Abstract
Bacillary dysentery caused by Shigella species is a public health problem in developing countries including Bangladesh. Although, shigellae-contaminated food and drinks are often the source of the epidemic's spread, the possible presence of the pathogen and transmission of it through environmental waters have not been adequately examined. We analyzed surface waters collected in Dhaka, Bangladesh, for the presence of shigellae by a combination of PCR assays followed by concentration and culturing of PCR-positive samples. Analysis of 128 water samples by PCR assays for Shigella-specific virulence genes including ipaBCD, ipaH, and stx1 identified 14 (10.9%) samples which were positive for one or more of these virulence genes. Concentration of the PCR-positive samples by filtration followed by culturing identified live Shigella species in 11 of the 14 PCR-positive samples. Analysis of rRNA gene restriction patterns (ribotype) showed that the environmental isolates shared ribotypes with a collection of clinical isolates, but in contrast to the clinical isolates, 10 of the 11 environmental isolates were either negative or carried deletions in the plasmid-encoded invasion-associated genes ipaB, ipaC, and ipaD. However, all environmental Shigella isolates were positive for the chromosomal multicopy invasion-associated gene ipaH and all Shigella dysenteriae type 1 isolates were positive for the stx1 gene in addition to ipaH. This study demonstrated the presence of Shigella in the aquatic environment and dispersion of different virulence genes among these isolates which appear to constitute an environmental reservoir of Shigella-specific virulence genes. Since critical virulence genes in Shigella are carried by plasmids or mobile genetic elements, the environmental gene pool may contribute to an optimum combination of genes, causing the emergence of virulent Shigella strains which is facilitated in particular by close contact of the population with surface waters in Bangladesh.
Shigellosis occurs as a disease endemic in Bangladesh, and at least three large epidemics caused by Shigella dysenteriae type 1 have occurred between 1972 and 1994, causing high morbidity and mortality, particularly in children (6, 16, 23). The most common underlying cause of death in fatal childhood shigellosis is severe colitis combined with septicemia and pneumonia (26). In Bangladesh, the predominant species of the genus Shigella are S. flexneri and S. dysenteriae type 1; infections due to S. dysenteriae 1 usually progress to the most severe stages of dysentery and life-threatening complications (6, 25). Factors affecting the emergence or decline of epidemic shigellosis are not clear, and shigellae are generally believed to have only a human or primate host. Shigellae-contaminated food and drinks are often the source of epidemic spread, and very little is known about its presence and possible spread through environmental waters. In many developing countries with inadequate sanitation, fecal contamination of environmental waters by enteric pathogens is very common. It is therefore important to understand whether Shigella can survive and persist in environmental waters in the absence of a primate host and the virulence characteristics of such environmental strains.
Identification of Shigella in environmental samples, where the number of organisms is likely to be small, is limited mainly by the lack of a suitable enrichment technique. Although DNA probes or PCR assays directed against the large invasion plasmid or genes encoding Shiga toxins (13, 20, 21, 29) can be used to detect the presence of the organism, isolation of the live bacteria is essential to characterize their pathogenic potential as well as their sensitivity to antimicrobial agents. Detailed analysis of a large number of water samples for the presence of Shigella by conventional culture methods is impractical, particularly because the number of non-lactose-fermenting colonies to be further analyzed may be too high (13). The present study was designed to isolate Shigella strains from the environment by a combination of PCR and culture methods and characterize them by appropriate biochemical and serological tests. Furthermore, molecular techniques were used to genetically characterize such environmental Shigella isolates and compare them with representative clinical isolates to understand the origin and pathogenic potential of the environmental Shigella isolates.
MATERIALS AND METHODS
Bacterial strains and water samples.
Water samples for the study were collected once every 2 weeks during a period of 6 months between June and November 2001 in different sampling sites in Dhaka. A total of 128 water samples, which included samples from different sites along two major rivers and a lake in Dhaka City, were analyzed in the study. All water samples were collected in sterile containers and transported to the laboratory for processing within 2 h of collection. Initially, all samples were immediately subjected to multiplex PCR assays as well as culture as described later in this paper. An environmental water sample mixed with different dilutions of a control, S. dysenteriae type 1 strain 33891 (15), was also included as a positive control to determine the detection limit of these assays. Non-lactose-fermenting colonies were picked from culture plates and were subjected to further analysis for the identification and isolation of possible Shigella colonies. After a review of these preliminary PCR and culture results, samples taken after August 2001 which were positive in an initial round of screening by PCR only were further analyzed by culture methods. Clinical Shigella strains used as controls in this study were either obtained from patients who reported to the treatment facilities of the International Centre for Diarrhoeal Disease Research, Bangladesh (ICDDR,B) or obtained from the culture collection of ICDDR,B.
PCR assays.
PCR reagents and kits were obtained from either the Perkin-Elmer Corporation (Norwalk, Conn.) or Bethesda Research Laboratories (Gaithersburg, Md.). Other enzymes and chemicals used in this assay were from Sigma Chemical Company (St. Louis, Mo.). Water samples (1.5 ml) were centrifuged initially for 2 min at low speed (1,000 rpm) in an Eppendorf microcentrifuge tube to precipitate debris. The supernatant was transferred into a fresh tube and centrifuged at high speed (14,000 rpm) to collect the suspended microorganisms. The pellet was resuspended in 100 μl of 50 mM Tris-HCl buffer (pH 7.5) containing 20% (wt/vol) sucrose, 50 mM EDTA, and 100 μg of lysozyme/ml. The suspension was incubated at 37°C for 30 min, and then 300 μl of a solution containing 50 mM NaCl, 1% (wt/vol) sodium dodecyl sulfate (SDS), and 200 μg of proteinase K/ml was added. The sample was incubated for an additional 60 min at 37°C, and then extraction was performed twice with an equal volume of phenol-chloroform (1:1, vol/vol) containing 2% isoamyl alcohol. The nucleic acids were precipitated by adding 2.5 volumes of absolute ethanol, harvested by centrifugation, and resuspended in 15 μl of TE buffer (10 mM Tris-Cl [pH 7.5], 1 mM EDTA). One to five microliters of the extracted nucleic acids was used in a multiplex PCR assay for the genes encoding invasive plasmid antigens (ipaB, ipaC, and ipaD) and Shiga toxin (stx1), as well as a second PCR assay for ipaH. For analyzing Shigella isolates by PCR, colonies were suspended in sterile distilled water and disrupted by boiling for 3 min, and aliquots of the suspension were directly used for PCR assays. Dilutions of a known Shigella strain were included with each batch of PCR assays as a positive control to determine the detection limit of the assay. PCR primers used in this study were either previously reported (22) or synthesized based on published sequences of the relevant genes (27, 28). Descriptions and sequences of the PCR primers used in this study are given in Table 1. Thermal cycling parameters for the PCR assays consisted of denaturation at 94°C for 2 min, annealing of primers at 55°C for 2 min, and primer extension at 72°C for 3 min. Amplification was performed for 35 cycles, and the expected sizes of the amplicons were ascertained by electrophoresis in 1.5% agarose gels with an appropriate molecular size marker (1-kb DNA ladder; Gibco BRL). The identities of all PCR amplicons were further confirmed by Southern blot hybridization with specific probes.
TABLE 1.
PCR primers used in this study to detect Shigella-specific virulence genes
| Primer | Description | Sequence |
|---|---|---|
| EVT-1 | Sense primer for stx1 | 5′-CAACACTGGATGATCTCAG |
| EVT-2 | Antisense primer for stx1 | 5′-CCCCCTCAACTGCTAATA |
| IpaBCD-u | Upstream primer for ipaBCD | 5′-GCTATAGCAGTGACATGG |
| IpaBCD-d | Downstream primer for ipaBCD | 5′-ACGAGTTCGAAGCACTC |
| IpaH-u | Upstream primer for ipaH | 5′-GCTGGAAAAACTCAGTGCCT |
| IpaH-d | Downstream primer for ipaH | 5′-CCAGTCCGTAAATTCATTCT |
Culturing of environmental samples.
Environmental water samples were analyzed for the presence of Shigella by modification of a previously described method (1). Briefly, 50 ml of water was centrifuged at low speed (3,000 rpm in a Sorval SS34 rotor) for 5 min to precipitate debris, the supernatant was filtered through a 0.22-μm-pore-size filter (Millipore Corporation, Bedford, Mass.), and the filters were removed and incubated in nutrient broth for 4 h at 37°C with shaking. Aliquots of this suspension were streaked on MacConkey agar plates (Difco; Becton Dickinson and Company) and xylose lysine desoxycholate agar plates (Plasmatec Laboratory Products Ltd., Dorset, United Kingdom) and incubated overnight at 37°C. Suspected colonies were picked and subjected to biochemical and serological tests to identify Shigella. Depending on the number of non-lactose-fermenting colonies appearing on each plate, between 5 and 15 colonies were tested by biochemical methods. Eventually, culture-confirmed Shigella isolates were further analyzed by PCR and probes for the presence of virulence genes.
Probes and hybridization.
The gene probe used in this study to detect the ipaB, ipaC, and ipaD genes was a 3.4-kb HindIII fragment of pWR1002 (28, 29). Probes for the ipaH and stx1 genes were PCR-generated amplicons derived from a control, S. dysenteriae type 1 strain 33891 (15), with specific primers (Table 1). The rRNA gene probe was a 7.5-kb BamHI fragment of the Escherichia coli rRNA clone pKK3535 described previously (4, 8). Southern blots were prepared using nylon filters (Hybond; Amersham International Plc., Ayelesbury, United Kingdom) and processed by standard methods (17). Probes were labeled by random priming (11) with a random primers DNA labeling kit (Gibco BRL) and [α-32P]dCTP (3,000 Ci/mmol; Amersham).
Restriction fragment length polymorphisms (RFLP) in different genes were analyzed as described previously (8, 9). Briefly, total DNA was isolated from strains grown overnight, and approximately 5 μg of the whole-cell DNA was digested with the appropriate restriction enzymes (Bethesda Research Laboratories) by using 5 U of enzyme per μg of DNA. The digested DNAs were electrophoresed in 0.8% agarose gels, and DNA fragments from the gels were blotted onto nylon membranes (Hybond-N; Amersham). Southern blots were prehybridized for 2 h and then hybridized with freshly denatured radioactively labeled probes at 68°C for 18 h in a solution containing 6× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 5× Denhardt solution (1× Denhardt solution is 0.02% polyvinylpyrrolidone, 0.02% Ficoll 400, and 0.02% bovine serum albumin), 0.5% SDS, and 100 μg of freshly denatured sheared salmon sperm DNA per ml. Hybridized blots were washed once in 2× SSC for 5 min at room temperature, two times in 2× SSC-0.1% SDS for 10 min at 65°C, and once in 0.2× SSC-0.1% SDS for 15 min at 65°C. Autoradiographs were developed from the hybridized filters with either Kodak (Rochester, N.Y.) X-Omat AR film or Fuji X-ray film at −70°C as described previously (9). RFLP in different genes and their flanking chromosomal sequences were studied from the bands that appeared in the corresponding autoradiographs. Banding profiles derived from different strains were compared based on the absence or presence of bands in different loci.
Antimicrobial susceptibility analysis.
All isolates were tested for antimicrobial resistance by the method of Bauer et al. (2) on standard antibiotic disks (Oxoid Ltd., Basingstoke, Hampshire, United Kingdom) at the following antibiotic concentrations (micrograms/disk): ampicillin, 10; chloramphenicol, 30; streptomycin, 10; tetracycline, 30; trimethoprim-sulfamethoxazole, 1.25 and 23.75, respectively; kanamycin, 30; erythromycin, 15; gentamicin, 10; ciprofloxacin, 5; norfloxacin, 10; rifampin, 5; and nalidixic acid, 30.
RESULTS AND DISCUSSION
Detection of Shigella in environmental water.
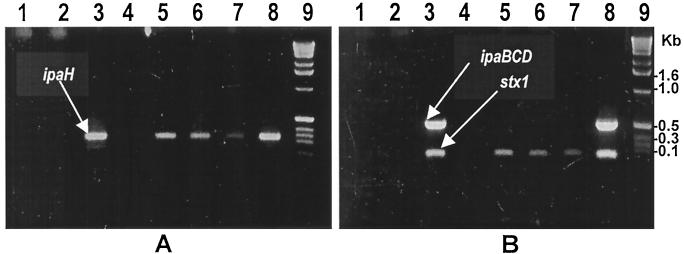
Understanding the ecology of Shigella had been limited mainly by the lack of suitable techniques to detect the presence of Shigella in environmental samples. In the present study, we used a combination of PCR and culture techniques to detect Shigella in surface waters. Of a total of 128 water samples tested, 14 (10.9%) were positive by PCR for one or more Shigella-specific virulence-associated genes (Fig. 1). Culturing of these 14 samples allowed isolation of live Shigella species from 11 samples (Table 2). Initially, we standardized the assays by simultaneously conducting PCR tests as well as culturing of environmental water samples and water samples mixed with different dilutions of control S. dysenteriae type 1 strain 33891. As expected, water samples which yielded live colonies on culture were invariably positive by PCR and none of the PCR-negative samples were positive by culturing. PCR assays were able to distinctly identify as few as 5 CFU of the bacterium/ml in water samples. Subsequently, for the environmental samples, we cultured only those samples which were found to be positive in an initial round of screening by PCR assays. However, in the present study, 3 of 14 PCR-positive samples did not yield any Shigella colonies on culture. These three samples were distinctly positive for the ipaH gene in repeated PCR assays. It is not clear whether the inability to culture Shigella from these three PCR-positive samples was due to the absence of live cells, since dead cells or their nucleic acids may also yield a positive PCR. An alternative explanation could be the possible presence of the ipaH gene in other species of environmental bacteria which escaped detection in our culture assay. Nevertheless, these results showed that PCR-based detection of Shigella-specific virulence genes can be used as an indicator for the possible presence of live Shigella strains in environmental waters. It is worth mentioning that it is impractical to test a large number of samples by culture followed by a detailed analysis of non-lactose-fermenting colonies. Hence, an initial screening by PCR may reduce the number of samples to be cultured and practically enhance the detection of Shigella in environmental samples.
FIG. 1.
PCR analysis of environmental surface water samples for Shigella-specific virulence genes. Products of a PCR assay for the ipaH gene (A) and a multiplex PCR assay for the ipaBCD and stx1 genes (B) are shown. Lanes 1 through 7 contain PCR products of environmental water samples, and lane 8 contains a positive control strain. Lane 9 shows the 1-kb DNA ladder (Bethesda Research Laboratories) used as molecular size markers.
TABLE 2.
Screening of surface water samples by PCR for Shigella-specific genes and by culture for Shigella spp.
| Source | No. of samples | No. positive by PCR (%) | No. confirmed by culture (%) |
Shigella species isolated
|
|
|---|---|---|---|---|---|
| S. dysenteriae 1 | S. flexneri | ||||
| River | 86 | 10 (11.6) | 7 (8.1) | 3 | 4 |
| Lake | 42 | 4 (9.5) | 4 (9.5) | 1 | 3 |
Molecular analysis of environmental Shigella strains.
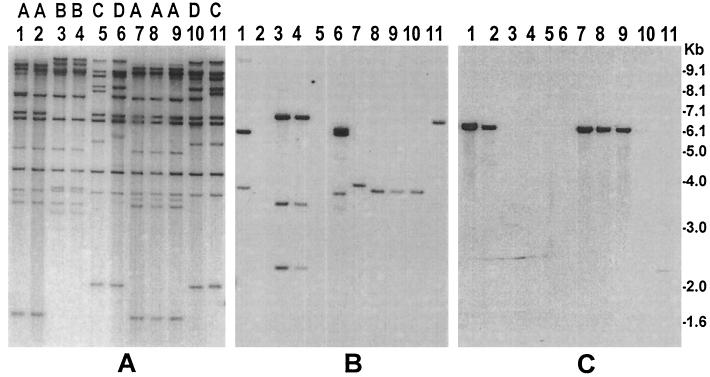
To understand the possible origin of the environmental Shigella strains and their potential to cause disease, we compared the environmental strains with virulent Shigella strains isolated from hospitalized patients. The most important components of virulence in Shigella infection include adherence, invasiveness, and toxigenicity (24, 25). The genes for these virulence factors reside on either plasmids or the bacterial chromosome or on lysogenic bacteriophages. Shigella strains have large plasmids that encode many virulence genes (5, 24), especially genes involved in invasion and intracellular movement, and these products are known as invasion plasmid-associated antigens. We examined the environmental strains for the presence of different virulence genes and compared them with the clinical strains by restriction analysis of conserved rRNA genes (ribotype). Ribotyping was used previously to analyze Shigella (8) and toxigenic Vibrio cholerae (9) strains and demonstrated that restriction patterns of conserved rRNA genes may be considered fairly stable markers for identifying different clones of enteric bacteria. In the present study, analysis of RFLP in the rRNA genes of the 11 environmental strains and 10 clinical strains showed a total of four different ribotype patterns, designated A through D (Fig. 2). While all S. dysenteriae type 1 isolates belonged to ribotype pattern A, S. flexneri strains produced two ribotypes, C and D, and the environmental strains shared ribotypes with the clinical strains, suggesting a clonal relationship. However, when analyzed for the presence of virulence genes by PCR, 10 out of the 11 environmental strains were found to be negative for the ipaB, ipaC, and ipaD genes, which encode factors involved in epithelial cell invasion by Shigella species (5). These strains were further analyzed by Southern blot hybridization using a DNA probe for the ipaB, ipaC, and ipaD genes. Strains which were negative in the PCR assay for ipaBCD produced a small-sized HindIII restriction fragment (∼7.0 to 2.5 kb) of the ipaB, ipaC, and ipaD genes and flanking sequences compared to the clinical strains, which produced multiple bands between 7.5 and 2.2 kb (Fig. 2). This suggested that these environmental strains carried a deletion in the ipaBCD gene cluster.
FIG. 2.
Molecular analysis of S. dysenteriae type 1 and S. flexneri strains isolated from environmental waters in Bangladesh and comparison with representative clinical Shigella strains. Genomic DNA was digested with HindIII, and corresponding Southern blots were hybridized with the rRNA gene probe (A), ipaBCD probe (B), and Shiga toxin gene probe (C). Ribotype designations A through D, which correspond to the rRNA gene restriction patterns of different strains, are shown on top of the corresponding lanes. Lanes 1, clinical S. dysenteriae type 1 strain (SD-16); lanes 2 and 7 through 9, S. dysenteriae type 1 strains (SD-477, SD-461, SD462, and SD-469) isolated from environmental waters; lanes 3 and 4, clinical S. dysenteriae type 2 strains (AF-8724 and AF-979); lanes 5, 10, and 11, S. flexneri strains (SF-525, SF-455, SF-526) isolated from environmental waters; lanes 6, clinical S. flexneri strain (SF-12163). Numbers indicating molecular sizes of bands correspond to the 1-kb DNA ladder (Gibco BRL) used as molecular size markers.
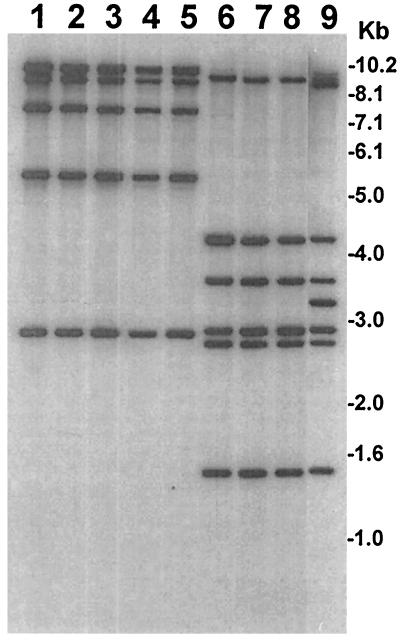
Fresh contamination of surface water by the fecal material of dysentery patients is a possibility in developing countries where sanitation is poor, resulting in the presence of shigellae in surface water. Several previous studies have also detected shigellae in surface water or sewage samples (7, 18) and have indicated that Shigella strains can possibly be transported by surface waters (18). Even a recreational spray fountain was found to be contaminated with Shigella and was associated with the spread of the pathogen (12). However, in the present study, the demonstration of the absence or deletion of crucial virulence genes in environmental Shigella isolates suggested that the presence of these strains was not possibly due to fresh fecal contamination of surface water, but instead, these strains may have survived and persisted in the aquatic environment. It appears that since in the environment the invasion-associated genes did not have any known function, the bacteria apparently lost some of these plasmid-encoded genes. However, a chromosomally located multicopy virulence gene, ipaH, which is also known to have a role in producing invasive characteristics (5, 21), was found to be more stable and was present in all the environmental strains analyzed (Fig. 3). Therefore, PCR screening of environmental samples for the ipaH gene should provide a better indicator of the possible presence of shigellae than screening for the plasmid-carrying ipaBCD genes.
FIG. 3.
Analysis of S. dysenteriae type 1 and S. flexneri strains isolated from environmental waters in Bangladesh for the presence of ipaH genes. Genomic DNAs were digested with HindIII, and corresponding Southern blots were hybridized with the ipaH probe. Identities of the environmental isolates (strain numbers) represented in different lanes are as follows. Lanes 1 and 9 represent clinical strains of S. dysenteriae 1 (SD-16) and S. flexneri (SF-12163), respectively, that were included in the study for comparison with environmental isolates. Lanes 2 through 5 represent S. dysenteriae 1 isolates (SD-461, SD-462, SD-469, and SD-479), and lanes 6 through 8 represent S. flexneri isolates (SF-455, SF-505, and SF-507). Numbers indicating molecular sizes of bands correspond to the 1-kb DNA ladder (Gibco BRL) used as molecular size markers.
Taxonomically, the genus Shigella appears closely related to E. coli, which is a normal inhabitant of the human and animal intestine. It has been suggested that the four species of Shigella are so closely related to E. coli that all these bacteria could be considered members of a single species (14). They share greater than 90% homology by DNA-DNA reassociation analysis and display colinearity of their chromosomes such that gene transfer by conjugation and transduction and formation of recombinant between Shigella and E. coli occur with high efficiency (14, 19). The major virulence genes in Shigella are encoded by accessory genetic elements such as plasmids or by chromosomal genes of bacteriophage origin (20, 24, 27, 28). This is suggestive of the possibility that shigellae may lose or acquire clusters of genes encoding virulence as well as other accessory functions and thus the transit between virulent and possible nonvirulent forms which are indistinguishable from normal gut flora (3, 14, 19). Conversion from a nonpathogenic to pathogenic form has been demonstrated for another enteric pathogen, V. cholerae, which also produces a phage-encoded enterotoxin (10).
Epidemiological implications.
Shigella epidemics spread through contaminated water and food, and transmission of the pathogen is believed to be facilitated by a very low infectious dose of Shigella (6, 23, 25). However, the mechanism associated with periodic outbreaks of shigellosis in areas of shigella endemicity is not clear. The presence of Shigella in surface waters, as demonstrated in the present study, may have public health implications. Although most of the environmental strains lacked one or more of the known virulence genes, the environmental strains shared a ribotype with the clinical strains (Table 3) and belonged to the two species which are associated with most cases of shigellosis in Bangladesh (6). Moreover, the serotypes of these environmental strains were the same as those of the clinical strains. The acquisition of clusters of virulence genes is recognized as an important element in the evolution of bacterial pathogens (14). Thus, plasmid- or bacteriophage-mediated horizontal transfer of genes may lead to the emergence of virulent Shigella strains from closely related avirulent precursors. The detection of Shigella strains in the surface water thus appears significant. It seems possible that these strains may participate in receiving or donating virulence genes, leading to the origination of virulent Shigella strains. Previously reported epidemics of shigellosis have always been preceded by the appearance of new antibiotic resistance among Shigella strains. The drug resistance patterns of the environmental and clinical Shigella strains are summarized in Table 3. It is important to note that most of the environmental strains were resistant to one or more antibiotics. These strains may thus also serve as reservoirs of drug resistance genes.
TABLE 3.
Characteristics of Shigella strains isolated from surface water samples and their comparison with clinical strains isolated from patients with shigellosis
| Strain no. | Description | Source | Ribotypea | PCR analysis of virulence genes
|
Antibiotic resistanceb | ||
|---|---|---|---|---|---|---|---|
| ipaH | ipaBCD | stx1 | |||||
| SD-16 | S. dysenteriae type I | Patient | A | + | + | + | A, T, Na, Rf, S, SXT, C, E |
| SD-37 | S. dysenteriae type I | Patient | A | + | + | + | A, T, Na, Rf, S, SXT, C, E |
| SD-39 | S. dysenteriae type I | Patient | A | + | + | + | A, T, Na, Rf, S, SXT, C, E |
| SD-2078 | S. dysenteriae type I | Patient | A | + | + | + | A, T, Na, Rf, S, SXT, C, E |
| SD-2146 | S. dysenteriae type I | Patient | A | + | + | + | A, T, Na, S, SXT, C, E |
| SD-2499 | S. dysenteriae type I | Patient | A | + | + | + | A, T, Na, S, SXT, C, E |
| AF-8724 | S. dysenteriae type II | Patient | B | + | + | − | T, Rf, E |
| AF-979 | S. dysenteriae type II | Patient | B | + | + | − | T, Rf, E |
| SF-5C592 | S. flexneri 2a | Patient | C | + | + | − | S, Rf, E |
| SF-12163 | S. flexneri 2b | Patient | D | + | + | − | T, S, Rf, C, E |
| SD-461 | S. dysenteriae type I | Surface water | A | + | − | + | A, T, Na, Rf |
| SD-462 | S. dysenteriae type I | Surface water | A | + | − | + | A, T, Na, Rf |
| SD-469 | S. dysenteriae type I | Surface water | A | + | − | + | A, T, Na, Rf, S |
| SD-477 | S. dysenteriae type I | Surface water | A | + | − | + | A, T, Na, Rf, S |
| SF-455 | S. flexneri 3a | Surface water | D | + | − | − | Rf |
| SF-505 | S. flexneri 3a | Surface water | D | + | − | − | Rf |
| SF-507 | S. flexneri 3a | Surface water | D | + | − | − | S, Rf |
| SF-508 | S. flexneri 3a | Surface water | D | + | − | − | Rf |
| SF-509 | S. flexneri 3a | Surface water | D | + | − | − | Rf |
| SF-525 | S. flexneri 2a | Surface water | C | + | − | − | Rf |
| SF-526 | S. flexneri 2b | Surface water | C | + | + | − | A, T, S, Rf |
Ribotypes are based on the HindIII restriction patterns of the rRNA genes and their flanking chromosomal sequences.
All strains were tested for resistance to ampicillin (A), tetracycline (T), chloramphenicol (C), erythromycin (E), gentamicin, ciprofloxacin, norfloxacin, nalidixic acid (Na), streptomycin (S), rifampin (Rf), and trimethoprim-sulfamethoxazole (SXT).
The environmental biology of Shigella, which is an almost exclusively human pathogen, is poorly understood, and animal or environmental reservoirs have not been identified. In the present study, we demonstrated the existence of Shigella in surface water in Bangladesh, where a large part of the rural and semiurban population consume surface water for daily needs. The present study also emphasizes the need to study the emergence of virulent Shigella strains from possible environmental nonvirulent progenitors by the acquisition of virulence genes and its relation to outbreaks of shigellosis.
Acknowledgments
This research was funded by a special research grant from the government of Japan to the ICDDR,B. The ICDDR,B is supported by countries and agencies which share its concern for the health problems of developing countries. Current donors providing unrestricted support include the aid agencies of the governments of Australia, Bangladesh, Belgium, Canada, Japan, the Kingdom of Saudi Arabia, The Netherlands, Sweden, Sri Lanka, Switzerland, and the United States of America.
We thank Afjal Hossain for secretarial assistance.
REFERENCES
- 1.American Public Health Association. 1992. Detection of pathogenic bacteria, p. 9-86-9-100. In A. E. Greenberg, L. S. Clesceri, and A. D. Eaton (ed.), Standard methods for the examination of water and wastewater, 18th ed. American Public Health Association, Washington, D.C.
- 2.Bauer, A. W., W. M. M. Kirby, J. C. Sherris, and M. Turk. 1966. Antibiotic susceptibility by a standardized single disk method. Am. J. Clin. Pathol. 45:493-496. [PubMed] [Google Scholar]
- 3.Brenner, D. J., G. R. Fanning, K. F. Johnson, R. V. Cutarella, and S. Falkow. 1969. Polynucleotide sequence relationships among members of Enterobacteriaceae. J. Bacteriol. 98:637-650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brosius, J., A. Ullrich, M. A. Raker, A. Gray, T. J. Dull, R. R. Gutell, and H. F. Noller. 1981. Construction and fine mapping of recombinant plasmids containing the rRNB ribosomal RNA operon of E. coli. Plasmid 6:112-118. [DOI] [PubMed] [Google Scholar]
- 5.Buysse, J. M., C. K. Stover, E. V. Oaks, M. Venkatesan, and D. J. Kopecko. 1987. Molecular cloning of invasion plasmid antigen (ipa) genes from Shigella flexneri: analysis of ipa gene products and genetic mapping. J. Bacteriol. 169:2561-2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen, L. C., M. Rahman, and A. M. Sarder. 1980. Epidemiology and causes of death among children in a rural area of Bangladesh. Int. J. Epidemiol. 9:25-33. [DOI] [PubMed] [Google Scholar]
- 7.De, A., P. C. Sen, and I. C. Tewari. 1993. Enteropathogenic bacteria in river Ganges in Varanasi. Indian J. Pathol. Microbiol. 36:425-432. [PubMed] [Google Scholar]
- 8.Faruque, S. M., K. Haider, M. M. Rahman, A. R. M. A. Alim, Q. S. Ahmad, M. J. Albert, and R. B. Sack. 1992. Differentiation of Shigella flexneri strains by rRNA gene restriction patterns. J. Clin. Microbiol. 30:2996-2999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Faruque, S. M., S. K. Roy, A. R. M. A. Alim, A. K. Siddique, and M. J. Albert. 1995. Molecular epidemiology of toxigenic Vibrio cholerae in Bangladesh studied by numerical analysis of rRNA gene restriction patterns. J. Clin. Microbiol. 33:2833-2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Faruque, S. M., Asadulghani, M. N. Saha, A. R. M. A. Alim, M. J. Albert, K. M. N. Islam, and J. J. Mekalanos. 1998. Analysis of clinical and environmental strains of nontoxigenic Vibrio cholerae for susceptibility to CTXΦ: molecular basis for the origination of new strains with epidemic potential. Infect. Immun. 66:5819-5825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feinberg, A., and B. Vogelstein. 1984. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal. Biochem. 137:266-267. [DOI] [PubMed] [Google Scholar]
- 12.Fleming, C. A., D. Caron, J. E. Gunn, M. S. Horine, B. T. Matyas, and M. A. Barry. 2000. An outbreak of Shigella sonnei associated with a recreational spray fountain. Am. J. Public Health 90:1641-1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Frankel, G., L. Riley, J. A. Giron, J. Valmassoi, A. Friedman, N. Strockbine, S. Falkow, and G. Schoolnik. 1990. Detection of Shigella in feces using DNA amplification. J. Infect. Dis. 161:1252-1256. [DOI] [PubMed] [Google Scholar]
- 14.Hacker, J., G. Blum-Ochler, I. Muhldorfer, and H. Tschape. 1997. Pathogenicity islands of virulent bacteria: structure, function and impact on microbial evolution. Mol. Microbiol. 123:1089-1097. [DOI] [PubMed] [Google Scholar]
- 15.Haider, K., A. Chatkaeomorakot, B. A. Kay, K. A. Talukder, D. N. Taylor, P. Echeverria, and D. A. Sack. 1990. Trimethoprim resistance gene in Shigella dysenteriae 1 isolates obtained from widely scattered locations of Asia. Epidemiol. Infect. 104:219-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Katz, S. L. 1986. The burden of disease resulting from diarrhea, p. 159-169. In S. L. Katz (ed.), New vaccine development: establishing properties. Diseases of importance in developing countries, vol. 2. National Academy Press, Washington, D.C. [Google Scholar]
- 17.Maniatis, T., E. F. Fritsch, and J. Sambrook. 1982. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 18.Mathan, V. I., P. Bhat, C. R. Kapadia, J. Ponniah, and S. J. Baker. 1984. Epidemic dysentery caused by Shiga bacillus in a southern Indian village. J. Diarrhoeal Dis. Res. 2:27-32. [PubMed] [Google Scholar]
- 19.Maurelli, A. T., E. Fernandez, C. A. Bloch, C. K. Rode, and A. Fasano. 1998. “Black holes” and bacterial pathogenicity: a large genomic deletion that enhances the virulence of Shigella spp. and enteroinvasive E. coli. Proc. Natl. Acad. Sci. USA 95:3943-3948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Newland, J. W., and R. J. Neill. 1988. DNA probes for Shiga-like toxins I and II and for toxin-converting bacteriophages. J. Clin. Microbiol. 26:1292-1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oberhelman, R. A., D. J. Kopecko, M. M. Venkatesan, E. Salazar-Lindo, E. Gotuzzo, A. Yi, E. Chea-Woo, R. Ruiz, C. Fernandez-Prada, R. León-Barúa, and R. B. Sack. 1993. Evaluation of alkaline phosphatase-labelled ipaH probe for diagnosis of Shigella infections. J. Clin. Microbiol. 31:2101-2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oyofo, B. A., Z. S. Mohran, S. H. El-Etr, M. O. Wasfy, and L. F. Peruski, Jr. 1996. Detection of enterotoxigenic Escherichia coli, Shigella, and Campylobacter spp. by miltiplex PCR assay. J. Diarrhoeal Dis. Res. 14:207-210. [PubMed] [Google Scholar]
- 23.Ronsmans, C., M. L. Bennish, and T. Wierzba. 1988. Diagnosis and management of dysentery by community health workers. Lancet ii:552-555. [DOI] [PubMed] [Google Scholar]
- 24.Salyers, A. A., and D. D. Whitt (ed.). 1994. Dysentery caused by Shigella species, p. 169-181. In Bacterial pathogenesis: a molecular approach. American Society for Microbiology, Washington, D.C.
- 25.Shears, P. 1996. Shigella infections. Ann. Trop. Med. Parasitol. 90:105-114. [DOI] [PubMed] [Google Scholar]
- 26.Speelman, P., I. Kabir, and M. Islam. 1984. Distribution and spread of colonic lesions in shigellosis: a colonoscopic study. J. Infect. Dis. 150:899-903. [DOI] [PubMed] [Google Scholar]
- 27.Unkmeir, A., and H. Schmidt. 2000. Structural analysis of phage-borne stx genes and their flanking sequences in Shiga toxin-producing Escherichia coli and Shigella dysenteriae type 1 strains. Infect. Immun. 68:4856-4864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Venkatesan, M. M., J. M. Buysse, and D. J. Kopecko. 1988. Characterization of invasion plasmid antigen genes (ipaBCD) from Shigella flexneri. Proc. Natl. Acad. Sci. USA 85:9317-9321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Venkatesan, M., J. M. Buysse, E. Vandendries, and D. J. Kopecko. 1988. Development and testing of invasion-associated DNA probes for detection of Shigella spp. and enteroinvasive Escherichia coli. J. Clin. Microbiol. 26:261-266. [DOI] [PMC free article] [PubMed] [Google Scholar]