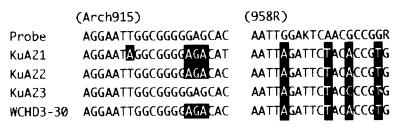Abstract
Fluorescence in situ hybridization has shown that cells labeled with an Archaea-specific probe (ARCH915) accounted for approximately 10% of the total cell count in oil-contaminated groundwater accumulated at the bottom of an underground crude oil storage cavity. Although chemical analyses have revealed vigorous consumption of nitrate in cavity groundwater, the present study found that the methane production rate was higher than the nitrate consumption rate. To characterize the likely archaeal populations responsible for methane production in this system, fragments of 16S ribosomal DNA (rDNA) were amplified by PCR using eight different combinations of universal and Archaea-specific primers. Sequence analysis of 324 clones produced 23 different archaeal sequence types, all of which were affiliated with the kingdom Euryarchaeota. Among them, five sequence types (KuA1, KuA6, KuA12, KuA16, and KuA22) were obtained in abundance. KuA1 and KuA6 were closely related to the known methanogens Methanosaeta concilii (99% identical) and Methanomethylovorans hollandica (98%), respectively. Although no closely related organism was found for KuA12, it could be affiliated with the family Methanomicrobiaceae. KuA16 and KuA22 showed substantial homology only to some environmental clones. Both of these branched deeply in the Euryarchaeota, and may represent novel orders. Quantitative competitive PCR showed that KuA12 was the most abundant, accounting for ∼50% of the total archaeal rDNA copies detected. KuA1 and KuA16 also constituted significant proportions of the total archaeal rDNA copies (7 and 17%, respectively). These results suggest that limited species of novel archaea were enriched in the oil storage cavity. An estimate of specific methane production rates suggests that they were active methanogens.
Underground cavities are used for the long-term storage of crude oil in several countries, and one such facility is situated at Kuji in Iwate, Japan. These cavities have been constructed in groundwater-rich rocky strata, where high groundwater pressure confines the stored oil in the cavities (42). Consequently, groundwater migrates into such cavities, accumulates at the bottom (called cavity groundwater), and is subsequently discharged to maintain the oil storage capacity. This flow of groundwater facilitates establishing a continuous culture of microorganisms at the bottom of these cavities. In this system, the total count of microorganisms in cavity groundwater was constantly more than 106 cells per ml (a density 100 times higher than that observed in the groundwater around the cavity). This habitat is characterized by immediate contact with a large quantity of crude oil and by an excess of electron donors (i.e., hydrocarbons) but also by a shortage of electron acceptors (42). These characteristics may be similar to those of microbial habitats in subterranean oil fields that have recently attracted strong microbiological attention (21, 23, 27, 31, 40). Since groundwater can easily be obtained from the bottom of an oil storage cavity by using installed sampling facilities without contamination by surface water (42), oil storage cavities are considered to represent good models for studying the microbial ecology of subterranean oil fields.
In our previous study, fluorescence in situ hybridization (FISH) showed that cells labeled with Bacteria- and Archaea-specific probes (EUB338 and ARCH915, respectively) accounted for approximately 50 and 10% of the total cell count in cavity groundwater (K. Watanabe, Y. Kodama, and N. Kaku, submitted for publication). Cloning and sequencing approaches for bacterial 16S ribosomal DNA (rDNA) and subsequent quantitative analyses have indicated that bacteria related to Desulfotomaculum, Acetobacterium, Desulfovibrio, Zoogloea, and Thiomicrospira denitrificans constituted significant populations in the cavity groundwater (42; Watanabe et al., submitted). These results suggest the coexistence of bacteria exhibiting different types of energy metabolism and were somewhat unexpected, since chemical analyses of the groundwater have suggested that nitrate reduction was the dominant terminal electron-accepting (TEA) process (42).
The purpose of the present study was to characterize the archaeal populations present in oil-contaminated cavity groundwater. Culture-independent molecular phylogenetic approaches have recently shown that archaeal populations are present in a variety of environments, including marine (7, 8, 13), freshwater (25, 35, 49), and soil (4, 5, 15, 16) ecosystems. In addition, archaeal populations have also been found in microbial communities engaged in anaerobic petroleum hydrocarbon degradation (9, 12, 45, 48). We therefore anticipated that archaeal populations would constitute important niches in the microbial community that had been established in cavity groundwater.
MATERIALS AND METHODS
Groundwater samples.
Samples of oil-contaminated cavity groundwater were obtained from a sampling facility of the TK101 underground crude oil storage cavity at Kuji in Iwate, Japan, in 1999, 2000, and 2001. The characteristics of cavity groundwater from this site have been described elsewhere (42). A groundwater sample used for measuring metabolic activities was placed on ice, immediately after it was collected in a bottle. By minimization of the headspace in the bottle, the groundwater could be kept anaerobic; its redox potential was below −100 mV when it was used for measuring metabolic activities. Groundwater samples used for molecular analyses were filtered through 0.22-μm-pore-size membrane filters (type GV; Nihon Millipore, Tokyo, Japan) immediately after collection.
TEA activities.
Approximately 3 h after sampling, cavity groundwater (100 ml) was infused into a 150-ml bottle under nitrogen atmosphere and treated with the Anaeropak reagent (Mitsubishi Gas Chemicals, Tokyo, Japan) to remove any trace amount of oxygen. The bottles were sealed with butyl rubber septa and aluminum crimp caps, and resazurin was added to a final concentration of 2 mg liter−1 to confirm anaerobicity. Sodium nitrate was then added to approximately 1 mg liter−1 (12 μM), and the bottle contents were incubated at 16°C (the in situ temperature [42]). The concentrations of nitrate and sulfate were determined by ion chromatography with an IA-100 ion analyzer (DKK Toa, Tokyo, Japan). The amount of methane was measured by headspace gas chromatography using a GC-14 gas chromatograph (Shimadzu, Kyoto, Japan) equipped with a flame ionization detector (Shimadzu) and a Porapak-Q column (80/100 mesh; Nihon Waters, Tokyo, Japan) as previously described (19, 38). The total amount of methane in the sample was estimated from a standard curve produced with bottles containing 100 ml of N2-purged sterile groundwater and known amounts of methane. TEA activities were calculated from changes in concentration of these compounds during the initial 12 h after commencement of the incubation.
Design of PCR primers.
Primers A341If, A1063Ir, and A533r were designed by modifying known ones (Table 1). The following archaeal 16S rDNA sequences were analyzed for the modification: Desulfurococcus mobilis (M36474), Thermosphaera aggregans (X99556), Pyrodictium occultum (M21087), Pyrodictium abyssi DSM 6158 (X99559), Sulfolobus acidocaldarius DSM 639 (D14053), Sulfolobus metallicus DSM 6482 (X90479), Sulfolobus shibatae (M32504), Sulfolobus yangmingensis (AB010957), Acidianus ambivalens (X90484), Metallosphaera prunae DSM 10039 (X90482), Stygiolobus azoricus DSM 6296 (X90480), Sulfurisphaera ohwakuensis IFO15161 (D85507), uncultured Sulfolobales strain MS1 (AF169011), Thermofilum pendens DSM 2475 (X14835), Pyrobaculum aerophilum (L07510), Thermoproteus tenax (M35966), Caldivirga maquilingensis (AB013926), Pyrobaculum aerophilum (L07510), Pyrobaculum islandicum (L07511), Pyrobaculum oguniense (AB029339), Thermocladium modestius (AB005296), Caldococcus noboribetus NC12 (D85038), Cenarchaeum symbiosum (U51469) unidentified archaeon SCA11 (U62820), Archaeoglobus fulgidus VC-16 (Y00275), Ferroglobus placidus DSM 10642 (X99565), F. placidus (AF220166), hyperthermophile strain 234 (AF220165), Haloarcula aidinensis (AB000563), Methanobacterium bryantii (AF028688), Methanothermus fervidus (M59145), Methanothermus sociabilis (AF095273), Methanococcus aeolicus (U39016), Methanococcus deltae (U38485), Methanococcus jannaschii (M59126), Methanocorpusculum bavaricum (AF095266), Methanocorpusculum labreanum (AF095267), Methanomicrobium mobile (M59142), Methanogenium cariaci DSM 1497 (M59130), Methanopyrus kandleri av19 (M59932), Methanosarcina siciliae (U89773), Thermococcus celer (M21529), Ferromonas metallovorans (AJ224936), Picrophilus oshimae (X84901), Thermoplasma acidophilum 122-1B2 (M38637), Methanogen sp. (AB008853), unidentified Korarchaeote strain pJP27 (L25852), and unidentified Korarchaeote strain pJP78 (L25303).
TABLE 1.
PCR primers used in this study
| Primera | Sequence (5′ to 3′)b | Positionc | Specificity (sequence type) | Reference or source |
|---|---|---|---|---|
| A25f | CYGGTYGATYCTGCCRG | 9-25 | Archaea | 9 |
| A341If | CCTAIGGGGIGCAICAG | 341-357 | Archaea | This studyd |
| A958r | YCCGGCGTTGAMTCCAATT | 958-976 | Archaea | 18 |
| A1063Ir | CGGCCATGCACCICCICTC | 1045-1063 | Archaea | This studye |
| A1391r | GACGGGCGGTGTGTRCA | 1375-1391 | Archaea | 3 |
| U1492r | GGTTACCTTGTTACGACTT | 1492-1510 | Universal | 9 |
| A533r | TTACCGCGGCGGCTG | 519-533 | Most archaea | This studyf |
| MS827f | TAGGTGTCGGCCACGGT | 827-843 | Methanosaeta (KuA1, KuA3) | This studyg |
| MH163f | GGGATAATTCCGGATAGAT | 163-182 | Methanomethylovorans (KuA6) | This study |
| MH441r | ATTTATACATATGGACAGCCAAC | 419-441 | Methanomethylovorans (KuA6) | This study |
| MM402f | GAGTGCCCGTAAAATCGG | 402-419 | Part of Methanomicrobiales (KuA12) | This study |
| MM837r | CGTTTCACGATTTCTCTTCG | 818-837 | Part of Methanomicrobiales (KuA10, KuA11, KuA12) | This study |
| K311f | TGAGATGGACTCTGAGACAT | 311-330 | Part of candidate division II (KuA15, KuA16) | This study |
| K463r | CAGTCAACACAATGTGCTG | 445-463 | Part of candidate division II (KuA15, KuA16) | This study |
| K780f | AGTAAACACTGCACACTAAACA | 780-811 | Part of candidate division IV (KuA22) | This study |
| T7W | TAATACGACTCACTATAGGGC | pGEM-T vector | 42 | |
| SP6W | ATTTAGGTGACACTATAGAATACTC | pGEM-T vector | 42 |
f, forward PCR primer; r, reverse PCR primer.
I, inosine.
Corresponding to the numbering in the sequence of the 16R rRNA gene of E. coli.
This primer is a modified form of S-D-Arch-0344-a-S-20 (39).
This primer is a modified form of 1068 Forward (1).
This primer is a modified form of 533F (9).
This primer is a modified form of MX825 (33).
PCR, cloning, and sequencing of 16S rDNA.
DNA was extracted from the cavity groundwater as described previously (42) with two modifications. First, cells in the cell suspension buffer were treated with 10 mg of proteinase K ml−1 at 37°C for 1 h. Second, three cycles of freezing at −80°C and thawing in a 70°C water bath were conducted following the hot-detergent treatment. PCR amplification of the 16S rDNA fragments was performed with eight pairs of primers, A25f and A958r, A25f and A1063Ir, A25f and A1391r, A25f and U1492r, A341If and A958r, A341If and A1063Ir, A341If and A1391r, and A341If and U1492r (Table 1). PCR mixtures (50 μl) contained 1.25 U of Taq DNA polymerase (Amplitaq Gold; Applied Biosystems Japan, Tokyo, Japan), 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 1.5 mM MgCl2, 0.001% (wt/vol) gelatin, each deoxynucleoside triphosphate at a concentration of 200 μM, 25 pmol of each primer, and 10 ng of template DNA. The amplification conditions were as follows: 10 min of activation of the polymerase at 94°C, followed by 30 cycles consisting of 1 min at 94°C, 1 min at the annealing temperature (described next) and 2 min at 72°C, and finally 10 min of extension at 72°C. The annealing temperature used was 50°C, except for primer set A341If and A1063Ir (55°C). Amplified fragments were purified by electrophoresis, ligated into the pGEM-T vector (Promega, Madison, Wis.), and cloned into Escherichia coli as described previously (42). Vector-harboring clones were selected on Luria-Bertani plates (34) supplemented with ampicillin (50 μg ml−1). PCR-amplified 16S rDNA fragments were recovered from each colony by PCR with primers T7W and SP6W (these primers target pGEM-T vector sequences flanking the insertion; see Table 1) as described previously (42). Clones containing appropriate insert sizes were selected by an electrophoretic analysis, and their nucleotide sequences were determined as described previously (42).
Nucleotide sequence analysis.
Database searches for related 16S rDNA sequences were conducted using the BLAST program (20) and the GenBank database. The profile alignment technique of ClustalW version 1.7 (37) was used to align the sequences. Each alignment was refined by visual inspection, and secondary structures were considered for the refinement analysis (17). A phylogenetic tree was constructed by the neighbor-joining method with the njplot program in ClustalW, version 1.7. Nucleotide positions at which any sequence had a gap or an ambiguous base were not included in the phylogenetic calculations. Checks for chimeric sequences were conducted by the chimera check program in the Ribosomal Database Project database (26).
Competitive PCR (cPCR).
The primers used for cPCR (Table 1) were designed by comparing the 16S rDNA sequences obtained in this study with those in the RDP database (26). The specificity of each primer was checked using the probe match program in the Ribosomal Database Project database. Competitor fragments were produced using the competitive DNA construction kit (Takara Shuzo, Ohtsu, Japan). The cPCR systems developed in this study are summarized in Table 2. The composition of PCR was as described above, except for the competitor fragment being added at a known copy number. The PCR conditions were as described above except that 35 cycles of amplification were performed and the annealing temperatures were as described in Table 2. Two microliters of the PCR product was analyzed by electrophoresis through 1.5% (wt/vol) agarose gels with Tris-borate-EDTA buffer, and the gels were photographed after staining with SYBR Gold (FMC Bioproducts, Vallensbaek Strand, Denmark). The band intensities of the target and competitor fragments were quantified by using the Multianalyst program supplied with Gel Doc 2000 (Bio-Rad, Hercules, Calif.). The copy number of the target 16S rDNA fragment was estimated by considering the band intensity, length of the fragment, and copy number of the competitor as described by Lee et al. (22).
TABLE 2.
Summary of cPCR assay conditions
| cPCR typea | Primer pair | Annealing temp (°C) | Length of DNA fragment (bp)
|
Specificity
|
||
|---|---|---|---|---|---|---|
| Target | Competitor | Sequence type | Phylogenetic group | |||
| AC | A341If and A533r | 60 | 150-160 | 232 | All | Most archaea |
| MS | MS827f and A1063Ir | 62 | 233 | 186 | KuA1, KuA3 | Methanosaeta |
| MH | MH163f and MH441r | 55 | 279 | 192 | KuA6 | Methanomethylovorans |
| MM | MM402f and MM837r | 55 | 434 | 338 | KuA12 | Part of Methanomicrobiales |
| CDII | cK311f and K463r | 55 | 116 | 239 | KuA15, KuA16 | Part of candidate division II |
| CDIV | K780f and A1063Ir | 55 | 248 | 191 | KuA22 | Part of candidate division IV |
AC, archaea; MS, Methanosaeta; MH, Methanomethylovorans hollandica; MM, Methanomicrobiales; CDII, candidate division II; CDIV, candidate division IV.
Nucleotide sequence accession numbers.
The nucleotide sequences reported in this paper have been deposited in the DDBJ, EMBL, and National Center for Biotechnology Information nucleotide sequence databases under accession nos. AB077211 to AB077233.
RESULTS
TEA activities.
Our previous studies have suggested the presence of Archaea in cavity groundwater in addition to bacteria related to sulfate reducers and nitrate reducers (42; Watanabe et al., submitted). This present study measured the rates of nitrate consumption, sulfate consumption, and methane production in cavity groundwater to compare the TEA activities (Table 3). It was found that the methane production rate was approximately fivefold higher than the nitrate consumption rate. The sulfate concentration in the bottles slowly increased during the incubation period.
TABLE 3.
TEA activities with endogenous substrates in cavity groundwatera
| Groundwater date
|
Nitrate data
|
Sulfate data
|
Methane data
|
|||
|---|---|---|---|---|---|---|
| Initial concn (μM) | Consumption rateb (μmol liter−1 h−1) | Initial concn (μM) | Consumption rate (μmol liter−1 h−1) | Initial concn (μM) | Production rate (μmol liter−1 h−1) | |
| July 2001 | NDc | 1.18 ± 0.22 | 51 ± 1.3 | −0.03 ± 0.01d | 34 ± 8.6 | 5.5 ± 0.9 |
| December 2001 | ND | 0.95 ± 0.26 | 45 ± 0.1 | −0.23 ± 0.11 | 27 ± 5.5 | 5.8 ± 1.1 |
Each datum value is the mean ± standard deviation (n = 3).
Nitrate (12 μM) was added prior to measuring the rate.
ND, not detected (the detection limit was 0.5 μM).
The sulfate concentration was increased.
Diversity of rDNA sequences.
A 16S rDNA approach was used to characterize the likely archaeal populations responsible for methane production in cavity groundwater. The 16S rDNA fragments were amplified by PCR from groundwater DNA obtained in March 1999 using eight pairs of primers (see Tables 1 and 4). The rationale for using multiple primer sets was to evaluate and obviate bias in the rDNA approach resulting from primer-specific amplification of preferential groups of rDNA. Among the primers used, A341If and A1063Ir were developed in this study by modifying known rDNA primers (Table 1). This modification was achieved by considering mismatches between the original sequences and the archaeal 16S rDNA sequences listed in Materials and Methods. Inosine residues were introduced at positions in these primers where mismatches with any of the aforementioned 16S rDNA sequences had been found. The introduction of inosine residues into PCR primers has been suggested to be useful for reducing the amplification bias caused by primer mismatches (6, 43).
TABLE 4.
rDNA sequence types obtained from cavity groundwater
| Sequence type or total | No. of sequence type found in the following librarya
|
Database match (>90% identity) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 25/958 | 25/1063 | 25/1391 | 25/1492 | 341/958 | 341/1063 | 341/1391 | 341/1492 | Total | ||
| KuA1 | 19 | 13 | 16 | 31 | 16 | 10 | 29 | 11 | 145 | 99% M. concilii (X16932) |
| KuA2 | 1 | 1 | 97% lake sediment clone Rot18 (Y18091) | |||||||
| KuA3 | 1 | 1 | 97% granular sludge clone TA05 (AF229778) | |||||||
| KuA4 | 1 | 1 | 94% M. hollandica (AF120163) | |||||||
| KuA5 | 1 | 1 | 96% M. hollandica (AF120163) | |||||||
| KuA6 | 17 | 11 | 10 | 1 | 18 | 3 | 60 | 98% M. hollandica (AF120163) | ||
| KuA7 | 4 | 1 | 5 | 93% groundwater clone WCHD3-07 (AF050617) | ||||||
| KuA8 | 1 | 1 | 99% rice paddy field clone AS08-03 (AF225635) | |||||||
| KuA9 | 3 | 3 | 97% Methanoculleus palmaeoli (Y16382) | |||||||
| KuA10 | 1 | 1 | 97% groundwater clone WCHD3-07 (AF050617) | |||||||
| KuA11 | 3 | 3 | 97% groundwater clone WCHD3-07 (AF050617) | |||||||
| KuA12 | 1 | 2 | 2 | 8 | 6 | 19 | 98% groundwater clone WCHD3-07 (AF050617) 91% M. liminatans DSM 4140 (Y16428) | |||
| KuA13 | 1 | 1 | 97% groundwater clone WCHA1-57 (AF050614) 99% sewerage clone 69-1 (AF424763) | |||||||
| KuA14 | 1 | 1 | 98% sewerage clone 69-1 (AF424763) | |||||||
| KuA15 | 1 | 1 | 98% groundwater clone WCHD3-33 (AF050619) | |||||||
| KuA16 | 4 | 12 | 1 | 1 | 2 | 20 | 99% groundwater clone WCHD3-33 (AF050619) | |||
| KuA17 | 1 | 1 | No homologous sequence | |||||||
| KuA18 | 1 | 1 | 2 | 97% lake sediment clone Rot13 (Y18089) | ||||||
| KuA19 | 2 | 2 | 97% lake sediment clone Rot13 (Y18089) | |||||||
| KuA20 | 1 | 1 | 90% lake sediment clone Rot13 (Y18089) | |||||||
| KuA21 | 2 | 2 | No homologous sequence | |||||||
| KuA22 | 10 | 10 | 95% groundwater clone WCHD3-30 (AF050612) | |||||||
| KuA23 | 1 | 1 | No homologous sequence | |||||||
| Bacteria | 5 | 32 | 37 | |||||||
| Chimera | 2 | 1 | 1 | 4 | ||||||
| Total | 42 | 42 | 35 | 38 | 38 | 43 | 42 | 44 | 324 | |
Names of the libraries were created according to the PCR primers used for amplifying 16S rDNA fragments from the groundwater DNA.
The PCR-amplified 16S rDNA fragments were cloned into E. coli, and eight libraries of clones were consequently made according to the primers used for PCR (Table 4). The nucleotide sequences of 324 clones in total were determined, which produced 23 different archaeal sequence types (classified as a unique clone or group of clones with sequence similarity of >0.98, named KuA; refer to Table 4). The number of archaeal sequence types found in one library ranged from 2 to 13, with one primer set (A341If and A1063Ir) producing the largest number of types. Primer set A341If and U1492r amplified many bacterial 16S rDNA fragments. One sequence type, KuA1, was found ubiquitously in all eight libraries, while 17 sequence types were specific for one library.
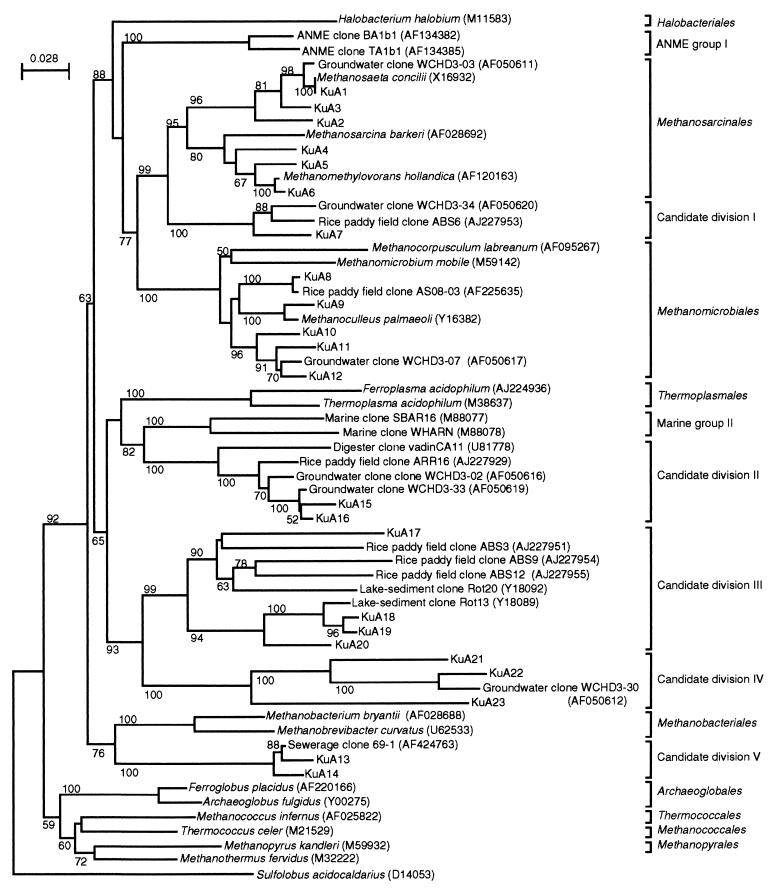
The database search (Table 4) and phylogenetic analysis (Fig. 1) indicated that all of the 23 sequence types were affiliated with Euryarchaeota. Among them, seven sequence types (KuA1, KuA2, KuA3, KuA4, KuA5, KuA6, and KuA9) were closely related to known methanogens. The other sequence types were related (>90%) only to unidentified environmental clones (13 sequence types) or were <90% identical to rDNA sequences found in the database (three sequence types). Eleven sequence types were affiliated with known euryarchaeotic orders: six with Methanosarcinales and five with Methanomicrobiales (Fig. 1). The other 12 sequence types were not affiliated with any of the known euryarchaeotic orders and were divided into five candidate divisions (order-level phylogenetic groups I to V, which include only 16S rRNA sequences of uncultured organisms; see Fig. 1).
FIG. 1.
A neighbor-joining tree showing the positions of KuA rDNA sequence types obtained from cavity groundwater. Sequences corresponding to nucleotide positions 341 to 976 (numbering based on the E. coli sequence) were aligned and used for this analysis. S. acidocaldarius was used as the outgroup. Accession numbers of the sequences retrieved from the databases are indicated in parentheses. The numbers at the branch nodes are bootstrap values (per 100 trials); only values greater than 50 are shown. The scale bar indicates 0.028 substitution per site.
Quantification of rDNA copies by cPCR.
The rDNA sequence analysis detected sequence types KuA1, KuA6, KuA12, and KuA16 in relative abundance in several libraries (Table 4). In addition, KuA22 constituted a large proportion in library 341/1063, although it was found only in this library (Table 4). In order to examine whether the archaeal populations represented by these sequence types actually constituted significant proportions in the total archaeal population of cavity groundwater, quantitative cPCR assays with specific primer sets (Table 1) were developed and are summarized in Table 2. The utility and limitation of cPCR assays for quantifying microbial populations in natural ecosystems have been described previously (22, 41; Watanabe et al., submitted).
When E. coli cells harboring these archaeal rDNA clones were used as templates, PCR products with the expected sizes were obtained only from those clones harboring the expected sequence types as listed in Table 2. Primers A341If and A533r were used in the cPCR for quantifying the total archaeal population. A533r was designed by comparing the rDNA sequences listed in Materials and Methods and had one mismatch (at nucleotide position 523 in the E. coli sequence) with typical 16S rDNA sequences affiliated with Methanococcales. However, this mismatch may not have affected the cPCR estimate of total archaeal rDNA in cavity groundwater, since no Methanococcales rDNA sequence was obtained from cavity groundwater (Fig. 1).
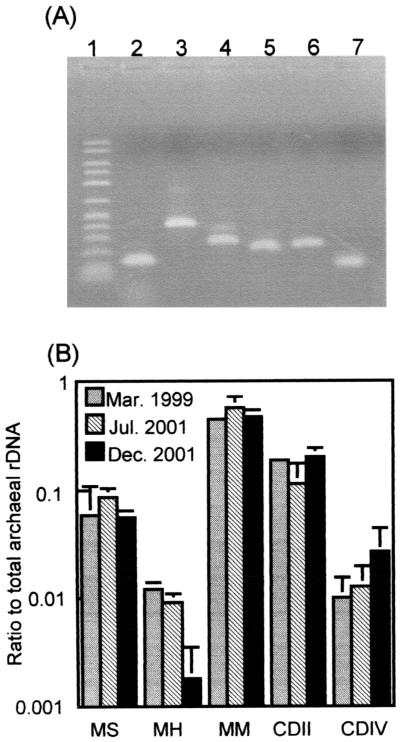
Figure 2A shows that the PCR successfully amplified DNA fragments of the expected sizes from the groundwater DNA (the March 1999 sample). The results of the cPCR assays summarized in Table 2 of three groundwater samples (obtained in March 1999, July 2001, and December 2001) are shown in Fig. 2B. KuA12 rDNA (detected by the MM cPCR assay) was the most abundant in these groundwater samples, constituting approximately 50% (the average of three samples) of the total archaeal rDNA copies (detected by the AC cPCR assay). The CDII and MS cPCR assays also detected relatively large proportions (approximately 17 and 9% on average, respectively) of the total archaeal rDNA. The rDNA detected by the MH cPCR assay that targeted KuA6, the sequence type abundantly identified in several libraries, was least abundant. The sum of the ratios of these five rDNA types accounted for 71% (the March 1999 sample), 74% (the July 2001 sample), and 80% (the December 2001 sample) of the total archaeal rDNA copies detected in these analyses.
FIG. 2.
cPCR assays for quantifying rDNA copies of the major sequence types. (A) Target fragments amplified from groundwater DNA (obtained in 1999). Lane 1, 50- to 2,500-Da molecular size marker (FMC Corp.); lane 2, AC cPCR; lane 3, MM cPCR; lane 4, MH cPCR; lane 5, MS cPCR; lane 6, CDIV cPCR; and lane 7, CDII cPCR. (B) Ratios of specific rDNA copies to the total archaeal rDNA copies (determined by the AC system in Table 2) in DNA extracted from the cavity groundwater. The mean of three determinations is shown, and an error bar indicates the standard deviation. See footnote a to Table 2 for definitions of abbreviations.
DISCUSSION
This study has characterized the archaeal populations that were present in an oil-contaminated cavity groundwater system and may have been involved in the decomposition of petroleum constituents, although no direct evidence of degradation processes was provided. Since methane production is a TEA process specifically catalyzed by a group of archaea called methanogens (2), we first measured the methanogenic activity (methane production rate) in the cavity groundwater. A comparison of the TEA activities in the cavity groundwater (Table 3) revealed active methane production that was much greater than the nitrate consumption rates measured. Recently, anaerobic methane oxidation coupled to sulfate reduction has been reported (4a, 18), suggesting the possibility that actual methanogenic activity may have been greater than the methane production rate measured.
Specific methane production rates were estimated for the total archaeal population in the Kuji cavity groundwater, and these values were compared with those of other environmental samples, enrichment cultures, and pure cultures (Table 5). It was found that the specific rates exhibited by the Kuji Archaea were quite high; surprisingly, the rates were higher than the rate of one of the most active thermophilic methanogens, Methanobacterium thermautotrophicum Marburg. Although we need to pay attention to possible bias caused by the methods employed for determining these values (e.g., the conditions for the activity measurements, the detection method for the archaeal cells, and the cell mass used for the calculation), the data suggest that quite active methanogens are present in this oil storage cavity. We are presently investigating how the methane is produced; further chemical analyses of the groundwater are under way to identify the specific substrate(s) for methane production.
TABLE 5.
Comparison of specific methane production rates exhibited by environmental samples, enrichments, and pure cultures
| Sample type or organism | Conditions | Specific methane production rate (μmol h−1 mg of dry cells−1) | Reference for original data |
|---|---|---|---|
| Environmental sample | |||
| Kuji groundwater | Endogenous substrates, 16°C | 79-83a,b | This study (Table 3) |
| Lake Rotsee sediment | Endogenous substrates, 6°C | 0.029a,b | 49 |
| Lake Mendota sediment | Acetate and H2/CO2, 30°C | 22a,b,c | 47 |
| Enrichment | |||
| UASB granular sludgef | H2-CO2, 35°C | 1.8d | 46 |
| EGSB aggregateg | Brewery wastewater, 30.5°C | 0.72d | 14 |
| Toluene-degrading enrichment | Toluene, 35°C | 1.45a | 10, 12 |
| Pure culture | |||
| M. concilii GP6 | Acetate, 35°C | 2.1 | 32 |
| M. hungatei GP1 | H2-CO2, 35°C | 23 | 32 |
| M. espanolae GP9 | H2-CO2, 35°C | 35 | 32 |
| M. bryantii M.o.H | H2-CO2, 35°C | 12 | 32 |
| M. thermautotrophicum Marburg | H2-CO2, 60°C | 60e | 28 |
The value was estimated for archaeal cells detected by FISH with the ARCH915 probe.
The cell mass for E. coli (2.8 × 10−13 g per cell [29]) was used for estimating the value per milligram of dry cells.
The archaeal cell content in the Rotsee sediment (49) was used for calculation.
The value was estimated for the total biomass determined as volatile suspended solids.
A value per milligram of protein was converted to that per milligram of dry cells by using the standard protein content of bacterial and archaeal cells (0.5 [30]).
UASB, upflow anaerobic sludge blanket.
EGSB, expanded granular sludge bed.
Although rDNA analyses have previously suggested the presence of sulfate-reducing bacteria as a significant proportion of the total bacterial populations in cavity groundwater (Watanabe et al., submitted), Table 3 shows that there was no net consumption of sulfate but that it was, in fact, slowly produced. In addition, sulfate was constantly detected in cavity groundwater at concentrations from 4 to 7 mg liter−1 (Table 3 and reference 42). We have recently isolated a chemolithotroph that oxidized sulfide to sulfate coupled to nitrate reduction (K. Watanabe, Y. Kodama, and S. Harayama, Abstr. 9th Int. Symp. Microb. Ecol., abstr. P.20.002, 2001). In addition, FISH (42) and cPCR studies (Watanabe et al., submitted) have shown that this bacterium constituted the major population in cavity groundwater. We found that groundwater migrating into the Kuji cavities contained substantial amounts of nitrate that were almost completely consumed in the oil storage cavities (42). It is therefore likely that a sulfur cycle involving sulfate-reducing bacteria and anaerobic sulfur-oxidizing bacteria equilibrated the sulfate concentration. This idea allows us to further suggest that sulfate reduction is also an active TEA process in the oil storage cavity. Together with the findings just described, our observations suggest that multiple TEA processes simultaneously operate in the cavity groundwater; it would be interesting to investigate how these TEA processes interact with each other.
Eight pairs of oligonucleotide primers were used in this study for PCR amplification of archaeal 16S rDNA fragments from groundwater DNA. The cloning and sequencing of the amplified fragments produced many primer-set specific sequence types, while only one sequence type (KuA1) was common to all libraries (Table 4). Although KuA1 included the largest number of clones (145 out of 324), the cPCR analysis showed that this rDNA type represented only 7% of total archaeal rDNA, indicating that this sequence type was preferentially cloned. We consider that less efficient amplification of KuA1 than of its competitor was not likely, because the MS cPCR assay was successfully used to estimate a copy number of the KuA1 fragment recovered by PCR with primers T7W, and SP6W from an E. coli clone. In contrast, KuA12, the rDNA type most abundantly detected by cPCR (sharing 50% of the total archaeal rDNA), included only 19 clones that were distributed in five libraries. KuA12 was not included in three libraries (25/958, 341/958, and 341/1492). We assume that rDNA analyses of other microbial communities using these primers may have failed to detect this group of 16S rDNA sequence types even if organisms represented by such sequence types constituted major populations.
The cPCR data (Fig. 2) indicated that a large percentage of the total archaeal rDNA (71 to 80%) was represented in the clone libraries. We suggest that the use of the multiple primer sets for retrieving the rDNA sequences enabled this wide detection, although this method has not been popular in previous studies. Ficker et al. have used a primer set corresponding to A25f/U1492r for PCR amplification of archaeal 16S rDNA fragments from a toluene-degrading methanogenic consortium (12). In their study, a subsequent FISH analysis with probes specific for the obtained 16S rDNA sequences detected only 29% of the total archaeal population (12), suggesting that a large portion of the total archaeal rDNA in the consortium remained uncharacterized. We suggest that the use of multiple primer sets is one possible way to obviate or at least mitigate the bias caused by primer-specific amplification in constructing rDNA libraries.
Environmental rDNA sequences have been used to infer some properties of the organisms that they represent (9, 12; Watanabe et al., submitted), although this type of analysis is possible only if some common properties are known for isolated organisms affiliated with the same phylogenetic group. In the present study, sequence type KuA1 was closely related to M. concilii (Table 4), for which the only known energy metabolism is acetoclastic methanogenesis (44). This suggests that the archaeon represented by this sequence type in our analysis also engages in acetoclastic methanogenesis. Ribosomal DNA sequences highly homologous to Methanosaeta have also been retrieved as major clones from hydrocarbon-contaminated groundwater (9) and from toluene-degrading (12) and hexadecane-degrading (48) methanogenic consortia, and acetoclastic methanogenesis has been suggested to be the major TEA process associated with anaerobic hydrocarbon degradation (9, 12). As already discussed, however, this study indicated the possibility of bias in the cloning and sequencing analysis toward the Methanosaeta rDNA type, implying that the importance of this group of archaea may have been overestimated. We assume that the contribution of the Methanosaeta archaeon to methane production in the cavity groundwater (Tables 3 and 5) was not large, because the methane production rates of this group of archaea are generally low (an example is presented in Table 5).
Other rDNA sequence types whose phylogenetic information could be used to predict their physiology include KuA6 (affiliated with the genus Methanomethylovorans, a methylotrophic methanogen [24]) and KuA9 (affiliated with Methanoculleus in Methanomicrobiales, known to utilize H2-CO2 and formate [44]). In addition, the phylogenetic analysis shown in Fig. 1 indicates that KuA12 could be affiliated with Methanomicrobiales (Fig. 1), although its phylogenetic relationship to known cultivated members of this order is rather weak; the highest match in the sequence was observed with Methanofollis liminatans (91%). We thus conducted a further rDNA analysis to identify the phylogenetic position of KuA12 (Fig. 3) and found that it belongs to the family Methanomicrobiaceae. Accordingly, we assume that KuA12 represents an archaeon that utilizes H2-CO2 and formate for methanogenesis (44).
FIG. 3.
A neighbor-joining tree showing that KuA12 is affiliated with the family Methanomicrobiaceae. Sequences corresponding to nucleotide positions 28 to 1488 (numbering based on the E. coli sequence) were aligned and used for this analysis. M. labreanum was used as the outgroup. Accession numbers of the sequences retrieved from the databases are indicated in parentheses. The numbers at the branch nodes are bootstrap values (per 100 trials); only values greater than 50 are shown. The scale bar indicates 0.02 substitution per site.
The phylogenetic analysis shown in Fig. 1 indicates that 12 rDNA sequence types could not be affiliated with the known orders in Euryarchaeota, these being grouped into five candidate divisions. Among them, candidate division I corresponds to rice cluster I (16), which forms a novel clade within the phylogenetic radiation characterized by members of the two orders Methanomicrobiales and Methanosarcinales. Phylogenetic distance analyses of the members of Methanomicrobiales, Methanosarcinales, and rice cluster I have indicated the latter to be an order-level phylogenetic group (16). Similarly, candidate divisions II and III, respectively, correspond to rice clusters III and V (16). Candidate division II includes the second most abundant rDNA type, KuA15/16. Candidate division III includes rather diverse rDNA sequences. For example, the sequence identity between ABS9 and Rot13 is 82%. This level of identity has in some cases been used to separate rDNA sequences into two order-level phylogenetic groups (16), suggesting that further studies are needed on this group. The rice clusters have originally been proposed for archaea inhabiting a rice paddy field (15, 16), while the results of the present study indicate that members of these groups of archaea are distributed more widely in anoxic environments, e.g., in subsurface aquifers, anaerobic digesters, and lake sediments.
Candidate divisions IV and V branched deeply in Euryarchaeota (Fig. 1). The branching points of these phylogenetic groups were almost unchanged when the maximum-likelihood method (11) was used as the treeing algorithm (data not shown). Candidate division IV includes three KuA sequence types together with one sequence obtained from a hydrocarbon-contaminated aquifer in the United States (9). In addition, these sequences also show homology to some rDNA sequences retrieved from a deep-sea hydrothermal vent (e.g., environmental clone pISA48 [AB019755] [36]). In the present study, this group of rDNA was obtained only when primer set A341If and A1063Ir was used for PCR amplification. We thus assume that these rDNA sequences have hardly ever been obtained when some conventional archaeal rDNA primers were used. This notion is based on sequence analyses of these rDNA types; several specific nucleotide substitutions were found in the regions that have been used for the archaeal probe and primer, e.g., probe ARCH915 and primer A958r (Fig. 4). The three mismatches in ARCH915 may have allowed this group of archaea to escape from detection by FISH. It is therefore likely that archaea represented by this group of rDNA may be distributed more widely than hitherto believed. Candidate division V includes two KuA sequence types and one sewerage-associated clone. In addition, these rDNA sequences show high homology to the rDNA sequence (WCHA1-57) obtained from a hydrocarbon-contaminated aquifer in Michigan (9). WCHA1-57 was not included in the dendrogram (Fig. 1), since its sequence region only partially overlaps the region of KuA14. These sequence analyses reveal that many KuA rDNA types are related to rDNA types that were obtained from the hydrocarbon-contaminated aquifer in the United States (9), suggesting that these rDNA types may be widely distributed in oil-contaminated subsurface environments.
FIG. 4.
Comparison of nucleotide sequences between the probes and rDNA sequences affiliated with candidate division IV. Mismatches with the probes are indicated by black boxes.
In conclusion, our molecular phylogenetic approach found substantial diversity in the archaeal populations present in oil-contaminated cavity groundwater and demonstrated that several species of Archaea were enriched in this groundwater. Results obtained in this study also suggest the need for careful selection of PCR primers and probes and, in some cases, use of multiple primer sets, when one truly wants to understand the molecular diversity of a microbial community. Besides, the estimated specific methane production rate suggests that hydrogenotrophic methanogens may have played a major role in the TEA process in cavity groundwater.
Acknowledgments
We thank Yoichi Matsumura (Japan Under-ground Oil Storage Co.) for help in the groundwater sampling and Ikuko Hiramatsu for technical assistance.
This work was supported in part by the New Energy and Industrial Technology Development Organization (NEDO).
REFERENCES
- 1.Achenbach, L., and C. Woese. 1995. 16S and 23S rRNA-like primers, p. 521-523. In K. R. Sowers and H. J. Schreier (ed.), Methanogens. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 2.Balch, W. E., G. E. Fox, L. J. Magnum, C. R. Woese, and R. S. Wolfe. 1979. Methanogens: reevaluation of a unique biological group. Microbiol. Rev. 43:260-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barns, S. M., C. F. Delwiche, J. D. Palmer, and N. R. Pace. 1996. Perspectives on archaeal diversity, thermophily and monophily from environmental rRNA sequences. Proc. Natl. Acad. Sci. USA 93:9188-9193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bintrim, S. B., T. J. Donohue, J. Handelsman, G. P. Roberts, and R. M. Goodman. 1997. Molecular phylogeny of Archaea from soil. Proc. Natl. Acad. Sci. USA 94:277-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4a.Boetius, A., K. Ravenschlag, C. J. Schubert, D. Rickert, F. Widdel, A. Gieseke, R. Amann, B. B. Jorgensen, U. Witte, and O. Pfannkuche. 2000. A marine microbial consortium apparently mediating anaerobic oxidation of methane. Nature 407:623-626. [DOI] [PubMed] [Google Scholar]
- 5.Buckley, D. H., J. R. Graber, and T. M. Schmidt. 1998. Phylogenetic analysis of nonthermophilic members of the kingdom Crenarchaeota and their diversity and abundance in soils. Appl. Environ. Microbiol. 64:4333-4339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Candrian, U., B. Furrer, C. Hofelein, and J. Luthy. 1991. Use of inosine-containing oligonucleotide primers for enzymatic amplification of different alleles of the gene coding for heat-stable toxin type I of enterotoxigenic Escherichia coli. Appl. Environ. Microbiol. 57:955-961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.DeLong, E. F. 1992. Archaea in coastal marine environments. Proc. Natl. Acad. Sci. USA 89:5685-5689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.DeLong, E. F., K. Y. Wu, B. B. Prezolin, and R. V. M. Jovine. 1994. High abundance of Archaea in Antarctic marine picoplankton. Nature 371:695-697. [DOI] [PubMed] [Google Scholar]
- 9.Dojka, M. A., P. Hugenholtz, S. K. Haack, and N. R. Pace. 1998. Microbial diversity in a hydrocarbon- and chlorinated-solvent-contaminated aquifer undergoing intrinsic bioremediation. Appl. Environ. Microbiol. 64:3869-3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Edwards, E. A., and D. Grbiæ-Galiæ. 1994. Anaerobic degradation of toluene and o-xylene by a methanogenic consortium. Appl. Environ. Microbiol. 60:313-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Felsenstein, J. 1989. PHYLIP, phylogeny inference package (version 3.2). Cladistics 5:164-166. [Google Scholar]
- 12.Ficker, M., K. Krastel, S. Orlicky, and E. Edwards. 1999. Molecular characterization of a toluene-degrading methanogenic consortium. Appl. Environ. Microbiol. 65:5576-5585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fuhrman, J. A., K. McCallum, and A. A. Davis. 1992. Novel major archaebacterial group from marine plankton. Nature 356:148-149. [DOI] [PubMed] [Google Scholar]
- 14.Gonzalez-Gil, G., P. N. L. Lens, A. Van Aelst, H. Van As, A. I. Versprille, and G. Lettinga. 2001. Cluster structure of anaerobic aggregates of an expanded granular sludge bed reactor. Appl. Environ. Microbiol. 67:3683-3692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Großkopf, R., P. H. Janssen, and W. Liesack. 1998. Diversity and structure of the methanogenic community in anoxic rice paddy soil microcosms as examined by cultivation and direct 16S rRNA gene sequence retrieval. Appl. Environ. Microbiol. 64:960-969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Großkopf, R., S. Stubner, and W. Liesack. 1998. Novel euryarchaeotal lineages detected on rice roots and in the anoxic bulk soil of flooded rice microcosms. Appl. Environ. Microbiol. 64:4983-4989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gutell, R. R. 1994. Collection of small subunit (16S and 16S-like) ribosomal RNA structures. Nucleic Acids Res. 22:3502-3507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hinrichs, K. U., J. M. Hayes, S. P. Sylva, P. G. Brewer, and E. F. DeLong. 1999. Methane-consuming archaebacteria in marine sediments. Nature 398:802-805. [DOI] [PubMed] [Google Scholar]
- 19.Kaku, N., A. Ueki, H. Fujii, and K. Ueki. 2000. Methanogenic activities on rice roots and plant residues and their contributions to methanogenesis in wetland rice field soil. Soil Biol. Biochem. 32:2001-2010. [Google Scholar]
- 20.Karlin, S., and S. F. Altschul. 1990. Methods for assessing the statistical significance of molecular sequence features by using general scoring schemes. Proc. Natl. Acad. Sci. USA 87:2264-2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Krumholz, L. R., J. P. McKinley, G. A. Ulrich, and J. M. Suflita. 1997. Confined subsurface microbial communities in Cretaceous rock. Nature 386:64-66. [Google Scholar]
- 22.Lee, S. Y., J. Bollinger, D. Bezdicek, and A. Ogram. 1996. Estimation of the abundance of an uncultured soil bacterial strain by a competitive quantitative PCR method. Appl. Environ. Microbiol. 62:3787-3793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.L'Haridon, S., A. L. Reysenbach, P. Glenat, D. Prieur, and C. Jeanthon. 1995. Hot subterranean biosphere in a continental oil reservoir. Nature 377:7223-7224. [PubMed] [Google Scholar]
- 24.Lomans, B. P., R. Maas, R. Luderer, H. J. M. O. den Camp, A. Pol, C. van der Drift, and G. D. Vogels. 1999. Isolation and characterization of Methanomethylovorans hollandica gen. nov., sp. nov., isolated from freshwater sediment, a methylotrophic methanogen able to grow on dimethyl sulfide and methanethiol. Appl. Environ. Microbiol. 65:3641-3650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.MacGregor, B. J., D. P. Moser, E. W. Alm, K. H. Nealson, and D. A. Stahl. 1997. Crenarchaeota in Lake Michigan sediment. Appl. Environ. Microbiol. 63:1178-1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maidak, B. L., J. R. Cole, C. T. Parker, Jr., G. M. Garrity, N. Larsen, B. Li, T. G. Lilburn, M. J. McCaughey, G. J. Olsen, R. Overbeek, S. Pramanik, T. M. Schmidt, J. M. Tiedje, and C. R. Woese. 1999. A new version of the RDP (Ribosomal Database Project). Nucleic Acids Res. 27:171-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Margot, M., B. Ollivier, and B. K. C. Patel. 2000. Microbiology of petroleum reservoirs. Antonie Leeuwenhoek 77:103-116. [DOI] [PubMed] [Google Scholar]
- 28.Mountfort, D. O., E. Morschel, D. B. Beimborn, and P. Schonheit. 1986. Methanogenesis and ATP synthesis in a protoplast system of Methanobacterium thermoautotrophicum. J. Bacteriol. 168:892-900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Neidhardt, F. C., and H. E. Umbarger. 1996. Chemical composition of Escherichia coli, p. 13-16. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, D.C.
- 30.Nester, E. W., C. E. Roberts, M. E. Lidstrom, N. N. Pearsall, and M. T. Nester. 1983. Microbiology. Saunders College Publishing, Philadelphia, Pa.
- 31.Nilsen, R. K., J. Beeder, T. Thorstenson, and T. Torsvik. 1996. Distribution of thermophilic marine sulfate reducers in North Sea oil field waters and oil reservoirs. Appl. Environ. Microbiol. 62:1793-1798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Patel, G. B., B. J. Agnew, and C. J. Dicaire. 1991. Inhibition of pure culture of methanogens by benzene ring compounds. Appl. Environ. Microbiol. 57:2969-2974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Raskin, L., J. M. Stromley, B. E. Rittmann, and D. A. Stahl. 1994. Group-specific 16S rRNA hybridization probes to describe natural communities of methanogens. Appl. Environ. Microbiol. 60:1232-1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 35.Schleper, C., W. Holben, and H.-P. Klenk. 1997. Recovery of crenarchaeotal ribosomal DNA sequences from freshwater lake sediments. Appl. Environ. Microbiol. 63:321-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Takai, K., and K. Horikoshi. 1999. Genetic diversity of archaea in deep-sea hydrothermal vent environments. Genetics 152:1285-1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ueki, A., K. Ueki, A. Oguma, and C. Ohtsuki. 1989. Partition of electrons between methanogenesis and sulfate reduction in the anaerobic digestion of animal waste. J. Gen. Appl. Microbiol. 35:151-162. [Google Scholar]
- 39.Vetriani, C., H. W. Jannasch, B. J. MacGregor, D. A. Stahl, and A. L. Reysenbach. 1999. Population structure and phylogenetic characterization of marine benthic Archaea in deep-sea sediments. Appl. Environ. Microbiol. 65:4375-4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Voordouw, G., S. M. Armstrong, M. F. Reimer, B. Fouts, A. J. Telang, Y. Shen, and D. Gevertz. 1996. Characterization of 16S rRNA genes from oil field microbial communities indicates the presence of a variety of sulfate-reducing, fermentative, and sulfide-oxidizing bacteria. Appl. Environ. Microbiol. 62:1623-1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Watanabe, K., M. Teramoto, and S. Harayama. 1999. An outbreak of non-flocculating catabolic populations caused the breakdown of a phenol-digesting activated-sludge process. Appl. Environ. Microbiol. 65:2813-2819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Watanabe, K., K. Watanabe, Y. Kodama, K. Syutsubo, and S. Harayama. 2000. Molecular characterization of bacterial populations in petroleum-contaminated groundwater discharged from underground crude-oil-storage cavities. Appl. Environ. Microbiol. 66:4803-4809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Watanabe, K., Y. Kodama, and S. Harayama. 2001. Design and evaluation of PCR primers to amplify bacterial 16S ribosomal DNA fragments used for community fingerprinting. J. Microbiol. Methods 44:253-262. [DOI] [PubMed] [Google Scholar]
- 44.Whitman, W. B., T. L. Bowen, and D. R. Boone. 1992. The methanogenic bacteria, p. 739-745. In A. Balows, H. G. Truper, M. Dworkin, W. Harder, and K. H. Schleifer (ed.), The prokaryotes, vol. 1. Springer-Verlag, New York, N.Y. [Google Scholar]
- 45.Widdel, F., and R. Rabus. 2001. Anaerobic biodegradation of saturated and aromatic hydrocarbons. Curr. Opin. Biotechnol. 12: 259-276. [DOI] [PubMed] [Google Scholar]
- 46.Wu, W., R. F. Hickey, and J. G. Zeikus. 1991. Characterization of metabolic performance of methanogenic granules treating brewery wastewater: role of sulfate-reducing bacteria. Appl. Environ. Microbiol. 57:3438-3449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zeikus, J. G., and M. R. Winfrey. 1976. Temperature limitation of methanogenesis in aquatic sediments. Appl. Environ. Microbiol. 31:99-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zengler, K., H. H. Richnow, R. R. Mora, W. Michaelis, and F. Widdel. 1999. Methane formation from long-chain alkanes by anaerobic microorganisms. Nature 401:266-269. [DOI] [PubMed] [Google Scholar]
- 49.Zepp Falz, K., C. Holliger, R. Großkopf, W. Liesack, A. N. Nozhevnikova, B. Muller, B. Wehrli, and D. Hahn. 1999. Vertical distribution of methanogens in the anoxic sediment of Rotsee (Switzerland). Appl. Environ. Microbiol. 65:2402-2408. [DOI] [PMC free article] [PubMed] [Google Scholar]