Abstract
Oxalate degradation by the anaerobic bacterium Oxalobacter formigenes is important for human health, helping to prevent hyperoxaluria and disorders such as the development of kidney stones. Oxalate-degrading activity cannot be detected in the gut flora of some individuals, possibly because Oxalobacter is susceptible to commonly used antimicrobials. Here, clarithromycin, doxycycline, and some other antibiotics inhibited oxalate degradation by two human strains of O. formigenes. These strains varied in their response to gut environmental factors, including exposure to gastric acidity and bile salts. O. formigenes strains established oxalate breakdown in fermentors which were preinoculated with fecal bacteria from individuals lacking oxalate-degrading activity. Reducing the concentration of oxalate in the medium reduced the numbers of O. formigenes bacteria. Oxalate degradation was established and maintained at dilution rates comparable to colonic transit times in healthy individuals. A single oral ingestion of O. formigenes by adult volunteers was, for the first time, shown to result in (i) reduced urinary oxalate excretion following administration of an oxalate load, (ii) the recovery of oxalate-degrading activity in feces, and (iii) prolonged retention of colonization.
Oxalate is formed in the liver by amino acid catabolism (18). It is also present in a wide range of food and drinks, including tea, coffee, chocolate, and fruit and vegetables (19). The concentration of oxalate voided in the urine plays an important role in urolithiasis, the formation of calcium oxalate kidney stones (27). The main known bacterial species involved in oxalate degradation in the gut is the anaerobe Oxalobacter formigenes. First isolated from ruminants (13), O. formigenes has been found in many animal species and in humans (reviewed by Allison et al. [3]). Almost all 6- to 8-year-old individuals tested possess O. formigenes, but this bacterium may later be lost and it is detected in the feces of 60 to 80% of adults (31). The therapeutic use of antibiotics and other drugs may contribute to the loss of O. formigenes (6; M. J. Allison, H. Sidhu, and A. B. Peck, Abstr. 22nd Int. Cong. Microb. Ecol. Dis., abstr. 5.002, 1997). Patients with cystic fibrosis often lack fecal oxalate degraders, perhaps because of intensive drug therapy (32).
Understanding the ecology of O. formigenes could aid our knowledge of oxalate metabolism in the gut. This might lead to the use of probiotics to combat hyperoxaluria. Here we describe experiments performed in vitro and in vivo which aimed to measure relevant physiological properties of O. formigenes and the consequences of its introduction in fermentors simulating conditions in the human colon and in human volunteers.
MATERIALS AND METHODS
Isolations, reference strains, and culture maintenance.
O. formigenes strains OxB (ATCC 35274) and HC1 (M. J. Allison, Department of Microbiology, Iowa State University) were isolated from sheep rumen (13) and human feces (2), respectively. Strain Va3 (Rowett Research Institute culture collection) was isolated by enrichment of the feces from an adult volunteer. The medium used was medium D2, which is medium D of Daniel et al. (11), modified by replacing the trace elements listed in reference 11 with those listed in reference 23. The main components were as follows (amounts are liter−1): di-sodium oxalate, 2.68 g; clarified rumen fluid, 200 ml; sodium acetate · 3H2O, 1.36 g; yeast extract (Difco), 1 g; minerals; and 1 g of cysteine HCl as a reductant. The final pH was adjusted to 6.8. Enrichment cultures (10 ml) were inoculated with approximately 0.5 g of feces provided by a healthy person who had not taken antibiotics for at least 3 months preceding sampling, and then the cultures were incubated for up to 48 h. After around 15 successive transfers, 0.5 ml of the serially diluted (15) culture was inoculated into anaerobic roll tubes containing Ca oxalate agar medium (described below). After incubation for 21 days, cultures were picked from colonies surrounded by cleared zones in medium that was partially opaque due to Ca oxalate. Culture medium (10 ml), diluting fluid (9 ml), and roll tubes (4.5 ml) were prepared under O2-free CO2 in glass Hungate tubes (16 by 125 mm; Bellco Glass Inc., Vineland, N.J.) (10). All incubations were at 38 ± 1°C.
Biotyping of strain Va3 and molecular detection of O. formigenes.
Genomic DNA was prepared, and the oxc gene was amplified using genus-specific primers as described by Sidhu et al. (30). The amplified DNA was run on an agarose gel, blotted onto a nylon membrane, and hybridized with group I- and group II-specific probes (31). Tests with feces were performed as described in reference 31, using genus-specific probes for Southern hybridizations, and fermentor samples were treated similarly.
Culture test for oxalate degradation.
The sample (swabbed feces, serially diluted fermentor contents, or batch culture as defined in the text) was incubated with 8 ml of medium D2 (described above) for 2 to 5 days and then was tested for residual oxalate using the Ca precipitation test (31).
Analytical methods.
Concentrations of oxalate and short-chain fatty acids (SCFA) in cultures were determined by capillary gas chromatography (GC) following conversion to t-butyldimethylsilyl derivatives (29), allowing accurate quantification of approximately ≥1 mM oxalate. When long-term survival of O. formigenes strain HC1 was discovered in a fermentor medium without added oxalate (see Results), retrospective tests for oxalate at low concentrations were conducted by isotope dilution. Samples (1 ml) were mixed with 0.25 ml of internal standard at 1.06 mM oxalic acid-13C2-sodium salt, 0.25 ml of distilled H2O, and 100 μl of concentrated HCl. The mixture was extracted twice with 2 ml of ether, and the extracts were dried at 45°C under N2. Eighty microliters of 1:1 N-methyl-N-t-butyldimethylsilyltrifluoro-acetamide-acetonitrile was added, followed by heating at 90°C for 15 min. GC-mass spectrometry analysis was essentially as previously described (33). The level of urinary oxalate was determined enzymatically (diagnostic kit no. 591; Sigma, Poole, United Kingdom). Creatinine was determined using picrate (21).
Sensitivity to antibiotics and bile salts.
Aliquots (10 ml) of medium D2 were amended to contain the relevant compound added as a filter-sterilized (0.2-μm-pore-size membrane filters) aqueous solution to a range of final concentrations. Triplicate incubations (38°C) were conducted at each concentration. After inoculation with 0.5 ml of a pregrown culture of O. formigenes and incubation for 3 days, the level of residual oxalate was determined by the Ca precipitation test, and the main findings were confirmed by GC analysis. Compounds listed in Table 1 were from Sigma, except for clarithromycin (Abbott, Queensborough, United Kingdom) and amoxicillin-clavulanic acid (1:2 [wt/wt] clavulanic acid-amoxicillin; Co-Amoxiclav; SmithKline Beecham, Slough, United Kingdom).
TABLE 1.
Effects of antibiotics, porcine bile salts, and deoxycholate on oxalate degradation by O. formigenes strains Va3 and HC1
| Antibiotic or other addition (concna) | Degradation byb
|
|
|---|---|---|
| Va3 | HC1 | |
| Antibiotic | ||
| None | + | + |
| Chloramphenicol (1) | − | + |
| Chloramphenicol (5) | − | − |
| Nalidixic acid (30) | + | + |
| Nalidixic acid (40) | − | + |
| Ampicillin (25) | + | + |
| Erythromycin (1) | + | − |
| Erythromycin (5) | − | − |
| Amoxicillin (50) | + | + |
| Clarithromycin (1) | − | − |
| Co-amoxiclav (25) | + | + |
| Co-amoxiclav (50) | − | + |
| Metronidazole (10) | + | + |
| Metronidazole (25) | − | − |
| Streptomycin (20) | + | + |
| Doxycycline (1) | − | − |
| Other additions | ||
| 0.2% bile salts | + | + |
| 0.5% bile salts | − | + |
| 1.0% bile salts | − | + |
| Deoxycholate (1) | + | + |
| Deoxycholate (5) | − | + |
| Deoxycholate (10) | − | − |
Concentrations are in micrograms per milliliter for antibiotics and mM for deoxycholate.
Symbols: +, tolerance (loss of oxalate not significantly different to that observed for controls without antibiotics or bile salts); −, no significant loss of oxalate and failure to grow at the given concentration. All tests were performed in triplicate.
Tolerance of low pH.
Anaerobic medium M2 (15) was modified by the omission of sugars and lactate, and the pH of aliquots was adjusted to 6.8, 3.0, and 2.0 with HCl. The pH-adjusted medium was sterilized by filtration (described above). Strains Va3, HC1, and OxB were grown for 24 h on medium D2, and the cells were deposited by centrifugation (2,000 × g, 10 min). The thick cell suspensions (0.75 ml) were resuspended in triplicate 7.5-ml aliquots of modified medium M2 for each of the different pH values. The number of viable cells present was determined using Ca oxalate roll tubes (below) at 0 and after 2 h of incubation (38°C).
Tolerance of air.
Pregrown cells were centrifuged (described above), resuspended in 5 ml of phosphate-buffered saline (pH 6.8) (PBS), and recentrifuged. The excess PBS was decanted, and the thick cell suspension, contained in a Hungate tube (above) fitted loosely with a plastic cap, was incubated in triplicate in air in a shaking incubator (38°C) at 200 rpm. Parallel triplicate anaerobic incubations were performed in PBS prepared and maintained in O2-free N2. Viable oxalate-degrading bacteria were enumerated using Ca oxalate roll tubes (below) at 0 and 24 h.
Fermentor studies.
Experiments with strains OxB and Va3 employed a single-stage continuous flow fermentor at 38 ± 1°C with a culture vessel with a 900-ml working volume (Gallenkamp, London, United Kingdom). The experiment with strain HC1 employed a similar but smaller device (250-ml culture volume). Mixing was achieved with a magnetic stirrer and by a stream of O2-free CO2. The fermentation was maintained at pH 6.8 ± 0.2 by a controller (Electrolab, Hemel Hempsted, United Kingdom). The colon habitat-simulating (CH) medium was based on that of Macfarlane et al. (24). It contained xylan, amylopectin, arabinogalactan, pectin, starch (potato), casein, peptone water (Unipath, Basingstoke, United Kingdom), hemin, bile salts, and minerals as listed in reference 24 with (liter−1) 3.0 g of Na2HCO3, 0.5 g of cysteine HCl, 0.6 mg of resazurin, and 0.5 ml of antifoam A (Sigma). Disodium oxalate (final concentration, 10 mM or 20 mM) was added when specified (CHOX10 and CHOX20 medium, respectively, in the text). The medium was sterilized by autoclaving at 121°C for 15 min, stirred while hot and filter-sterilized vitamin and trace element solutions (1) were then added.
Inoculation with mixed human fecal bacteria employed freshly voided feces from volunteers lacking detectable fecal oxalate-degrading activity (Ox negative). For each milliliter of fermentor contents, approximately 10 mg of feces was dispersed in 40 μl of 50 mM phosphate buffer, pH 6.8, with 0.5% (wt/vol) cysteine HCl and then inoculated through a port in the fermentor. At least 4 days was allowed with continuous flow of medium (with dilution rate [D] as defined in the text) for establishment of fecal bacteria and confirmation of the absence of oxalate degradation. This was judged by determining the concentration of SCFA and oxalate by GC (above). O. formigenes was then inoculated (defined as day 0 in the text). Cultures of O. formigenes for inoculation (10 ml) were pregrown on medium D2 for 24 h. Counts on Ca oxalate roll tubes (below) showed that the inoculum provided approximately 106 viable cells of strain OxB and 3 × 105 viable cells of strains Va3 and HC1 ml of fermentor contents−1.
When D was changed, the time equivalent to at least two, normally five, turnovers elapsed before measurements began. The interval between samples (taken on different days) was at least one turnover. Following changes in the concentration of oxalate in the medium reservoir, at least 7 days elapsed before sampling, and samples were then taken on different days.
Bacterial enumeration.
Total counts were performed using roll tubes (10) of medium M2 (15) (without lactate) (three tubes per dilution), and oxalate degraders were enumerated as colonies surrounded by clear zones using Ca oxalate roll tubes of medium D2 containing 20 g of agar liter−1 (three tubes per dilution).
Oxalate load experiments.
All research involving humans complied with all relevant institutional and federal guidelines. Adult volunteers lacking detectable oxalate-degrading activity in feces fasted overnight and then consumed a standard meal consisting of a sandwich with two rounds of buttered bread and 60 g of turkey meat. Sodium oxalate equivalent to 2 mmol 70 kg of body weight−1 was dissolved in H2O and taken with the meal. All urine voided during the following 6 h was collected, and 100 ml of water was taken at intervals of 1 h.
Human colonization with O. formigenes.
Pregrown biomass (500 mg [wet weight]; approximately 108 viable cells mg−1) of O. formigenes strain HC1 was spread on a sandwich (above) and taken with an oxalate load as described above. Oxalate-degrading activity of swabbed feces was determined at the intervals defined in the text.
RESULTS
Isolation and tolerance of antibiotics, bile salts, low pH, and air.
O. formigenes strain Va3 was isolated from human feces which were positive in the cultural test for oxalate degradation. The cells were gram-negative rods, approximately 3.5 by 1 μm. Preparation of genomic DNA, PCR amplification of the oxc gene, and Southern hybridization with group-specific probes demonstrated that the strain belonged to group II of this species (30) (data not shown). Antibiotic resistance testing showed that strains Va3 and HC1 were sensitive to clarithromycin (Klaracid) and doxycycline (Vibramycin) at 1 μg ml−1 (Table 1). Higher concentrations of the other antibiotics tested were tolerated, except for chloramphenicol for strain Va3 and erythromycin for strain HC1. At 50 μg ml−1, amoxicillin plus clavulanic acid was more inhibitory to strain Va3 than was amoxicillin alone (Table 1). The two strains also differed slightly in their resistance to porcine bile salts and deoxycholate (Table 1).
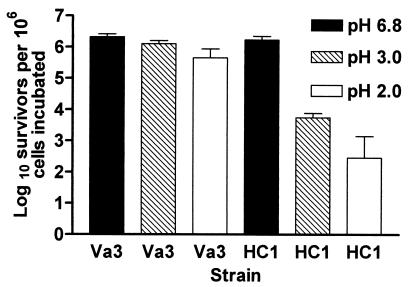
When strain Va3 was incubated for up to 2 h in modified medium M2 at pH 6.8 and at pH 3.0, no decrease in the viable cell count was recorded. At pH 2.0, however, the viable cell count declined by around 60% (Fig. 1). The type strain OxB behaved similarly (data not shown). Strain HC1 showed no decrease in cell viability upon incubation at pH 6.8, but for this strain less than 1 and 0.1% of the cells were viable after 2 h at pH 3.0 and pH 2.0, respectively.
FIG. 1.
Effect of incubation at pH 6.8, 3, and 2 for 2 h upon survival of O. formigenes strains. Values are the averages of three separate incubations + SD (bars).
Incubation of strain Va3 in anaerobic PBS for 24 h resulted in the recovery of (2.0 ± 1.5) × 105 ml−1 (mean ± standard deviation [SD]; n = 3) for every 106 viable cells incubated. After incubation under air, only around 0.1% of the starting cells were recovered ([1 ± 3] × 103; n = 3). In comparable tests, the number of viable strain HC1 cells recovered after incubation under anaerobic and aerobic conditions was (7.0 ± 5.0) × 104 (n = 3) and none, respectively.
Establishment of O. formigenes strains in fermentor simulations of the colonic fermentation.
A (single) fermentor was supplied with medium CHOX20 (20 mM oxalate) at a dilution rate (D) of 0.04 h−1 and inoculated with feces from an Ox-ngative adult as described in Materials and Methods. Following establishment of fecal bacteria, a pure culture of O. formigenes strain OxB was introduced (day 0). Oxalate, detected in the fermentor at 20 mM immediately before inoculation with strain OxB, could not be detected by day 2 and was not found subsequently (data not shown). The experiment was terminated on day 7. Parallel batch incubations (up to 10 days) of cultures inoculated with the same Ox-negative feces showed no oxalate degradation.
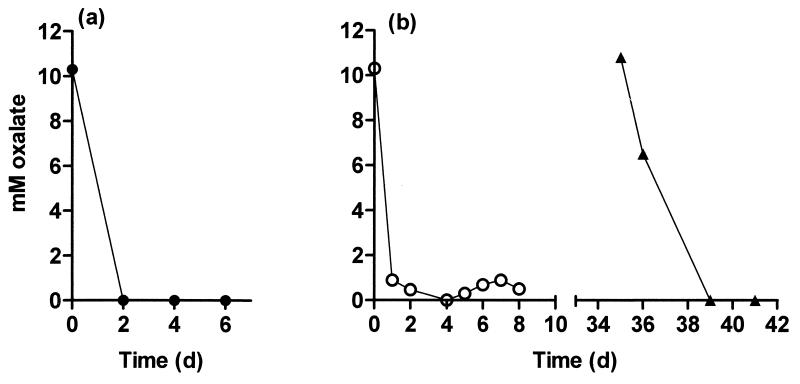
An experiment with O. formigenes strain Va3 tested the effect of changing the dilution rate and oxalate concentration. The (single) fermentor was continuously supplied, at a D of 0.015 h−1, with medium CHOX10 (10 mM oxalate) and inoculated with feces from a further Ox-negative adult, and this was followed by inoculation with O. formigenes strain Va3 on day 0. By day 1, oxalate was not detectable (Fig. 2a, vessel V). During the following 101 days, the fermentor was operated at the following dilution rates: 0.015 h−1 (for 38 days), 0.02 h−1 (for 37 days), 0.125 h−1 (for 6 days), and 0.036 h−1 (for 20 days). Except at a D of 0.125 h−1 (below), oxalate was not present at concentrations quantifiable by GC. Cultivable anaerobes were present at (3.8 ± 3.4) × 109 viable cells ml−1 (mean ± SD; n = 18) throughout and were unaffected by changes in D. The numbers of oxalate degraders detected using roll tubes averaged (2.2 ± 1.0) ×107 ml−1 (n = 6) at a D of 0.015 h−1 and (1.7 ± 0.02) × 107 ml−1 (n = 4) at a D of 0.02 h−1. Pooled values for concentrations of strain Va3 obtained with the culture test procedure (performed in triplicate on each sample) for a D of 0.02 and 0.036 h−1 ranged from 105 to 107 ml−1 (n = 11). Concentrations of total SCFA were not significantly affected by changes in D from 0.036 to 0.015 h−1 (Table 2), but the butyrate concentration tended to be higher at the lower flow rate (Table 2). Increasing D to 0.125 h−1 decreased the SCFA concentration and increased the concentrations of formate and oxalate (Table 2).
FIG. 2.
The effect of introduction of O. formigenes on oxalate concentrations in fermentor simulations of the human colon operated at different dilution rates. (a) Vessel V inoculated on day 0 with O. formigenes strain Va3; D = 0.015 h−1. (b) Vessel H1 (○) inoculated on day 0 with O. formigenes strain HC1; D = 0.0625 h−1. Also shown are data for vessel H2 (▴), inoculated on day 35 with the mixed population from vessel H1; D = 0.02 h−1.
TABLE 2.
Effects of dilution rate on concentrations of oxalate and SCFA in a fermentor containing mixed human fecal bacteria and O. formigenes strain Va3
| D (h−1)a | Mean concn (mM± SD)
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Acetate | Prop | i-But | But | i-Val | Val | Form | Total SCFA | Oxalate | |
| 0.125 | 30.8 ± 4.5 | 21.0 ± 2.9 | 0.9 ± 0.2 | 2.1 ± 0.3 | 0.4 ± 0.1 | 4.4 ± 0.65 | 1.9 ± 1.6 | 61.5 ± 6.9 | 2.4 ± 4.2 |
| 0.036 | 40.4 ± 2.0 | 25.7 ± 1.3 | 1.5 ± 0.1 | 2.6 ± 0.1 | 1.0 ± 0.1 | 5.6 ± 0.5 | ND | 78.2 ± 1.9 | ND |
| 0.015 | 37.7 ± 3.6 | 26.6 ± 2.8 | 1.6 ± 0.1 | 3.6 ± 0.7 | 1.2 ± 0.2 | 5.1 ± 0.4 | ND | 75.7 ± 5.9 | ND |
Dilution rates 0.125, 0.036, and 0.015 h−1 correspond to turnover times of approximately 8, 28, and 67 h, respectively. Abbreviations: ND, not detected; Prop, propionate; But, butyrate; Val, valerate; Form, formate; i, iso.
Decreasing the oxalate concentration in the medium decreased the numbers of O. formigenes cells. When, on day 102 and with a D of 0.02 h−1, the concentration of oxalate in the medium was decreased to 1 mM, the viable cell numbers (Ca oxalate roll tubes) of O. formigenes subsequently fell to (1.5 ± 0.4) × 105 ml−1 (n = 4). Further reduction of the medium oxalate concentration to 0.1 mM on day 113 decreased the numbers of O. formigenes detected to (7.85 ± 0.5) × 104 ml−1 (n = 4). The experiment was terminated on day 126. Parallel batch incubations (50 days) inoculated with the same Ox-negative feces showed no oxalate degradation (data not shown).
Feces from the same Ox-negative donor were also used to inoculate two more fermentors continuously supplied with CHOX10 medium at a D of 0.02 h−1. After establishment of fecal bacteria and confirmation of the absence of oxalate degradation, D was adjusted to 0.0625 h−1, and one vessel (vessel H1, Fig. 2b) was inoculated with a pure culture of strain HC1. This increase in D was done to extend the range of D values at which initial establishment of O. formigenes was studied, to reproduce the variation that might be encountered in different individuals. The concentration of oxalate fell following introduction of strain HC1 (Fig. 2b, vessel H1). Vessel H1 was operated with CHOX10 medium for 13 days at a D of 0.0625 h−1, 41 days at a D of 0.02 h−1, and 9 days at a D of 0.125 h−1. Oxalate was not detected at quantifiable concentrations when D was 0.02 h−1 (data not shown). At a D of 0.125 h−1, the average concentration of oxalate detected was 2.8 ± 1.7 mM (n = 5). The number of viable cells of strain HC1 (Ca oxalate roll tubes) at a D of 0.02 h−1 averaged (1.25 ± 0.45) × 107 ml−1 (n = 6).
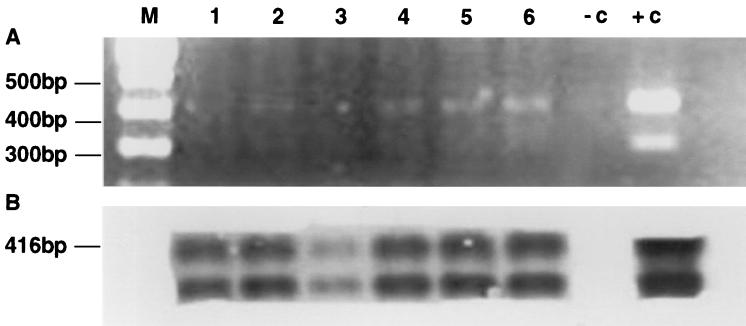
On day 35, with a D of 0.02 h−1, vessel H2 was inoculated with 10 ml of the contents from vessel H1. Oxalate, still detected at around 10 mM in vessel H2 despite prolonged incubation with mixed fecal bacteria, then disappeared by day 39 (Fig. 2b, vessel H2). Finally, on day 63, the medium for vessels H1 and H2 was replaced with CH medium, to which sodium oxalate was not added. D was maintained at 0.02 h−1. The numbers of viable O. formigenes cells found using Ca oxalate roll tubes (n = 2, one sample from each vessel) then fell to 106, 2.5 × 105, and 2.6 × 103 ml−1 when measured on days 71, 79, and 91, respectively. On day 108, no viable cells of strain HC1 were detected on Ca oxalate roll tubes inoculated with 0.05-ml samples. However, PCR and Southern blotting (Fig. 3) demonstrated the continued presence of the strain in both vessels on day 107, some 44 days after the switch to the oxalate-free CH medium. Following the switch back to CHOX10 medium on day 148, oxalate disappeared by day 152 (data not shown). O. formigenes was subsequently found using roll tubes at 107 ml−1 (n = 2, one sample from each vessel). The 85 days for which CH medium was fed should have ensured washout of O. formigenes (28); however, retrospective isotope dilution measurements revealed the presence of approximately 15 μmol of oxalate liter−1 as a contaminant in the complex CH medium.
FIG. 3.
PCR-based detection of O. formigenes in fermentor samples. A genus-specific oligonucleotide probe was used that hybridizes to the PCR amplification product of the oxc gene. (A) Agarose gel electrophoresis of the PCR products. (B) Southern blotting performed on the same gel. Lanes 1, 2, and 3 are replicate samples from vessel H1 and lanes 4, 5, and 6 are replicate samples from vessel H2, 44 days after the start of perfusion with medium to which oxalate was not added. Lane +c, amplification of genomic DNA of the type strain OxB; lane −c, negative control; lane M, molecular size marker.
Experiments in humans.
Tests in humans required gram quantities of cell biomass. Strain HC1 was used because this strain could grow in media containing up to 100 mM oxalate, providing more biomass than strain Va3, which grew poorly at oxalate concentrations of ≥60 mM. Ingesting 500 mg of O. formigenes strain HC1 resulted in detection of oxalate-degrading activity in the feces of two individuals (consuming standard omnivorous Western diets) who were consistently Ox negative (based on the culture test for oxalate degradation) for a period of 2 to 4 years prior to the experiment. Here, subject 1 showed no fecal oxalate-degrading activity when tested 10 days and 1 day prior to ingestion of strain HC1, despite incubation of fecal samples for more than 20 days in batch culture with medium D2 and in continuous culture (D = 0.02 h−1) with CHOX10 medium (data not shown). However, this activity was detected both by the CaCl2 test and by GC determination of residual oxalate in batch cultures incubated with feces voided 1, 2, 4, 8, 9, 14, 21, 25, 59, 86, and 120 days after ingesting O. formigenes. A PCR test performed on a separate stool sample taken around 90 days after ingestion of O. formigenes was positive for this species (data not shown). Stool samples from subject 2 tested negative by PCR for O. formigenes on two separate occasions preceding its ingestion (data not shown). No oxalate-degrading activity was found in these samples or in samples taken 3 days before ingestion of strain HC1. The culture test confirmed that stool samples taken 1, 2, 4, 6, 12, and 106 days after ingestion of the bacterium possessed oxalate-degrading activity, and the PCR test performed on a stool sample taken 9 months after ingestion of biomass was positive for O. formigenes (data not shown).
The ingestion of 500 mg of strain HC1 by four subjects reduced the amount of oxalate excreted during the 6 h immediately following ingestion of an oxalate load (paired t test, P < 0.025). Results were similar for all four subjects tested; the mean rate of oxalate excretion during the 6-h test period without ingestion of O. formigenes was 3.0 ± 0.6 mg h−1 (n = 4), and this decreased to 1.9 ± 0.1 mg h−1 when the oxalate load was accompanied by O. formigenes. In the same tests, the ingestion of O. formigenes was accompanied by a decrease (paired t test, P < 0.025) in the urinary oxalate/creatinine ratio from 45.2 ± 9.9 to 27.0 ± 4. 2 mg g−1.
DISCUSSION
It remains questionable whether O. formigenes is the only, or most-important, species involved in oxalate degradation in the human gut. Other oxalate-degrading bacteria that have been isolated from human stool samples include Eubacterium lentum WYH-1 (20) and Enterococcus faecalis (17). In contrast to O. formigenes, these species are able to use substrates other than oxalate for growth, and whether they will degrade oxalate in the gut where alternative substrates are provided has not been determined. We found that O. formigenes strains can maintain significant oxalate degradation when cultivated in vitro with mixed human fecal bacteria and that they can establish resident gastrointestinal populations, measured by oxalate-degrading capacity in feces, when ingested orally. Molecular studies show that many human fecal bacteria have not yet been cultivated (34), so it is possible that as-yet-uncultivated species may contribute to oxalate degradation in the gut. It seems pertinent, however, that we found no evidence for selection of functional oxalate-degrading bacteria in either batch culture or in continuous fermentors prior to inoculation with O. formigenes.
A second issue relates to how oxalate-degrading activity may be lost by a significant proportion of adults. The favored explanation is that oxalate-degrading bacteria are susceptible to antibiotics and other therapeutic drugs (5). Here, clarithromycin and doxycycline were particularly inhibitory; the effective inhibitory concentration of ca. 1 μg ml−1 would presumably be exceeded in the gut of individuals receiving standard therapeutic doses of ≥300 mg. Doxycycline is used to combat respiratory tract and urinary infections (35). Among other applications, clarithromycin may be used, in combination with other drugs, to treat gastroduodenal infections with Helicobacter pylori (6, 16). Two strains of oxalate-degrading bacteria from rats were sensitive to chloramphenicol, colistin, and tetracycline, and one strain was sensitive to clindamycin. The strains were resistant to erythromycin, vancomycin, rifampin, streptomycin, penicillin, carbenicillin, ampicillin, cephalothin, and neomycin (11). Whether the practical use of antibiotics is responsible for the loss of oxalate-degrading activity requires urgent investigation. This could enable the selection of substances and/or doses causing minimal disturbance of the gut microbial flora, or posttherapy inoculation with O. formigenes to reestablish oxalate-degrading populations.
Gastric acidity and the action of bile salts are considered to act as barriers to the survival of ingested bacteria in the gut (8, 14). The survival rates of O. formigenes strains at low pH in this study were comparable to those of ingested Bifidobacterium strains in vivo (7). Others (5) found that ≥0.8 mM deoxycholate reduced oxalate degradation by human and guinea pig strains of O. formigenes, consistent with the present data. Up to approximately 15 mM bile salts may be encountered in the gut, though their antibacterial activity may be reduced by the presence of biliary phospholipids (25).
Although strain HC1 was the least robust, aerotolerant, and acid resistant of the strains tested here, it was successfully used to establish oxalate degradation in the gut of individuals lacking this activity. Our work showed that small populations of strain HC1 in fermentors, detectable by PCR but not by counting of viable cells, could recover to metabolize large amounts of oxalate. Thus, only small numbers of the bacterium may have to reach the colon in order to colonize. Complete washout was not achieved here, presumably due to traces of oxalate in the complex CH medium, which contains many plant-derived components. Oxalate is so ubiquitous in plants (vegetables and fruits) that “practical” oxalate-free diets for humans are difficult, if not impossible, to formulate (19). Colonization of humans by strain HC1 was persistent since detectable activity was observed months after ingestion.
Aerotolerance could favor survival of a probiotic strain during its production and distribution. The aerotolerance of strain Va3 is high compared to that of most gut anaerobes, but not unprecedented. Porphyromonas gingivalis was shown to survive exposure to air for 5 h without loss of viability (26) and some black-pigmented Bacteroides are highly aerotolerant (4).
SCFA concentrations in the fermentors were similar to those in other colonic fermentor studies (1), and increasing D, in accord with reference 1, reduced the butyrate concentration. These findings suggest that the Ox-negative feces used provided an otherwise representative fermentation. Here, O. formigenes maintained its population size and oxalate-degrading capacity in the presence of human fecal flora at D values from 0.036 to 0.015 h−1, consistent with colonic transit times in healthy adults (9). Even when D was 0.0625 h−1, more than 90% of the oxalate in the fermentor medium disappeared shortly after introduction of strain HC1. When D was 0.125 h−1, larger amounts of oxalate were detected in the fermentor, suggesting that this is approaching the D at which the bacterium would wash out. Persistent diarrhea, which may accompany disorders such as inflammatory bowel disease, might result in loss of Oxalobacter and thus contribute to the increase in oxalate absorption that characterizes enteric hyperoxaluria (M. Ghosh, H. Sidhu, R. Roses, and D. R. Cave, Abstr. 100th Annu. Conf. Am. Gastroenterol. Assoc., Gastroenterology 116:A551, 1999).
The ability of O. formigenes to establish oxalate degradation in fermentors suggests that colonization of the gut epithelium surface may not be necessary in vivo. Little is known about the location of O. formigenes in the human gut. However, it seems pertinent that oxalate-degrading activity was not detected in contents from the small intestine of laboratory rats that had been inoculated with O. formigenes, while rapid rates of oxalate degradation were measured in cecal and colonic contents from these rats (12). This fits with the current concept that anaerobic conditions are needed for growth of O. formigenes.
The values for urinary oxalate and creatinine concentrations in the four individuals tested fell within the range reported for healthy individuals following administration of similar oxalate loads (22). The reductions in urinary oxalate excretion and oxalate/creatinine ratios soon after ingestion of O. formigenes cells with the oxalate load together suggest that the ingested cells may have degraded oxalate in the small intestine, even though growth of this anaerobe in this relatively nonanaerobic environment is doubtful.
In summary, the present work suggests that intestinal oxalate degradation provides an ecological niche which O. formigenes may be uniquely fitted to occupy. Studies with colonized and noncolonized individuals of different ages and health status will advance our understanding of oxalate metabolism in the human gastrointestinal tract, and may ultimately provide practical approaches for the prevention or alleviation of hyperoxaluria.
Acknowledgments
We thank Harmeet Sidhu for guidance and for molecular analysis of strain Va3, Graham Calder for GC-mass spectrometry, Greitje Zuur for statistical advice, and Herb Cook and Gerald Lobley for valuable assistance.
The Scottish Executive Environment and Rural Affairs Department finance the Rowett Research Institute. M.J.A. is supported by U.S. Public Health Service grant DK-53556.
REFERENCES
- 1.Allison, C., C. McFarlan, and G. T. Macfarlane. 1989. Studies on mixed populations of human intestinal bacteria grown in single stage and multistage continuous culture systems. Appl. Environ. Microbiol. 55:672-678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allison, M. J., and N. S. Jensen. 1996. Are there two species of intestinal oxalate degrading bacteria?, p. 152-153. In C. Y. C. Pak, M. I. Resnik, and G. M. Preminger (ed.), Urolithiasis. Millet the Printer, Dallas, Tex.
- 3.Allison, M. J., S. L. Daniel, and N. A. Cornick. 1995. Oxalate-degrading bacteria, p. 131-168. In S. R. Khan (ed.), Calcium oxalate in biological systems. CRC Press, Boca Raton, Fla.
- 4.Amano, A., H. Tamagawa, M. Takegaki, Y. Murakami, S. Shizuishi, and A. Tsunemitsu. 1988. Relationship between enzyme activities involved in oxygen-metabolism and oxygen tolerance in black-pigmented Bacteroides. J. Dent. Res. 67:1196-1199. [DOI] [PubMed] [Google Scholar]
- 5.Argenzio, R. A., J. A. Liacos, and M. J. Allison. 1988. Intestinal oxalate-degrading bacteria reduce oxalate absorption and toxicity in guinea-pigs. J. Nutr. 118:787-792. [DOI] [PubMed] [Google Scholar]
- 6.Bardhan, K. D., C. Dallaire, H. Eisold, and A. E. Duggan. 1997. Ranitidine bismuth citrate with clarithromycin for the treatment of duodenal ulcer. Gut 41:181-186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berrada, N., J.-F. Lemeland, G. Laroche, P. Thuvenot, and M. Piaia. 1991. Bifidobacterium from fermented milks: survival during gastric transit. J. Dairy Sci. 74:409-413. [DOI] [PubMed] [Google Scholar]
- 8.Bezkorovainy, A. 2001. Probiotics: determinants of survival and growth in the gut. Am. J. Clin. Nutr. 73(Suppl.):399S-405S. [DOI] [PubMed] [Google Scholar]
- 9.Bouchoucha, M., and S. R. Thomas. 2000. Error analysis of classic colonic transit time estimates. Am. J. Physiol. Gastrointest. Liver Physiol. 279:G520-G527. [DOI] [PubMed] [Google Scholar]
- 10.Bryant, M. P. 1972. Commentary on the Hungate technique for cultivation of anaerobic bacteria. Am. J. Clin. Nutr. 25:1324-1328. [DOI] [PubMed] [Google Scholar]
- 11.Daniel, S. L., P. A. Hartman, and M. J. Allison. 1987. Microbial degradation of oxalate in the gastrointestinal tracts of rats. Appl. Environ. Microbiol. 53:1793-1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Daniel, S. L., P. A. Hartman, and M. J. Allison. 1987. Intestinal colonization of laboratory rats with Oxalobacter formigenes. Appl. Environ. Microbiol. 53:2767-2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dawson, K. A., M. J. Allison, and P. A. Hartman. 1980. Isolation and some characteristics of anaerobic oxalate degrading bacteria from the rumen. Appl. Environ. Microbiol. 40:833-839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Floch, M. H., H. J. Binder, B. Filburn, and W. Gershengoren. 1972. The effect of bile acids on intestinal microflora. Am. J. Clin. Nutr. 25:1418-1426. [DOI] [PubMed] [Google Scholar]
- 15.Hobson, P. N. 1969. Rumen bacteria. Methods Microbiol. 3b:133-149. [Google Scholar]
- 16.Hoffman, J. S., and D. R. Cave. 2001. Treatment of Helicobacter pylori. Curr. Opin. Gastroenterol. 17:30-34. [DOI] [PubMed] [Google Scholar]
- 17.Hokama, S., Y. Honma, C. Toma, and Y. Ogawa. 2000. Oxalate-degrading Enterococcus faecalis. Microbiol. Immunol. 44:235-240. [DOI] [PubMed] [Google Scholar]
- 18.Holmes, R. P., and D. G. Assimos. 1998. Glyoxylate synthesis, and its modulation and influence on oxalate synthesis. J. Urol. 160:1617-1624. [PubMed] [Google Scholar]
- 19.Holmes, R. P., and M. Kennedy. 2000. Estimation of the oxalate content of foods and daily oxalate intake. Kidney Int. 57:1662-1667. [DOI] [PubMed] [Google Scholar]
- 20.Ito, H., N. Miura, M. Masai, K. Yamamoto, and T. Hara. 1996. Reduction of oxalate content of foods by the oxalate degrading bacterium, Eubacterium lentum WYH-1. Int. J. Urol. 3:31-34. [DOI] [PubMed] [Google Scholar]
- 21.Larsen, K. 1972. Creatinine assay by the reaction kinase principle. Clin. Chim. Acta 41:209-217. [DOI] [PubMed] [Google Scholar]
- 22.Liebman, L., E. Harvey, and W. Chai. 1999. Olestra and fat inhibit oxalate absorption. Nutr. Res. 19:1277-1285. [Google Scholar]
- 23.Lowe, S. E., M. K. Theodorou, A. J. P. Trinci, and R. B. Hespell. 1985. Growth of anaerobic rumen fungi on defined and semi-defined media lacking rumen fluid. J. Gen. Microbiol. 131:2225-2229. [Google Scholar]
- 24.Macfarlane, G. T., S. Hay, and G. R. Gibson. 1989. Influence of mucin on glycosidase, protease and arylamidase activities of human gut bacteria grown in a 3-stage continuous culture system. J. Appl. Bacteriol. 67:521-527. [DOI] [PubMed] [Google Scholar]
- 25.Marteau, P., M. Minekus, R. Havenaar, and J. H. J. Huis in't Veld. 1997. Survival of lactic acid bacteria in a dynamic model of the stomach and small intestine: validation and the effects of bile. J. Dairy Sci. 80:1031-1037. [DOI] [PubMed] [Google Scholar]
- 26.Nakayama, K. 1994. Rapid viability loss on exposure to air in a superoxide dismutase-deficient mutant of Porphyromonas gingivalis. J. Bacteriol. 176:1939-1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Neuhaus, T. J., T. Belzer, N. Blau, B. Hoppe, H. Sidhu, and E. Leumann. 2000. Urinary oxalate excretion in urolithiasis and nephrocalcinosis. Arch. Dis. Child. 82:322-326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pirt, S. J. 1985. Principles of microbe and cell cultivation, p. 37-38. Blackwell Scientific Publications, Oxford, United Kingdom.
- 29.Richardson, A. J., A. G. Calder, C. S. Stewart, and A. Smith. 1989. Simultaneous determination of volatile and non-volatile acidic fermentation products of anaerobes by capillary gas chromatography Lett. Appl. Microbiol. 9:5-8. [Google Scholar]
- 30.Sidhu, H., M. J. Allison, and A. B. Peck. 1997. Identification and classification of Oxalobacter formigenes strains by using oligonucleotide probes and primers. J. Clin. Microbiol. 35:350-353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sidhu, H., L. Enatska, S. Ogden, W. N. Williams, M. J. Allison, and A. B. Peck. 1997. Evaluating children in the Ukraine for colonization with the intestinal bacterium Oxalobacterium formigenes, using a polymerase chain reaction detection system. Mol. Diagn. 2:89-97. [DOI] [PubMed] [Google Scholar]
- 32.Sidhu, H., B. Hoppe, H. Albrecht, K. Tenbrock, S. Bromme, E. Rietschel, and A. B. Peck. 1998. Absence of Oxalobacter formigenes in cystic fibrosis patients: a risk factor for hyperoxaluria. Lancet 352:1026-1029. [DOI] [PubMed] [Google Scholar]
- 33.Stewart, C. S., S. H. Duncan, A. J. Richardson, G. Calder, and P. J. S. Dewey. 1995. The effect of the presence of glucose on the fermentation of mannose by the anaerobic fungus Neocallimastix frontalis strain RE1. FEMS Microbiol. Lett. 127:57-63. [Google Scholar]
- 34.Wilson, K. H., and R. B. Blitchington. 1996. Human colonic biota studied by ribosomal DNA sequence analysis. Appl. Environ. Microbiol. 62:2273-2278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ziegler, T., C. Winkler, K. Wege, and H. Schmechel. 2000. Doxycycline—the forgotten antimicrobial drug. Med. Klin. 95:629-631. [DOI] [PubMed] [Google Scholar]