHLs. Also, Dong et al. (4, 5) cloned a novel lactonase gene from a Bacillus sp. that is closely related to the Bacillus cereus group. This gene encodes an enzyme that renders AHLs biologically inactive.
In our company, the microbial genome project of Bacillus thuringiensis subsp. morrisoni has been ongoing, and 90% of the genome has been sequenced. In the database of DNA sequences, an aiiA homologue gene was found in B. thuringiensis subsp. morrisoni. These findings led to consideration of the possibility of the existence of the aiiA gene in the species of B. thuringiensis, a spore-forming gram-positive bacterium. This bacterium produces various insecticidal crystal proteins encoded by cry genes (1, 24). B. thuringiensis has been developed and is presently used as a biological control agent against insect pests in the agriculture and forestry industries (3). In this paper, the distribution of aiiA-homologous genes in the insecticidal B. thuringiensis is reported and the possibility of biological control of plant-pathogenic bacteria by B. thuringiensis is discussed.
MATERIALS AND METHODS
Bacterial strains, culture media, and conditions.
Escherichia coli DH5α and E. coli BL21(DE3) were used as a cloning host and an expression host, respectively. Subspecies of B. thuringiensis were kindly supplied by Institut Pasteur, Paris, France. Bacillus sp. strain IBN35 was isolated from a soil sample, and its partial 16S rDNA sequence was shown to be 99.0% identical to that of B. cereus. Red pepper soft-rot-causative Erwinia carotovora IBN98 was kindly supplied by K.-S. Han (Chung-Nam Agricultural Research and Extension Services, Daejon, Korea). Chromobacterium violaceum CV026 (11, 26) and Agrobacterium tumefaciens NT1 (pDCI41E33) (2) were used as reporter strains for bioassay. Escherichia coli and Bacillus spp. were cultivated in Luria-Bertani medium (1% bactotryptone, 0.5% yeast extract, 1% sodium chloride) at 37°C. If necessary, the medium was supplemented with 5 μg of ampicillin/ml. Cell growth was monitored by measurement of the optical densities at 600 nm (OD600) of culture media. If needed, IPTG (isopropyl-β-d-thiogalactopyranoside) was added to a final concentration of 1 mM at an OD600 of 0.6.
DNA manipulations.
Genomic DNAs were isolated from B. thuringiensis using a modification of a method by Schraft and Griffiths (25). Bacterial cells were washed with 0.85% (wt/vol) NaCl and then suspended in 400 μl of a 200 mM sucrose solution supplemented with lysozyme (400 μg) and RNase A (100 μg). After phenol-chloroform-isoamyl alcohol extractions, 2 volumes of absolute ethanol were added for DNA concentration. Then, the pelleted DNA was dissolved in 50 μl of distilled water. aiiA homologue genes were amplified using chromosomal DNAs of 16 strains of B. thuringiensis as a template and the oligonucleotide primers AIF (5′-TAAATGTAAAGGTGGATACATAATGACAGT [start codon underlined]) and AIR (5′-AGCTCATGACTTTTTGCACTATATATA [start codon underlined]). The PCR conditions involved denaturation at 94°C for 3 min followed by 28 cycles at 94°C for 30 s, 50°C for 30 s, and 72°C for 1 min with PCR Master Mix (Roche). The PCR product was ligated to the T7Blue T-vector (TaKaRa) and sequenced by an ABI3700 automatic sequencer (Applied Biosystems). The DNA sequences were analyzed using the Lasergene sequence analysis program (DNASTAR, Inc.). The aiiA genes were amplified using the chromosomes of B. thuringiensis subsp. morrisoni and B. thuringiensis subsp. kyushuensis as templates and the primer pair 5′-CCCCATATGACAGTAAAAAAGCTTTA and 5′-GGGCTCGAGTATATACTCCGGGAACA (the NdeI and XhoI restriction sites are underlined). The PCR products were digested with NdeI and XhoI, ligated with the NdeI-XhoI-digested pET22b(+) vector (Novagen), and introduced into E. coli BL21(DE3). The resulting aiiA expression vectors were named pETAIM and pETAIK, respectively.
SDS-PAGE analysis.
Sodium dodecyl sulfate-15% polyacrylamide gel electrophoresis (SDS-15% PAGE) was carried out by standard protocols (23). Molecular weight markers (M-3913; Sigma) were purchased from Sigma-Aldrich. Recombinant E. coli BL21(DE3) carrying pETAIM or pETAIK was cultivated for 3 h after IPTG induction, and the cells harvested from 10 ml of culture broth were resuspended in 1 ml of 50 mM Tris-HCl (pH 8.0). The resuspended cells were disrupted by sonication and centrifuged (5,000 × g for 5 min). Subsequently, 20 μl of supernatant was loaded in the polyacrylamide gel.
Bioassay of AHL-degrading activity.
N-octanoyl-l-homoserine lactone (OHL), N-hexanoyl-l-homoserine lactone (HHL), and N-3-oxohexanoyl-l-homoserine lactone (OHHL) were purchased from Quorum Sciences, Inc. Whole cells of B. thuringiensis and E. coli were used for bioassay of AHL-degrading activity. Various strains of B. thuringiensis were cultivated, harvested, and resuspended in 200 mM morpholineethanesulfonic acid buffer (pH 6.5) containing 2 mM ZnSO4 until an OD600 of 1.0 was achieved. Then, 40 μl of the cell resuspension mixture and 40 μl of OHHL (final concentration, 20 μM) were mixed and incubated at 37°C for 1 h with gentle shaking, followed by 95°C for 5 min to stop the reaction. Twenty microliters of sample was loaded in the hole of a CV026-overlaid plate. Recombinant E. coli BL21(DE3) carrying pETAIM or pETAIK was cultivated for 3 h after IPTG induction, and the cells harvested from 3 ml of culture broth were resuspended in 300 μl of morpholineethanesulfonic acid buffer. Forty- microliter volumes of OHHL (final concentration, 10 μM), HHL (final concentration, 10 μM), and OHL (final concentration, 10 μM) were added to 40-μl volumes of the cell suspensions and mixed. The sample was incubated at 37°C for 1 h, followed by incubation at 95°C for 5 min to stop the reaction. After heating, the original AHLs in the samples were diluted until they reached the appropriate concentrations. Subsequently, 5 μl of OHHL (0.1 μM) and 25 μl of OHL (1 μM) were loaded in the holes of NT1 (pDCI41E33)-overlaid plates and 50 μl of OHHL (5 μM) and 30 μl of HHL (1 μM) were loaded in the holes of CV026-overlaid plates. The plates were incubated at 30°C for 16 h for color development. The residual amounts of AHLs were evaluated according to the decrease in size of purple- and blue-colored areas around the holes in the CV026 and NT1 (pDCI41E33) plates, respectively.
Virulence tests.
Erwinia carotovora strain IBN98 was cultivated until the OD600 was 1.0 and was then diluted with saline solution (0.15 M NaCl). Recombinant E. coli carrying pETAIK was harvested 3 h after IPTG induction and resuspended in saline solution to a final OD600 of 5. Equal volumes of Erwinia carotovora IBN98 (OD600 = 0.05) and recombinant E. coli (OD600 = 5) were mixed. Twenty microliters of the mixture was loaded onto potato slices and incubated at 30°C for 18 h. Watery rotten lesions around inoculation sites were observed as evidence of the activation of virulence.
Nucleotide accession numbers.
The nucleotide sequences of aiiA homologue genes reported in this paper have been submitted to the GenBank-EMBL database under the following accession numbers (the B. thuringiensis subspecies names are shown in parentheses): AF478045 (aizawai HD11), AF478046 (alesti HD4), AF478047 (canadensis HD224), AF478048 (darmstadiensis HD146), AF478049 (galleriae HD29), AF478050 (indiana HD521), AF478051 (israelensis HD567), AF478052 (kyushuensis HD541), AF478053 (morrisoni HD12), AF478054 (ostriniae HD501), AF478055 (pakistani HD395), AF478056 (subtoxicus HD109), AF478057 (thompsoni HD542), AF478058 (toumanoffi HD201), AF478059 (kurstaki HD263), AF478060 (tolworthi HD537), and AF478061 (soil-isolated Bacillus sp. strain IBN35).
RESULTS
aiiA homologous genes in the subspecies of B. thuringiensis.
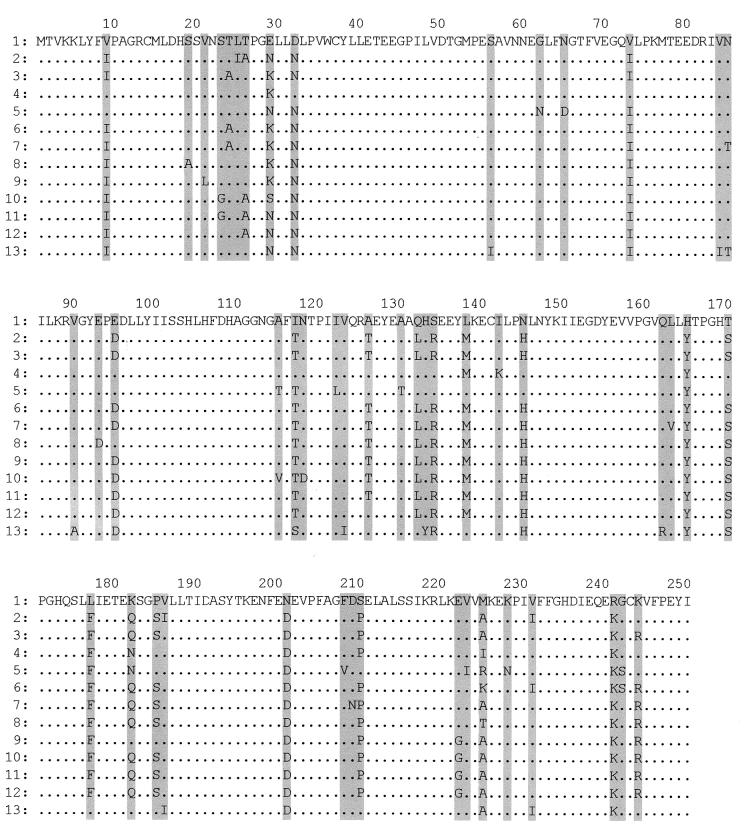
The aiiA gene, first cloned from Bacillus sp. strain 240B1, consists of 750 nucleotides encoding a polypeptide of 250 amino acids (5). In the incomplete genome database of Bacillus anthracis in The Institute for Genomic Research, an aiiA homologue was found which has 90% similarity in nucleotide sequence with the aiiA gene of Bacillus sp. strain 240B1. An aiiA homologue was searched for in our company's incomplete genome database of B. thuringiensis subsp. morrisoni HD12. As a result, a 3.7-kb contig containing an aiiA homologue was also found. Three aiiA homologue genes were aligned, and the most conserved oligonucleotide pair (AIF and AIR) was selected to investigate the presence of aiiA genes in other subspecies of B. thuringiensis. PCR amplification of the aiiA gene was carried out using the chromosomal DNAs of 16 strains of B. thuringiensis and one strain of soil-isolated Bacillus sp. strain IBN35 as templates. As a result, PCR products from all the strains of B. thuringiensis were obtained and subcloned into the T vector, and the sequences of each of the PCR products were determined. Figure 1 shows the deduced amino acid sequences of aiiA homologue genes and their alignments. Comparison of the Bacillus sp. strain 240B1 aiiA gene with the B. thuringiensis aiiA homologue genes revealed high homologies of 89 to 95% and 90 to 96% in nucleotide sequence and deduced amino acid sequence, respectively. Reportedly, two small conserved regions, 103SHLHFDH109 and 165HTPGHTPGH173, exist in AiiA, glyoxalase II, metallo-lactamase, and arylsulfatase (5). In the sequence alignment shown in Fig. 1, 103SHLHFDH109 is completely conserved between AiiA families, while 165HTPGHTPGH173 is not completely conserved. In a search using the BLAST program, 96DLLYIISSHLHFDHAGGNG114 is closely related to glyoxalase II in Arabidopsis thaliana (data not shown). The sequence alignment among the AiiA family might yield information in the study of AiiA specificity for different AHLs and in the engineering of enzymes with even greater activity or more refined specificity.
FIG. 1.
Sequence alignment of deduced amino acids of putative AHL-degrading enzyme genes from B. anthracis, Bacillus sp. isolates, and various subspecies of B. thuringiensis. Sequences of the following strains were used (accession numbers in parentheses): 1, Bacillus sp. strain 240B1 (AF196486); 2, B. anthracis Ames strain (contig number 4847 from unfinished B. anthracis genome; The Institute for Genomic Research); 3, Bacillus. sp. strain IBN35 isolated from soil (AF478061); 4, B. thuringiensis subsp. toumanoffi HD201 (AF478058); 5, B. thuringiensis subsp. kyushuensis HD541 (AF478052); 6, B. thuringiensis subsp. aizawai HD11 (AF478045), B. thuringiensis subsp. indiana HD521 (AF478050), and B. thuringiensis subsp. kurstaki HD263 (AF478059); 7, B. thuringiensis subsp. galleriae HD29 (AF478049); 8, B. thuringiensis subsp. ostriniae HD501 (AF478054); 9, B. thuringiensis subsp. subtoxicus HD109 (AF478056); 10, B. thuringiensis subsp. pakistani HD395 (AF478055); 11, B. thuringiensis subsp. tolworthi HD537 (AF478060); 12, B. thuringiensis subsp. canadensis HD224 (AF478047); 13, B. thuringiensis subsp. alesti HD4 (AF478046), B. thuringiensis subsp. darmstadiensis HD146 (AF478048), B. thuringiensis subsp. israelensis HD567 (AF478051), B. thuringiensis subsp. morrisoni HD12 (AF478053), and B. thuringiensis subsp. thompsoni HD542 (AF478057). Numbers 6 and 13 contain multiple conserved sequences. Shaded regions indicate all amino acid residues that were not completely conserved.
Figure 2 shows the aiiA homologue gene dendrogram from various Bacillus species. The aiiA gene of B. thuringiensis subsp. toumanoffi HD201 is most similar to that of Bacillus sp. strain 240B1. There are two Bacillus groups that have the same amino acid sequences in their AiiA proteins. One group is composed of B. thuringiensis subsp. aizawai HD11, indiana HD521, and kurstaki HD263. The other group is composed of B. thuringiensis subsp. alesti HD4, darmstadiensis HD146, israelensis HD567, morrisoni HD12, and thompsoni HD542. Although they have different serotypes against H1 flagella antigens, they have the same amino acid sequence as AiiA. As a result of analysis of a contig containing the aiiA gene of B. thuringiensis subsp. morrisoni HD12, a 293-amino acid alginate lyase homologous gene was located upstream of the aiiA gene (data not shown). The alginate lyase gene is homologous to that of B. halodurans. We could not find the aiiA homologue gene around the alginate lyase gene BH0738. However, there is a predicted protein sequence (GenBank accession no. BAB06979) that is significantly related to AiiA within the B. halodurans genome.
FIG. 2.
Phylogenetic dendrogram of AHL-degrading enzymes, constructed using the neighbor-joining method. The bar indicates 2% estimated sequence divergence.
Degradation of AHL by B. thuringiensis.
To determine whether B. thuringiensis strains with the aiiA gene can degrade AHL, we designed and performed a whole-cell bioassay. A mixture of B. thuringiensis and OHHL was incubated at 37°C for 1 h with gentle shaking, followed by heating at 95°C. Then, the sample was loaded in the holes of C. violaceum CV026 plates. After overnight incubation at 30°C, biodegradation of OHHL by B. thuringiensis could be observed. Figure 3 shows that some strains of B. thuringiensis can strongly degrade OHHL and others can weakly degrade OHHL. In the case of a longer incubation time for biodegradation (>12 h), residual OHHL was not detected in the CV026 plates (data not shown). This result shows that some strains of B. thuringiensis effectively degrade AHL, a signal molecule of gram-negative bacteria.
FIG. 3.
Relative AHL-degrading activity of various B. thuringiensis subspecies by bioassay. Chromobacterium violaceum CV026 was used as a reporter strain, and OHHL (N-[3-oxohexanoyl]-l-homoserine lactone) was used as a substrate of AHL. C, reaction buffer with OHHL; B, reaction buffer only. *Bacillus sp. strain IBN35 was isolated from soil.
AHL-degrading activity of recombinant AiiA proteins.
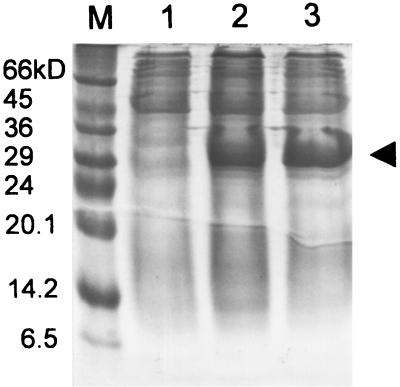
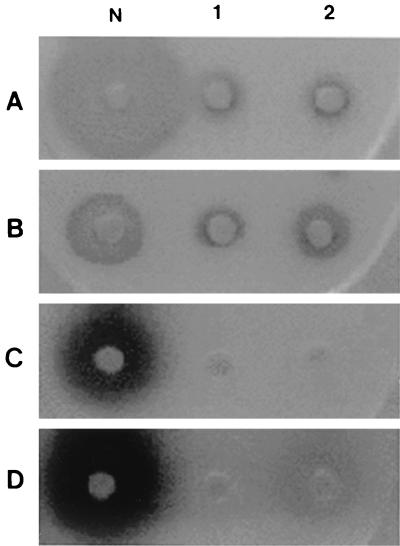
In order to verify that the AiiA homologues of B. thuringiensis have AHL-degrading activity, the aiiA genes were expressed in E. coli and the activity of the recombinant AiiA proteins was tested. Two aiiA genes were amplified from the chromosomes of B. thuringiensis subsp. morrisoni and subsp. kyushuensis and were then overexpressed in E. coli. We confirmed the overexpression of two recombinant AiiAs of correct size (about 28 kDa) as major bands in E. coli by SDS-PAGE analysis (Fig. 4). The enzyme activity was confirmed by whole-cell bioassay. AHLs with different acyl side chains (OHHL, HHL, and OHL) were used for the substrates of AiiA proteins. Recombinant E. coli expressing AiiA could effectively degrade all the substrate, regardless of the N-acyl side chain (Fig. 5). This result indicates that the aiiA gene in B. thuringiensis indeed encodes AHL-degrading enzymes having broad substrate specificity.
FIG. 4.
SDS-15% PAGE analysis of AiiA overexpression in E. coli. M, marker; 1, E. coli carrying pET22b(+); 2, E. coli carrying pETAIK; 3, E. coli carrying pETAIM. The arrow indicates the bands of recombinant AiiA proteins.
FIG. 5.
Bioassay for recombinant AHL-degrading enzyme activity in recombinant E. coli. OHL, OHHL, HHL, and OHHL were used as substrates for A, B, C, and D, respectively. The reporter strain for A and B was A. tumefaciens NT1 (pDCI41E33); that for C and D was C. violaceum CV026. N, E. coli carrying pET22b(+); 1, E. coli carrying pETAIK; 2, E. coli carrying pETAIM.
Attenuation of plant pathogenicity of Erwinia carotovora by E. coli producing an AHL-degrading enzyme.
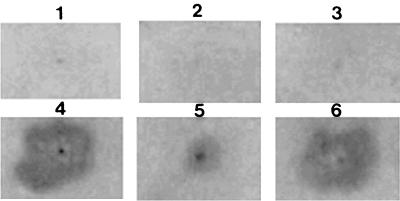
The change in plant pathogenicity of E. carotovora by recombinant AHL-degrading enzyme-overproducing E. coli was tested. Recombinant E. coli carrying pETAIK and E. carotovora IBN98 were mixed and inoculated onto potato slices. A decrease of the watery rotten lesions in the potato slices treated with the mixture of E. carotovora and the recombinant E. coli was observed (Fig. 6). This result showed that the potato virulence of E. carotovora was attenuated by AHL-degrading enzyme-overproducing E. coli. In contrast to the result with recombinant E. coli, attenuation was not observed with the wild type of B. thuringiensis (data not shown). This might be due to the lower level of expression of the aiiA gene in B. thuringiensis than in recombinant E. coli. These data strongly suggest that AHL-degrading enzyme-overproducing microbes can be used for the control of gram-negative plant-pathogenic bacteria in the production of crops.
FIG. 6.
Attenuation of potato pathogenicity of Erwinia carotovora by recombinant AiiA-producing E. coli. 1, saline solution; 2, E. coli carrying pETAIK; 3, E. coli carrying pET22b(+); 4, E. carotovora; 5, mixture of E. carotovora and E. coli carrying pETAIK; 6, mixture of E. carotovora and E. coli carrying pET22b(+).
DISCUSSION
Gram-negative microorganisms commonly use AHLs to regulate the expression of diverse phenotypic traits, such as bioluminescence (17), antibiotic production (16), and virulence factor synthesis (20). In the plant pathogen E. carotovora, virulence-related exoenzyme genes are regulated through an AHL-dependent quorum-sensing system (10). Because many animal and plant pathogens use quorum sensing to regulate virulence, the quorum-sensing system is an attractive target for novel antiinfective therapy (27). One strategy is to disrupt the signal generation process. This could be achieved by the inhibition of synthesis of metabolic precursors of AHL or by the direct inhibition of AHL synthesis (9). Secondly, the active efflux and diffusion system of AHLs could be a target for drug development (21). Thirdly, small AHL-mimetic antagonists such as halogenated furanones could compete with AHL for LuxR homologues (14, 15). Fourthly, an AHL-degrading enzyme or AHL-sequestering antibody might interfere with AHL-mediated cell-to-cell communications. Recently, a novel AHL-degrading enzyme gene (aiiA) from Bacillus sp. strain 240B1 was cloned (5). Transgenic tobacco and potato expressing the bacterial gene aiiA inactivated exogenous AHL and reduced the virulence of gram-negative E. carotovora (4).
As stated above, there has been increasing interest in AHL degradation in the control of quorum sensing. To our knowledge, this is the first publication to report the existence of aiiA homologue genes in many subspecies of B. thuringiensis and to identify aiiA homologue genes in B. anthracis and Bacillus species closely related to B. cereus (Fig. 1). Helgason et al. (8) reported the possibility of close relations among food-poisoning-inducing B. cereus, anthrax-inducing B. anthracis, and insecticidal B. thuringiensis, though they have widely different phenotypes and pathological effects. The sequences of aiiA genes in B. anthracis and Bacillus sp. strain IBN35 belonging to the B. cereus group were phylogenetically very close to those in various subspecies of B. thuringiensis (Fig. 2). aiiA homologue genes might be widespread not only in B. thuringiensis but also in other strains in the B. cereus group.
Wild-type strains of B. thuringiensis show various AHL-degrading activities (Fig. 3). Some strains show strong AHL-degrading activity, and others have weak AHL-degrading activity. When the aiiA genes of B. thuringiensis subsp. morrisoni and kyushuensis, showing weak and strong AHL-degrading activity, respectively, were overexpressed in E. coli (Fig. 4), both of the recombinant E. coli strains efficiently degraded AHLs having various side chains (Fig. 5). This shows that aiiA homologue genes in B. thuringiensis encode active AHL-degrading enzymes and indicates that the recombinant AiiA proteins could be applied for control of different kinds of AHL-producing gram-negative bacteria.
In the virulence attenuation test, the recombinant E. coli could effectively attenuate the virulence of E. carotovora in potatoes (Fig. 6). The attenuation of plant pathogenicity shows the possibility of biocontrol of gram-negative bacteria by the use of recombinant AiiA-overproducing microbes or genetically engineered B. thuringiensis. The discovery of the existence of aiiA genes in many subspecies of B. thuringiensis, presently used as an insecticidal agent in agriculture, indicates the possibility of developing a B. thuringiensis strain having additional functions, including the regulation of gram-negative pathogenic bacteria.
The reason AHL-degrading enzyme genes are widespread in subspecies of B. thuringiensis is not known. It could be supposed that the AHL-degrading activity of AiiA might give insecticidal B. thuringiensis the potential to compete with gram-negative bacteria in natural ecosystems, which could explain the frequent occurrence of B. thuringiensis on the phylloplane. Further studies should address the identification of the function of AiiA homologue proteins in B. thuringiensis.
Acknowledgments
The authors thank In-Kyu Hwang (Seoul National University, Korea) for providing C. violaceum CV026 and A. tumefaciens NT1 (pDCI41E33).
This work was supported by a grant from the Rural Development Administration of Korea (BioGreen21 Project).
REFERENCES
- 1.Angus, T. A. 1954. A bacterial toxin paralysing silkworm larvae. Nature (London) 173:545-546. [DOI] [PubMed] [Google Scholar]
- 2.Cook, D. M., P. L. Li, F. Ruchaud, S. Padden, and S. K. Farrand. 1997. Ti plasmid conjugation is independent of vir: reconstitution of the tra functions from pTiC58 as a binary system. J. Bacteriol. 179:1291-1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Diaz-Gomez, O., J. C. Rodriguez, A. M. Shelton, A. Lagunes, and R. Bujanos. 2000. Susceptibility of Plutella xylostella (L.) (Lepidoptera: Plutellidae) populations in Mexico to commercial formulations of Bacillus thuringiensis. J. Econ. Entomol. 93:963-970. [DOI] [PubMed] [Google Scholar]
- 4.Dong, Y. H., L. H. Wang, J. L. Xu, H. B. Zhang, X. F. Zhang, and L. H. Zhang. 2001. Quenching quorum-sensing-dependent bacterial infection by an N-acyl homoserine lactonase. Nature (London) 411:813-817. [DOI] [PubMed] [Google Scholar]
- 5.Dong, Y. H., J. L. Xu, X. Z. Li, and L. H. Zhang. 2000. AiiA, an enzyme that inactivates the acylhomoserine lactone quorum-sensing signal and attenuates the virulence of Erwinia carotovora. Proc. Natl. Acad. Sci. USA 97:3526-3531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fray, R. G., J. P. Throup, M. Daykin, A. Wallace, P. Williams, G. S. Stewart, and D. Grierson. 1999. Plants genetically modified to produce N-acylhomoserine lactones communicate with bacteria. Nat. Biotechnol. 17:1017-1020. [DOI] [PubMed] [Google Scholar]
- 7.Fuqua, C., and E. P. Greenberg. 1998. Self perception in bacteria: quorum sensing with acylated homoserine lactones. Curr. Opin. Microbiol. 1:183-189. [DOI] [PubMed] [Google Scholar]
- 8.Helgason, E., O. A. Økstad, D. A. Caugant, H. A. Johansen, A. Fouet, M. Mock, I. Hegna, and A.-B. Kolstø. 2000. Bacillus anthracis, Bacillus cereus, and Bacillus thuringiensis—one species on the basis of genetic evidence. Appl. Environ. Microbiol. 66:2627-2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hoang, T. T., and H. P. Schweizer. 1999. Characterization of Pseudomonas aeruginosa enoyl-acyl carrier protein reductase (FabI): a target for the antimicrobial triclosan and its role in acylated homoserine lactone synthesis. J. Bacteriol. 181:5489-5497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jones, S., B. Yu, N. J. Bainton, M. Birdsall, B. W. Bycroft, S. R. Chhabra, A. J. Cox, P. Golby, P. J. Reeves, S. Stephens, et al. 1993. The lux autoinducer regulates the production of exoenzyme virulence determinants in Erwinia carotovora and Pseudomonas aeruginosa. EMBO J. 12:2477-2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Latifi, A., M. K. Winson, M. Foglino, B. W. Bycroft, G. S. Stewart, A. Lazdunski, and P. Williams. 1995. Multiple homologues of LuxR and LuxI control expression of virulence determinants and secondary metabolites through quorum sensing in Pseudomonas aeruginosa PAO1. Mol. Microbiol. 17:333-343. [DOI] [PubMed] [Google Scholar]
- 12.Leadbetter, J. R., and E. P. Greenberg. 2000. Metabolism of acyl-homoserine lactone quorum-sensing signals by Variovorax paradoxus. J. Bacteriol. 182:6921-6926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mae, A., M. Montesano, V. Koiv, and E. T. Palva. 2001. Transgenic plants producing the bacterial pheromone N-acyl-homoserine lactone exhibit enhanced resistance to the bacterial phytopathogen Erwinia carotovora. Mol. Plant-Microbe Interact. 14:1035-1042. [DOI] [PubMed] [Google Scholar]
- 14.Manefield, M., M. Welch, M. Givskov, G. P. Salmond, and S. Kjelleberg. 2001. Halogenated furanones from the red alga, Delisea pulchra, inhibit carbapenem antibiotic synthesis and exoenzyme virulence factor production in the phytopathogen Erwinia carotovora. FEMS Microbiol. Lett. 205:131-138. [DOI] [PubMed] [Google Scholar]
- 15.Maximilien, R., R. de Nys, C. Holmstrom, L. Gram, M. Givskov, K. Crass, S. Kjelleberg, and P. Steinberg. 1998. Chemical medication of bacterial surface colonisation by secondary metabolites from the red alga Delisea pulchra. Aquat. Microb. Ecol. 15: 233-246. [Google Scholar]
- 16.McGowan, S., M. Sebaihia, S. Jones, B. Yu, N. Bainton, P. F. Chan, B. Bycroft, G. S. Stewart, P. Williams, and G. P. Salmond. 1995. Carbapenem antibiotic production in Erwinia carotovora is regulated by CarR, a homologue of the LuxR transcriptional activator. Microbiology 141:541-550. [DOI] [PubMed] [Google Scholar]
- 17.Nealson, K. H., and J. W. Hastings. 1979. Bacterial bioluminescence: its control and ecological significance. Microbiol. Rev. 43:496-518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Passador, L., J. M. Cook, M. J. Gambello, L. Rust, and B. H. Iglewski. 1993. Expression of Pseudomonas aeruginosa virulence genes requires cell-to-cell communication. Science 260:1127-1130. [DOI] [PubMed] [Google Scholar]
- 19.Pearson, J. P., L. Passador, B. H. Iglewski, and E. P. Greenberg. 1995. A second N-acylhomoserine lactone signal produced by Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 92:1490-1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pearson, J. P., E. C. Pesci, and B. H. Iglewski. 1997. Roles of Pseudomonas aeruginosa las and rhl quorum-sensing systems in control of elastase and rhamnolipid biosynthesis genes. J. Bacteriol. 179:5756-5767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pearson, J. P., C. Van Delden, and B. H. Iglewski. 1999. Active efflux and diffusion are involved in transport of Pseudomonas aeruginosa cell-to-cell signals. J. Bacteriol. 181:1203-1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pirhonen, M., D. Flego, R. Heikinheimo, and E. T. Palva. 1993. A small diffusible signal molecule is responsible for the global control of virulence and exoenzyme production in the plant pathogen Erwinia carotovora. EMBO J. 12:2467-2476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 24.Schnepf, E., N. Crickmore, J. Van Rie, D. Lereclus, J. Baum, J. Feitelson, D. R. Zeigler, and D. H. Dean. 1998. Bacillus thuringiensis and its pesticidal crystal proteins. Microbiol. Mol. Biol. Rev. 62:775-806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schraft, H., and M. W. Griffiths. 1995. Specific oligonucleotide primers for detection of lecithinase-positive Bacillus spp. by PCR. Appl. Environ. Microbiol. 61:98-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Throup, J. P., M. Camara, G. S. Briggs, M. K. Winson, S. R. Chhabra, B. W. Bycroft, P. Williams, and G. S. Stewart. 1995. Characterisation of the yenI/yenR locus from Yersinia enterocolitica mediating the synthesis of two N-acylhomoserine lactone signal molecules. Mol. Microbiol. 17:345-356. [DOI] [PubMed] [Google Scholar]
- 27.Whitehead, N. A., M. Welch, and G. P. Salmond. 2001. Silencing the majority. Nat. Biotechnol. 19:735-736. [DOI] [PubMed] [Google Scholar]
- 28.Zhang, L., P. J. Murphy, A. Kerr, and M. E. Tate. 1993. Agrobacterium conjugation and gene regulation by N-acyl-L-homoserine lactones. Nature (London) 362:446-448. [DOI] [PubMed] [Google Scholar]