Abstract
The effect of signal molecules on the cultivation efficiency of bacteria from the Gotland Deep in the central Baltic Sea was investigated. Numbers of cultivated cells were determined by the most-probable-number (MPN) technique. Artificial brackish water supplemented with different carbon substrates at low concentrations (200 μM each) was employed as the growth medium. Compared to the results of previous studies, this approach yielded significantly higher cultivation efficiencies (up to 11% in fluid media). A further and pronounced increase in cultivation success was accomplished by the addition of cyclic AMP (cAMP), N-butyryl homoserine lactone, or N-oxohexanoyl-dl-homoserine lactone at a low concentration of 10 μM. The most effective inducer was cAMP, which led to cultivation efficiencies of up to 100% of total bacterial counts. From the highest positive dilutions of these latter MPN series, several strains were isolated in pure culture and one strain (G100) was used to study the physiological effect of cAMP. Dot blot hybridization revealed, however, that strain G100 represented only a small fraction of the total bacterial community. This points towards an inherent limitation of the MPN approach, which does not necessarily recover abundant species from highly diverse communities. Bacterial cells of strain G100 that were starved for 6 weeks attained a higher growth rate and a higher biomass yield when resuscitated in the presence of cAMP instead of AMP.
Generally, only a small fraction of the bacteria present in natural samples multiply in laboratory media. Thus, the cultivation efficiency for marine bacteria varies between 0.001 and 0.1% of total cell counts for open ocean and coastal environments, respectively (2). However, culture-independent methods like microautoradiography or the direct viable count technique have revealed that up to 50%, and in some cases even 90%, of the bacterial cells may be metabolically active (17, 29, 31).
Possible reasons for the low cultivation success include (i) cell damage caused by oxidative stress preventing growth (for instance, by the SOS response mechanism [51]) until repair has been completed, (ii) the formation of viable but nonculturable, or dormant, cells, (iii) inhibition by high concentrations of those substrates which have been exhausted in a previous growth phase (substrate-accelerated death), (iv) the induction of lysogenic phages during starvation, and (v) a lack of cell-to-cell communication in laboratory media (6). Indeed, the addition of pyruvate or catalase to reduce oxidative stress yields increased colony counts in some instances (10), and the addition of carbon substrates at only low concentrations increases the cultivation success (7). Other factors, which potentially could stimulate the growth of bacteria from natural samples, have not been explored in a systematic fashion. This is especially true for extracellular signal molecules.
In enterobacteria, cyclic AMP (cAMP) is involved in the regulation of the majority of genes expressed under starvation (43), including those coding for high-affinity sugar transport systems (15). The addition of extracellular cAMP has been shown to prevent substrate-accelerated death in starved laboratory cultures (11). Furthermore, extracellular cAMP occurs in seawater at concentrations between 1 and 35 pM (3). Unlike Escherichia coli, marine planktonic bacteria express high-affinity uptake systems for cAMP and, despite the low ambient concentrations, are therefore capable of increasing intracellular cAMP to levels (up to 2.8 μM) typical for E. coli and other prokaryotes (3). The above observations make cAMP a promising candidate for improving the cultivation success of natural bacterioplankton.
In gram-negative bacteria, N-acyl homoserine lactones are major autocrine molecular signal molecules which trigger processes as diverse as resuscitation from the lag phase (8), bioluminescence (28), and virulence (46). In contrast, the typical signal molecules of gram-positive bacteria are posttranslationally processed peptides. Since gram-negative bacteria dominate in the planktonic environment (20), we focused on the effect of N-acyl homoserine lactones on the cultivation success of natural bacterioplankton.
In the present study, the influence of cAMP and two different N-acyl homoserine lactones on the cultivation efficiency of natural bacterial communities was assessed. We chose N-(oxohexanoyl)-dl-homoserine lactone (OHHL), since it represents the homoserine lactone which is used by the majority of gram-negative bacteria capable of quorum sensing. In addition, we tested N-(butyryl)-dl-homoserine lactone (BHL), which carries a shorter acyl side chain and therefore is more diffusible than the other known N-acyl homoserine lactone signal compounds. As a sampling site we selected the Gotland Deep, a depression in the central Baltic Sea between Latvia and the Swedish island of Gotland. Because of the pronounced vertical gradients of its physicochemical parameters, this system provides the opportunity to study very different bacterioplankton assemblages and sediment bacterial communities at one site.
MATERIALS AND METHODS
Study site.
The sampling site was located in the Gotland Deep in the central Baltic Sea (57°18′N, 20°05′E). The maximum water depth at this position was 240 m.
Sampling procedure.
Samples were obtained between 14 and 17 July 2000 during the cruise of the RV Alexander von Humboldt. Water samples were collected by means of a rosette sampler (Hydrobios, Altenholz, Germany) connected to a Sea-Brid 911 plus conductivity-temperature-depth profiler (Sea-Brid Electronics Inc., Bellevue, Wash.). Concentrations of dissolved oxygen were determined by Winkler titration (22). Based on the vertical stratification pattern (see Fig. 1A and B), depths of 5, 60, 95, and 200 m were chosen for sampling. A multicorer (Wuttke, Hamburg, Germany) was used for sampling of the upper 50 cm of the bottom sediment. Microbial biomass for direct DNA extraction was collected by tangential flow filtration (0.16-μm pore size, ProVario 3; Filtron, Karlstein, Germany).
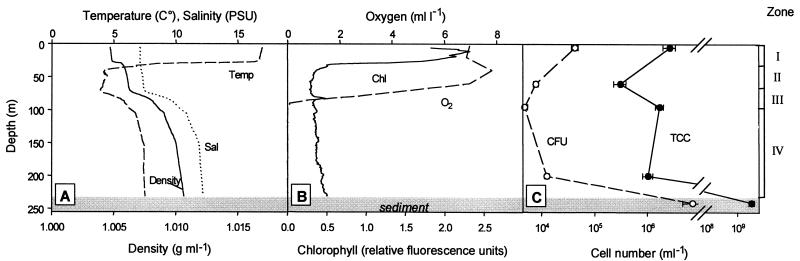
FIG. 1.
Vertical profiles of physicochemical and microbiological parameters in the Gotland Deep on 14 July 2000. (A) Temperature, salinity, and density; (B) oxygen and chlorophyll concentrations (in relative fluorescence units); (C) vertical distribution of total cell counts (TCC) and CFU on agar solidified media. Zone I: warm, photic zone; zone II: cold, aphotic zone; zone III: halocline-oxycline; zone IV: anoxic zone with high salinity.
Total bacterial numbers.
Water and sediment samples were fixed in 2% glutaraldehyde and stored at 4°C. Total cell counts were determined by epifluorescence after staining with 4′,6-diamidino-2-phenylindole (DAPI). After fixation, water samples or aliquots of the sediment suspensions were filtered onto 0.1-μm-pore-size polycarbonate filters (Nuclepore Track-Etch Membrane; Whatman, Springfield Mill, United Kingdom). The filters were then stained by floating on a solution containing 2% (vol/vol) paraformaldehyde, 0.05% (vol/vol) Triton X-100, and 0.36 μg of DAPI ml−1 in 0.5× phosphate-buffered saline (PBS) (1× PBS is 130 mM NaCl and 10 mM sodium phosphate, pH 7.25). Afterwards, the filters were dried, embedded in DABCO antifading solution (25 mg of 1,4-diazabicyclo[2.2.2]octane in 1 ml of PBS buffer plus 9 ml of glycerol), and subsequently examined by epifluorescence microscopy (Zeiss Axiolab microscope; filter set, 01; excitation, 365 nm; emission, 397 nm).
Cultivation and determination of most probable numbers (MPN).
The artificial brackish water medium (ABW) used for cultivation of heterotrophic bacteria consisted of 94.7 mM NaCl, 13.6 mM MgCl2 • 6H2O, 2.3 mM CaCl2 • 2H2O, 2.0 mM KCl, 6.4 mM Na2SO4, 2.4 mM NaHCO3, 192 μM KBr, 92 μM H3BO3, 34 μM SrCl2, 92 μM NH4Cl, 9 μM KH2PO4, and 16 μM NaF and was buffered with 10 mM HEPES. The pH was adjusted to 7.3. After autoclaving, 1 ml of trace element solution SL 10 (52), 1 ml of a selenite-tungstate solution (52), and 10 ml of a 10-vitamin solution (5) were added per liter of medium. The basal medium was supplemented with two different substrate mixtures to yield two modifications of the growth medium. The polymer medium, ABWP, contained 0.5 g each of agarose, starch, laminarin, xylan, and chitin per liter, whereas the monomer medium, ABWM, contained the 20 amino acids at a final concentration of 100 μM and glucose at a final concentration of 200 μM.
cAMP (Sigma Chemical Co., St. Louis, Mo.), N-(ketocaproyl)-dl-homoserine lactone (synonymous with OHHL) (Sigma Chemical Co.), and BHL (Fluka Chemie AG, Buchs, Switzerland) were added to the ABWP and ABWM media to a final concentration of 10 μM. Basal media (ABWP and ABWM) without inducers served as controls.
Aliquots of 180 μl of growth medium were dispensed into sterile polystyrene deep-well microtiter plates (Beckman Coulter Inc., Fullerton, Calif.). Directly after sampling, 20-μl subsamples of the water samples, or 20 μl of a 1:100 dilution of the surface sediment, were immediately inoculated into five wells of a microtiter plate. The inoculation of anoxic MPN series was performed in nitrogen-filled Atmosbags (Aldrich Chemicals, Milwaukee, Wis.). MPN series were generated in five parallels by 1:10 dilutions in consecutive microtiter wells. Incubation under anoxic atmosphere was carried out by employing the Anaerocult C mini system (Merck, Darmstadt, Germany). Microtiter plate cultures were incubated at 15°C for 6 weeks.
Subsequently, bacterial growth was monitored by epifluorescence microscopy. From each well, a 20-μl aliquot was transferred to a well of epoxy-coated slides (Omnilab, Bremen, Germany), and the samples were allowed to dry at 65°C. A 1.6-μl aliquot of a 1:1 mixture of DAPI solution (1 μg • ml−1) and DABCO (see above) was added to each slide well, and the slide was covered by a coverslip. The presence or absence of bacterial cells was monitored for each well, and most probable cell numbers were calculated by using a computer program (30). The statistical significance of differences in MPN between samples was calculated according to the method of Jones (26).
Isolation of bacterial strains.
Cells of the highest positive dilutions of each MPN microtiter plate were collected into one tube, yielding 600 μl of total volume. A 20-μl subsample was inoculated into fresh growth medium and incubated for 7 days at 15°C. For isolation of pure cultures of heterotrophic bacteria, the liquid subcultures were spread onto solid agar media containing both the polymer mix (agarose, starch, laminarin, xylan, and chitin) and monomer mix (all 20 amino acids and glucose) in the same concentrations as in the liquid media.
Starvation experiments.
A preculture of strain G100 was grown for 2 days in the monomer medium (ABWM). Afterwards, cells were pelleted at 10,000 × g for 20 min and the pellet was resuspended in sterile ABW to remove excess carbon substrates. After a second centrifugation, the supernatant was discarded and cells were resuspended in a small volume of sterile ABW devoid of carbon substrates. Subsequently, bacteria were incubated under four different conditions: oxic conditions at room temperature or at 4°C and anoxic conditions at room temperature or at 4°C. The anoxic ABW was prepared under N2, received 600 μM H2S, and was distributed in 100-ml screw-cap bottles in order to ensure reducing conditions (4).
After 6 weeks of incubation, the starved cells were collected by centrifugation (20 min at 9,000 × g) and bacterial pellets were resuspended by using sterile glass homogenizers. The homogenized cell suspensions were transferred to tubes containing ABWM supplemented with 10 μM cAMP or BHL. Media supplemented with either AMP or butyric acid plus homoserine lactones (10 μM each) served as negative controls. Incubation was carried out at 20°C, and growth was monitored as the optical density with a Ratio/XR turbidimeter (Hach Company, Loveland, Colo.). For calibration, cell numbers were determined at regular intervals by employing DAPI staining and epifluorescence microscopy.
DNA extraction.
The remaining 580 μl of cell suspensions (see “Isolation of bacterial strains”) was centrifuged for 30 min at 43,700 × g at 4°C. DNA was extracted by a freeze and thaw procedure (24). Cell concentrates of the natural bacterioplankton which had been obtained by tangential flow were centrifuged for 30 min at 43,700 × g and 4°C and then lysed by adding 250 μl of lysis buffer (200 mM Tris-HCl, 50 mM EDTA [pH 8.0]), 0.25 mg of proteinase K, and 25 μl of 20% (wt/vol) sodium dodecyl sulfate (SDS; pH 7.2) to 250 μl of cell pellet. After mixing, samples were incubated for 1 h at 65°C in a water bath and then centrifuged for 30 min at 12,000 × g. The samples were extracted with 1 volume of buffered phenol, then with phenol-chloroform-isoamyl alcohol (25:24:1, vol/vol/vol), and finally with chloroform. DNA was precipitated by adding 0.1 volume of sodium acetate (3 M, pH 5.2) and 2.5 volumes of ice-cold ethanol and finally quantified with the fluorescent dye PicoGreen (Molecular Probes, Eugene, Oreg.). For DNA extraction of the natural bacterial community in the surface sediment, 1 cm3 of wet sediment from the surface of a sediment core was centrifuged at 43,700 × g and 4°C. The pellet was then treated as described above for the cell pellets from the water samples.
DNA extraction of pure cultures was done by the freeze and thaw technique.
PCR amplification.
Approximately 620-bp-long 16S ribosomal DNA (rDNA) fragments for denaturing gradient gel electrophoresis (DGGE) analysis were amplified by using conditions described previously (40) and primer pair GC341f/907r (38). Each reaction mixture received 1 μl of the freeze and thaw DNA preparations, or 50 ng of genomic DNA in the case of genomic DNA from mixed bacterial communities.
DGGE analysis.
PCR products were applied onto 6% (wt/vol) polyacrylamide gels which contained a linear gradient of 30 to 70% denaturant (40). Electrophoresis proceeded for 30 min at 50 V, followed by 7 h at 150 V. After staining with ethidium bromide, gel images were captured with a charge-coupled device camera (CF 8/1 RCC; KAPPA Messtechnik, Gleichen, Germany) by employing the image analysis program Image P2 (H+K Messsysteme, Berlin, Germany). DNA bands of interest were excised with a sterile scalpel, and the DNA was eluted overnight at 4°C in 20 μl of sterile double-distilled water. After reamplification, the DNA was purified and quantified.
Cloning.
One DGGE band of a natural bacterioplankton community which did not yield an unambiguous sequence was suspected to contain more than one 16S rRNA gene sequence. The reamplification products of this band were therefore ligated into the pGEM-T vector (Promega, Madison, Wis.), and the products were transformed into competent E. coli JM109 cells (Promega). Ligation and transformation were performed according to the instructions of the manufacturer. After plating, 15 white colonies were picked at random, and the clones were subjected to PCR with primers pUC/M13 forward (5′-GTT TTC CCA GTC ACG AC-3′) and pUC/M13 reverse (5′-CAG GAA ACA GCT ATG AC-3′) (36). Amplification conditions comprised a denaturation at 96°C for 4 min followed by 30 thermal cycles with the melting temperature set to 95°C for 30 s, annealing at 50°C for 1 min, and extension at 72°C for 2 min. A final extension was performed at 72°C for 10 min. The resulting PCR products were reanalyzed by DGGE, and products with correct melting behavior were finally sequenced.
Sequencing and phylogenetic analysis.
The 16S rRNA gene fragments were sequenced as described previously (25) by using the SequiTherm Excel II DNA sequencing kit (Epicentre Technologies, Madison, Wis.) and an automated infrared laser fluorescence sequencer Li-Cor Model 4000 (Li-Cor Inc., Lincoln, Nebr.). The 16S rDNA sequences were compared to the sequences in the GenBank database by using the program BLASTN 2.0 (1), which is available through the National Center for Biotechnology Information website. Sequences of closely related bacteria were aligned with the aid of the ClustalX program (49). Evolutionary distances were then calculated with the algorithm of Jukes and Cantor (27), and a phylogenetic tree was constructed by the neighbor-joining method (42) by using the program MEGA version 2.0 of Kumar et al. (32).
Dot blot hybridization.
The abundance of one bacterial isolate obtained from the highest positive dilutions of MPN series was quantified by employing a dot blot procedure (12). The 16S rDNA fragments from the natural bacterioplankton community were amplified by using primers 341f and 907r (38), with an initial denaturing step at 96°C for 4 min followed by 25 cycles with melting at 94°C for 30 s, annealing at 58°C for 45 s, and extension at 72°C for 1 min and a final extension at 72°C for 10 min. PCR products were purified with the QIAquick purification kit (Qiagen), denatured for 10 min at 96°C, and vacuum blotted onto positively charged nylon membranes (Roche, Mannheim, Germany). For calibration, different amounts of genomic DNA of strain G100 were amplified, purified, and blotted in parallel. DNA was fixed by baking the membranes at 120°C for 30 min.
A probe for the detection of strain G100 was generated from its genomic DNA by PCR by using random labeling with digoxigenin (DIG)-11-dUTP (PCR DIG probe synthesis kit; Roche) and the conditions described above. Membranes were prehybridized in 8 ml of DIG Easy Hyb buffer (Roche) at 68°C. Hybridization was carried out for 16 h at 68°C in 10 ml of Easy Hyb buffer containing the denatured G100 probe. After hybridization, the blot was washed twice at room temperature for 15 min in 2× SSC (1× SSC is 150 mM NaCl plus 15 mM sodium citrate [pH 7.0]) containing 0.1% SDS, followed by two stringent washing steps (15 min in 1× SSC and 0.5× SSC plus 0.1% SDS at 85°C). The hybridization signal was detected with the DIG luminescence detection kit (Roche) and Lumi-Film (Roche) according to the instructions of the manufacturer. For quantification of individual spots, the image was digitized and analyzed by using ZERO-Dscan software (Scanalytics, Billerica, Mass.).
Sequence accession number and deposition of strain G100.
The nearly full-length 16S rDNA sequence of strain G100 has been submitted to the GenBank database and deposited under accession number AY043327. Strain G100 has been deposited at the German Collection of Microorganisms and Cell Cultures (Deutsche Sammlung von Mikroorganismen und Zellkulturen) under strain number DSMZ 14993.
RESULTS
Vertical stratification of the water column.
The water column of the Gotland Deep was characterized by pronounced gradients of temperature, salinity, and oxygen concentrations. The thermocline was located between 27 and 38 m, where water temperatures decreased from 14 to 4°C (Fig. 1A). Within the thermocline, chlorophyll concentrations dropped by a factor of 5 (Fig. 1B). A pronounced increase in salinity was detected between 75 and 110 m in depth. Within this halocline, water temperature increased again to 6°C. The vertical position of the halocline coincided with a pronounced decrease in oxygen concentration (oxycline) (Fig. 1A and B). The water layers below 100 m in depth were anoxic (Fig. 1B). Since the steep vertical density gradients caused by the thermocline and halocline limit convective mixing, four distinct water bodies can be discerned in the water column of the Gotland basin: (i) a warm, photic zone with an active phytoplankton assemblage; (ii) a cold, aphotic zone of low salinity; (iii) the halocline-oxycline; and (iv) an anoxic zone of high salinity (Fig. 1). Based on this vertical stratification pattern, four different sampling depths (5, 60, 95, and 200 m) were chosen for cultivation assays in order to cover the most diverse bacterioplankton communities. In addition, samples from the sediment surface at 241 m in depth were included in the analysis.
Vertical distribution of bacteria and cultivation efficiency.
In the water column, total bacterial cell counts varied between 2.7 × 106 cells • ml−1 at 5 m and 3.1 × 105 cells • ml−1 at 60 m in depth. Total bacterial cell counts reached 2.8 × 109 cells • cm−3 in the surface layer of the sediment (Fig. 1C). Direct plating onto solid agar media containing both monomers and polymers and incubation under oxic conditions yielded between 4.8 × 103 and 4.2 × 104 CFU per ml for water samples and 7.2 × 106 CFU • cm−3 for the sample from the sediment surface. The corresponding cultivation efficiencies thus ranged between 0.25 and 2.5% (Fig. 1C).
Cultivation in liquid medium in the absence of signal molecules led to cultivation efficiencies of up to 11% (Fig. 2, solid white bars). However, liquid media containing 10 μM cAMP, BHL, or OHHL yielded significantly (P < 0.05) enhanced values in 14 out of 80 MPN series, despite the high standard deviations, which are characteristic for MPN assays. In nine of the experiments, the difference from the controls was highly significant (P < 0.01; bars marked by ** in Fig. 2A to D). This stimulation was not correlated with the oxygen supply during incubation (oxic or anoxic conditions) or with the type of carbon substrates used (monomers or polymers). Rather, a correlation with the sampling depth was observed, since an increase in cultivation efficiency was observed most frequently for water samples from 60 and 200 m in depth.
FIG. 2.
Effect of signal compounds (cAMP, OHHL, and BHL), oxygen, and carbon substrates on the cultivation efficiency of bacteria from different depths of the Gotland basin. Cultivation efficiency was determined by the MPN technique and is given as a percentage of total cell counts. (A) Growth in carbon monomer medium, oxic conditions; (B) growth in polymer medium, oxic conditions; (C) growth in monomer medium, anoxic conditions; (D) growth in polymer medium, anoxic conditions. Controls consisted of the corresponding basal medium containing monomers or polymers but no signal compounds. Horizontal bars indicate 95% confidence intervals. Significant increases in cultivation success versus controls are marked by asterisks (*, P < 0.05; **, P < 0.01). Numbers on the right denote different water bodies (see Fig. 1 legend for zone definitions).
cAMP was the most effective signal compound for increasing MPN counts and led to a cultivation success of up to 100% (Fig. 2B). The latter value was attained with the water sample from 200 m in depth when incubated under oxic conditions. Positive effects on bacterial growth were also observed in media containing BHL, which yielded maximum cultivation efficiencies of 20% (Fig. 2A). An increased cultivation success by OHHL was observed only rarely, for anoxic water and sediment samples in media containing polymers (Fig. 2B and D). Interestingly, only low numbers (<3% of total cell counts) of bacteria from the oxycline grew in an oxic atmosphere, while values were often significantly higher (reaching 7%; in 4 out of 8 cases, values were significant at P < 0.05) when samples were incubated under the same conditions but under an anoxic atmosphere.
Molecular fingerprinting of cultured bacteria.
The extraordinarily high MPN values reached in liquid media containing cAMP and BHL suggested that the bacteria growing in the highest positive dilutions may be numerically dominant in the natural bacterioplankton community. This hypothesis was tested by two different molecular techniques, namely, DGGE fingerprinting and dot blot hybridization.
16S rDNA fragments were amplified from the highest positive dilutions of MPN series, and fingerprint patterns of MPN series with signal compounds were compared to those of the control dilution series and to those amplified directly from the natural community by DGGE (Table 1 and Fig. 3). This 16S rDNA fingerprinting was performed for all liquid dilution series in which MPN values were significantly increased by the addition of inducers.
TABLE 1.
Number of 16S rDNA fingerprints of natural bacterial communities which correspond to those of bacteria grown in the highest positive MPN dilutions
| Depth (m) | No. of 16S rDNA fingerprints:
|
||
|---|---|---|---|
| In natural bacterial community | Recovered exclusively with signal compound | Identical to control, but with significantly enhanced MPN (P < 0.01) | |
| 5 | 9 | 2a | 0 |
| 60 | 4 | 0 | 1b |
| 95 | 7 | 0 | 0 |
| 200 | 13c | 4d | 1e |
| 241 (sediment) | 11 | 0 | 2f |
Conditions: cAMP, ABWM, oxic and BHL, ABWP, anoxic.
Conditions: cAMP, ABWP, anoxic.
Compare Fig. 3, right lane.
Conditions: cAMP, ABWP, oxic (compare Fig. 3, band 2); BHL, ABWM, oxic; cAMP, ABWM, anoxic; and OHHL, ABWP, anoxic.
Conditions: cAMP, ABWP, oxic (compare Fig. 3, band 1).
Conditions: cAMP, ABWP, oxic and OHHL, ABWP, oxic.
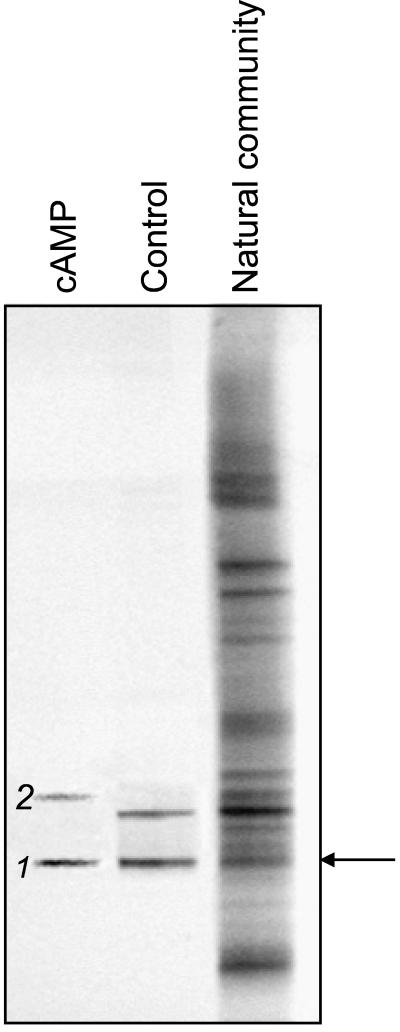
FIG. 3.
DGGE analysis of 16S rDNA fragments of the natural community at 200 m in depth and of bacteria cultured in liquid dilution series (ABWP medium, oxic incubation; compare results shown in Fig. 2B) from the same sample. The arrow indicates the band of the natural community which corresponds to band 1, the fingerprint of the isolated strain G100. Band 2 denotes a fingerprint which was obtained in the presence of cAMP but not with the control.
Our analyses revealed that two of the fingerprints of natural bacterioplankton from 5 m in depth and four of the fingerprints from 200 m in depth (e.g., band 2 in Fig. 3) were recovered only after cultivation in the presence of cAMP, BHL, or OHHL (Table 1). Furthermore, four fingerprints of the bacterial communities at water depths of 60 and 200 m (e.g., band 1 in Fig. 3) and at the sediment surface were detected in the control incubations as well, but the addition of signal compounds significantly increased MPN values (Table 1).
Bacterial isolates and their occurrence in the natural bacterial community.
The recovery of fingerprints of the natural bacterioplankton community and the high MPN values attained in two MPN series from the 200-m water sample prompted us to choose the highest positive dilutions of these samples for the isolation of bacterial strains. Two strains, designated G100 and G200, were isolated from the highest positive dilution of media supplemented with cAMP and BHL, respectively. Both strains yielded 16S rDNA fingerprints identical to those in the original highest positive dilution (e.g., band 1 in Fig. 3), and both had the same 16S rDNA sequence. A strain with a 16S rRNA gene sequence corresponding to fingerprint band 2 (Fig. 3) could not be isolated from the highest positive dilution of media supplemented with cAMP.
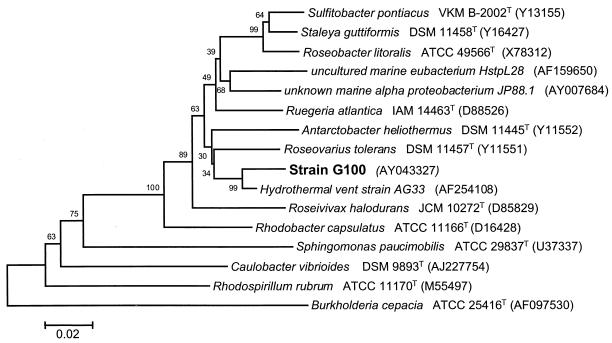
Sequence comparison identified the isolated strains as members of the α subclass of Proteobacteria. Their closest relative is strain AG33, isolated from a hydrothermal vent near the Galapagos Islands (48); the sequence similarity between G100 and AG33 is 97.6% (Fig. 4). The phylogenetic analysis clearly indicates an affiliation of strain G100 and G200 with the new family Rhodobacteraceae, as established in the second edition of Bergey's Manual of Systematic Bacteriology (18). The most closely related validly described species is Roseovarius tolerans, with a sequence similarity of 94.5%.
FIG. 4.
Phylogenetic affiliation of strain G100 within the α subclass of Proteobacteria. GenBank accession numbers are shown in parentheses. The bar represents 2% sequence divergence. Bootstrap values are based on 1,000 resamplings.
The 16S rRNA genes of strains G100 and G200 were also compared to the sequence of the corresponding band of the natural community (marked by the arrow in Fig. 3). Direct sequencing of the DNA eluted from this band did not result in an unambiguous sequence. Therefore, the DNA fragments were cloned. Four of the environmental clones had DGGE fingerprints matching those of strains G100 and G200 and thus were sequenced. All four had identical sequences but, unexpectedly, were affiliated with the cyanobacterium Synechococcus sp. strain PCC7001 (GenBank accession no. AB015058).
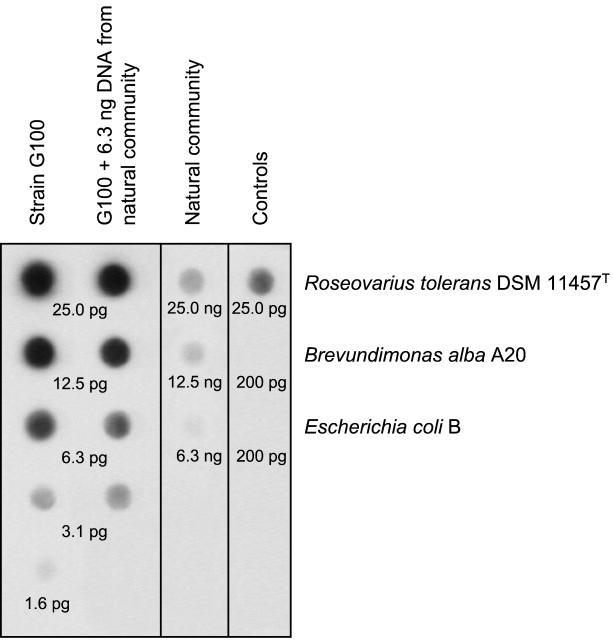
The failure to detect the sequences of strains G100 and G200 in the natural bacterioplankton community appeared to be inconsistent with the high MPN values determined. Therefore, the abundance of genomic DNA of strain G100 in the natural bacterioplankton community at 200 m in depth was quantified by dot blot hybridization. Initially, genomic DNA from bacterioplankton was blotted directly and hybridized with a probe for strain G100. However, no signal above the detection limit (0.1% of total genomic DNA) could be obtained with this technique (data not shown). Consequently, 16S rRNA fragments of the bacterioplankton community were amplified prior to blotting and subsequently quantified. Several controls were included to validate the latter analytical procedure. The addition of 1,000-fold excess genomic DNA from the natural bacterioplankton community did not interfere with the detection of DNA of strain G100 (Fig. 5, second lane). Our probe detected neither DNA of E. coli nor that of another α-proteobacterium, Brevundimonas alba strain A20 (E. Jaspers and J. Overmann, submitted for publication) (Fig. 5, right lane). Despite the stringent hybridization conditions applied, however, some cross-hybridization occurred with DNA from Roseovarius tolerans DSM11457T, the closest cultured relative of strain G100 (Fig. 5). Cross-hybridization could not be decreased further without a loss of the signal for strain G100. Consequently, our estimate represents an upper limit for the abundance of strain G100 in the natural bacterioplankton community. Based on the results of our dot blot procedure, genomic DNA of strain G100 represents ≤0.02% of the total genomic DNA of the bacterioplankton community (Fig. 5). This independent molecular analysis thus fully confirms the results of the DGGE fingerprinting. Clearly, strains G100 and G200, despite their origin from the highest positive dilutions, which corresponds to a cultivation success of up to 100%, do not represent abundant members of the bacterioplankton community in the 200-m-depth water sample of the Gotland Deep.
FIG. 5.
Quantification of genomic DNA of strain G100 in the natural bacterial community of 200 m in depth by dot blot hybridization. Different concentrations of template DNA of strain G100 served as standards. As a control, DNA of strain G100 was mixed with a constant amount (6.3 ng) of template DNA from the natural community. A digitized image of an exposed Lumi-Film is shown. Values below dots indicate the concentration of genomic DNA employed.
Effect of signal compounds on the regrowth of starved cells.
Strain G100 was the most promising candidate for studying the effect of signal compounds on resuscitation and growth. Cells were grown until stationary phase, washed, and starved in the absence of carbon substrates at either 4 or 20°C and under oxic or anoxic conditions, yielding a total of four different incubation conditions (Table 2). After 6 weeks of starvation, cells were inoculated into fresh monomer medium (ABWM) and growth was monitored at 6- to 8-h intervals (Fig. 6 and Table 2).
TABLE 2.
Effect of signal molecules on maximum growth rates, doubling times, and cell yields of starved cells of strain G100
| Signal molecule | Starvation conditions | Maximum growth rate (h−1) | Doubling time (h) | Cell yield (NTUa) |
|---|---|---|---|---|
| cAMP | Oxic, 4°C | 0.1073 ± 0.0081b | 6.46b | 12.75 ± 1.04b |
| Anoxic, 4°C | 0.0761 ± 0.0153b | 9.11b | 12.02 ± 1.11b | |
| Oxic, 20°C | 0.1191 ± 0.0060b | 5.82b | 12.34 ± 1.17 | |
| Anoxic, 20°C | 0.0709 ± 0.0080b | 9.78b | 12.69 ± 0.67 | |
| AMP | Oxic, 4°C | 0.0936 ± 0.0104b | 7.41b | 10.65 ± 1.03b |
| Anoxic, 4°C | 0.0453 ± 0.0069b | 15.30b | 8.27 ± 0.76b | |
| Oxic, 20°C | 0.0709 ± 0.0118b | 9.79b | 11.36 ± 0.43 | |
| Anoxic, 20°C | 0.0578 ± 0.0065b | 11.99b | 12.31 ± 0.67 | |
| BHL | Oxic, 4°C | 0.0879 ± 0.0075 | 7.89 | 9.16 ± 0.67 |
| Anoxic, 4°C | 0.0927 ± 0.0202 | 7.48 | 9.69 ± 0.74 | |
| Oxic, 20°C | 0.1196 ± 0.0135 | 8.02 | 9.63 ± 0.50 | |
| Anoxic, 20°C | 0.0869 ± 0.0095 | 7.98 | 8.97 ± 0.65 | |
| Butyric acid plus homoserine lactone | Oxic, 4°C | 0.0851 ± 0.0097 | 8.15 | 9.51 ± 0.82 |
| Anoxic, 4°C | 0.0864 ± 0.0373 | 8.02 | 9.19 ± 1.10 | |
| Oxic, 20°C | 0.1402 ± 0.0073 | 4.94 | 10.44 ± 0.85 | |
| Anoxic, 20°C | 0.1042 ± 0.0071 | 6.65 | 8.58 ± 0.52 |
NTU, nephelometric turbidity units.
Value was significantly (P < 0.001) increased in the presence of signal compound compared to control level.
FIG. 6.
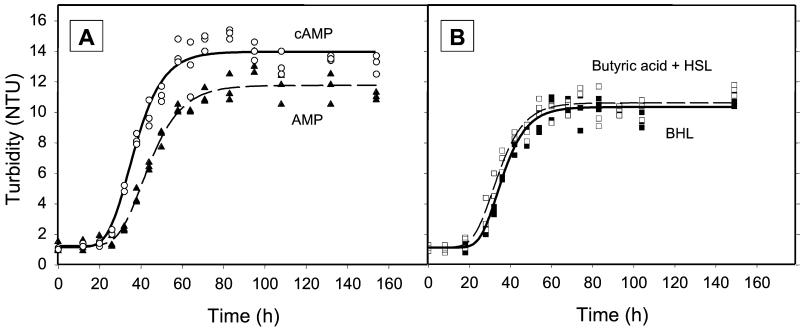
Resuscitation of strain G100 after starvation at 4°C under oxic conditions, measured as the increase in nephelometric turbidity units (NTU). (A) Regrowth after the addition of 10 μM cAMP (○) or AMP (▴); (B) regrowth after the addition of BHL (▪) or butyric acid plus homoserine lactone (□).
In all cases, batch cultures growing in the presence of cAMP attained doubling times which were significantly shorter than those of the controls containing AMP (Fig. 6A and Table 2). In contrast, the addition of BHL did not consistently lead to growth faster than that observed for the control media containing butyric acid plus homoserine lactone (Fig. 6B and Table 2); rather, BHL exerted an inhibitory effect in two of the experiments (Table 2). Cells starved at 4°C also reached a significantly higher biomass yield in the presence of cAMP, independent of the presence or absence of oxygen (Fig. 6 and Table 2). Repeated checks by epifluorescence cell counting confirmed that the observed increases in turbidity were accompanied by parallel increases in cell numbers. Cell yields were not increased, however, after cells had been starved at room temperature.
Since the monomer medium contained several carbon substrates, we investigated the growth response of strain G100 in synthetic ABW containing only individual organic carbon substrates. Poor growth was observed on single amino acids, leading to only one doubling of cells over 3 to 8 days. In contrast, growth on glucose alone was comparable to that in the complete monomer medium. In none of the cases, however, did the addition of cAMP result in higher growth responses or higher cell yields.
DISCUSSION
Generally, the cultivation efficiency of bacteria in seawater samples ranges from 0.001 to 0.1% (2) and the in situ abundance of culturable bacteria is very low (45; for a more optimistic view, see reference 41). The majority of bacterial species have not been cultured yet, as indicated by the high numbers of species which have been detected by culture-independent approaches in natural samples (25, 50). It therefore appears likely that many as-yet-uncultured bacteria possess unknown physiological properties and hence require novel techniques for isolation.
Means to increase cultivation success.
With our mineral brackish water medium, which contains low concentrations of organic carbon substrates, the very low cultivation efficiency of planktonic bacteria from the Gotland Deep on nutrient-rich solid media (≤0.18% [19]) could be increased by 1 order of magnitude. In liquid MPN series, our medium increased the cultivation success further by another order of magnitude, which is in line with the observation that some bacteria do not form colonies at an air-solid interface (13).
However, the most pronounced increase in viable counts was achieved by the addition of the signal molecules cAMP and BHL. So far, only the growth of a marine Pelagiobacter strain under the specific condition of iron limitation has been shown to be stimulated by the addition of synthetic N-octanoyl-homoserine lactone (21). Our present results suggest that signal compounds may be of relevance for the growth of other marine bacteria, as well.
Effect of cAMP on resuscitation and growth.
In enteric bacteria, cAMP is part of the pleiotropic crp activation system, which regulates most peripheral catabolic operons and carbohydrate transport systems and typically mediates carbon catabolite repression (34). Sugars transported by the phosphotransferase system decrease intracellular cAMP levels, which in turn prevents the expression of genes necessary for the uptake of non-phosphotransferase system sugars. After the depletion of glucose, intracellular cAMP concentrations rise from 0.3 up to 3 μM, and other sugars can be taken up and metabolized after a lag period required for the induction of the necessary genes (biphasic growth curve). The intermediate lag phase can be abolished by the addition of extracellular cAMP at millimolar concentrations (14) without changing the growth rate itself (39). The involvement of cAMP in the regulation of catabolic enzymes has also been demonstrated for a wide range of nonenteric bacteria, including α-proteobacteria (9). Other roles of cAMP in E. coli are known (23, 33, 47).
Of all signal compounds, cAMP yielded by far the highest numbers of viable cells in our liquid MPN series. Strain G100, isolated from the highest positive dilutions, exhibited a novel response to exogenous cAMP which clearly differed from diauxic growth in E. coli: (i) biphasic growth could never be detected in media containing several carbon substrates, (ii) cells exhibited a clear response to cAMP concentrations as low as 10 μM (rather than millimolar concentrations), and (iii) growth rates as well as cell yields increased significantly after the addition of exogenous cAMP.
Our data confirm the presence of high-affinity cAMP uptake systems in marine planktonic bacteria (3), enabling them to take up dissolved cAMP even at picomolar (rather than millimolar) concentrations and accumulate it to an intracellular concentration of 2.8 μM (3), a value known to be sufficient for gene activation. E. coli cells in continuous cultures excrete >99.9% of the intracellularly synthesized cAMP into the culture medium (35). Similarly, the cAMP present in seawater (3) may originate from dividing planktonic bacteria. The cAMP excreted could then serve as a signal to stimulate the growth of accompanying dormant cells.
Implications for the application of the MPN technique.
It has been shown that cellular isolates which correspond to abundant 16S rDNA sequences can indeed be isolated if the inocula are diluted in MPN series prior to enrichment (16). This effect has been attributed to the presence of rare but rapidly growing bacteria which are eliminated by the dilution step. Consequently, the MPN technique currently represents the method of choice to obtain cultures of abundant bacteria (16).
Unexpectedly, isolates obtained in the present study from the highest positive dilutions, despite the very high cultivation success of up to 100%, could not be detected by DGGE fingerprinting and yielded only a very faint signal during dot blot hybridization. Based on the relatively large inherent statistical uncertainty of the MPN technique, the fraction of strain G100 in the natural bacterioplankton community could also be as low as 12% (the lower limit of the MPN confidence interval). Nevertheless, this minimum value is still orders of magnitude higher than the abundance of strain G100 determined independently by the dot blot technique (0.02%). Obviously, bacteria growing at very high dilution steps may actually represent phylotypes which occur at a low frequency in the natural bacterioplankton community. Independent support for a low abundance of strain G100 comes from the notion that DGGE fingerprinting (which did not yield a signal of G100 from the natural bacterioplankton community) is known to detect only those 16S rDNA sequences which represent more than 1 to 9% of all phylotypes (37, 44). Finally, our present results corroborate recent results from the bacterioplankton community of a eutrophic freshwater lake, in which a strain isolated from an MPN dilution step corresponding to an abundance of 15% of the total cell count actually represented only 0.07% of the natural bacterial community (Jaspers and Overmann, submitted).
It appears unlikely that the observed discrepancy is due to a strong amplification bias selecting against the 16S rDNA of strain G100, since the addition of small amounts of its genomic DNA to the DNA from the entire bacterioplankton community yielded the expected signals during dot blot hybridization. Rather, our results may be explained by a high diversity and at the same time a high evenness of the bacterial community, in which many different phylotypes occur at similar but low frequencies. Indeed, DNA reassociation kinetics indicate that bacterioplankton communities consist of a high number (on the order of 550) of different species (25) and an even higher number of different phylotypes. Under such conditions, the type of bacteria isolated from the highest positive dilutions in MPN series would largely be determined by chance, and consequently, a high number of different isolates could potentially be obtained by cultivation.
A principal limitation of the MPN method is that only a small number (usually 3 to 10) of parallels are available, such that the number of strains which can be potentially isolated from the highest positive dilutions is on the order of 10 at most. Therefore, the MPN approach does not appear to be appropriate if the cultivated fraction is considerably diverse and different types of bacteria occur at low frequencies. In these cases, the generation of diluted inocula by automated methods and the screening of large numbers of isolates by molecular fingerprinting appear to constitute a more promising approach.
Acknowledgments
We thank Günter Jost for the invitation to participate in the cruise of the RV Alexander von Humboldt. Thanks are due to Henrik Sass for assistance in performing the MPN series and helpful discussions, to Peter Hirsch for providing Roseovarius tolerans strain EL-172, and to Thorsten Brinkhoff for assistance in the cloning work.
This work was funded by the BMBF (Bundesministerium für Bildung, Wissenschaft, Forschung und Technologie) to J. Overmann and H. Cypionka, grant no. 0311949.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amann, R., W. Ludwig, and K. Schleifer. 1995. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 59:143-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ammerman, J. W., and F. Azam. 1981. Dissolved cyclic adenosine monophosphate (cAMP) in the sea and uptake of cAMP by marine bacteria. Mar. Ecol. Prog. Ser. 5:85-89. [Google Scholar]
- 4.Babenzien, H. D., and H. Sass. 1999. Desulfurikation, p. 435-444. In W. von Tümpling and G. Friedrich (ed.), Biologische Gewässeruntersuchung, vol. 2. Gustav Fischer Verlag, Jena, Germany. [Google Scholar]
- 5.Balch, W. E., G. E. Fox, L. J. Magrum, C. R. Woese, and R. S. Wolfe. 1979. Methanogens: reevaluation of a unique biological group. Microbiol. Rev. 43:260-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barer, M. R., and C. R. Harwood. 1999. Bacterial viability and culturability. Adv. Microb. Physiol. 41:93-137. [DOI] [PubMed] [Google Scholar]
- 7.Bartscht, K., H. Cypionka, and J. Overmann. 1999. Evaluation of cell activity and of methods for the cultivation of bacteria from a natural lake community. FEMS Microbiol. Ecol. 28:249-259. [Google Scholar]
- 8.Batchelor, S. E., M. Cooper, S. R. Chhabra, L. A. Glover, G. S. A. B. Stewart, P. Williams, and J. I. Prosser. 1997. Cell density-regulated recovery of starved biofilm populations of ammonia-oxidizing bacteria. Appl. Environ. Microbiol. 63:2281-2286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Botsford, J. L., and J. G. Harman. 1992. Cyclic AMP in prokaryotes. Microbiol. Rev. 56:100-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brewer, D. G., S. H. Martin, and Z. J. Ordal. 1977. Beneficial effects of catalase or pyruvate in a most-probable-number technique for the detection of Staphylococcus aureus. Appl. Environ. Microbiol. 34:797-800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Calcott, P. H., and J. R. Postgate. 1972. On substrate-accelerated death in Klebsiella aerogenes. J. Gen. Microbiol. 70:115-122. [DOI] [PubMed] [Google Scholar]
- 12.Coolen, M. J. L., and J. Overmann. 1998. Analysis of subfossil molecular remains of purple sulfur bacteria in a lake sediment. Appl. Environ. Microbiol. 64:4513-4521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eilers, H., J. Pernthaler, F. O. Glöckner, and R. Amann. 2000. Culturability and in situ abundance of pelagic bacteria from the North Sea. Appl. Environ. Microbiol. 66:3044-3051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Epstein, W., L. B. Rothman-Denes, and J. Hesse. 1975. Adenosine 3′:5′-cyclic monophosphate as mediator of catabolite repression in Escherichia coli. Proc. Natl. Acad. Sci. USA 72:2300-2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ferenci, T. 1996. Adaptation to life at micromolar nutrient levels: the regulation of Escherichia coli glucose transport by endoinduction and cAMP. FEMS Microbiol. Rev. 18:301-317. [DOI] [PubMed] [Google Scholar]
- 16.Ferris, M. J., A. L. Ruff-Roberts, E. D. Kopczynski, M. M. Bateson, and D. M. Ward. 1996. Enrichment culture and microscopy conceal diverse thermophilic Synechococcus populations in a single hot spring microbial mat habitat. Appl. Environ. Microbiol. 62:1045-1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fry, J. C. 1990. Direct methods and biomass estimation, p. 41-85. In R. Grigorova and J. R. Norris (ed.), Methods in microbiology, vol. 22. Academic Press, London, United Kingdom. [Google Scholar]
- 18.Garrity, G. M., M. Winters, and D. B. Searles. Release 1.0, April 2001, posting date. Taxonomic outline of the procaryotic genera—Bergey's manual of systematic bacteriology. Bergey's Manual Trust 2. [Online.] http://www.cme.msu.edu/bergeys/april2001-genus.pdf.
- 19.Gast, V., and K. Gocke. 1988. Vertical distribution of number, biomass and size-class spectrum of bacteria in relation to oxic/anoxic conditions in the central Baltic Sea. Mar. Ecol. Prog. Ser. 45:179-186. [Google Scholar]
- 20.Glöckner, F. O., B. M. Fuchs, and R. Amann. 1999. Bacterioplankton compositions of lakes and oceans: a first comparison based on fluorescence in situ hybridization. Appl. Environ. Microbiol. 65:3721-3726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guan, L. L., H. Onuki, and K. Kamino. 2000. Bacterial growth stimulation with exogenous siderophore and synthetic N-acyl homoserine lactone autoinducers under iron-limited and low-nutrient conditions. Appl. Environ. Microbiol. 66:2797-2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hansen, H.-P. 1999. Determination of oxygen, p. 75-89. In K. Grasshoff, K. Kremling, and M. Ehrhardt (ed.), Methods of seawater analysis. Wiley-VCH, Weinheim, Germany.
- 23.Hughes, P., A. Landoulsi, and M. Kohiyama. 1988. A novel role for cAMP in control of the activity of the E. coli chromosome replication initiator protein, DnaA. Cell 55:343-350. [DOI] [PubMed] [Google Scholar]
- 24.Jaspers, E., and J. Overmann. 1997. Separation of bacterial cells by isoelectric focusing, a new method for analysis of complex microbial communities. Appl. Environ. Microbiol. 63:3176-3181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jaspers, E., K. Nauhaus, H. Cypionka, and J. Overmann. 2001. Multitude and temporal variability of ecological niches as indicated by the diversity of cultivated bacterioplankton. FEMS Microbiol. Ecol. 36:153-164. [DOI] [PubMed] [Google Scholar]
- 26.Jones, J. G. 1979. A guide to methods for estimating microbial numbers and biomass in fresh water, vol. 39. The Freshwater Biological Association, Cumbria, United Kingdom.
- 27.Jukes, T. H., and C. R. Cantor. 1969. Evolution of protein molecules, p. 21-132. In H. N. Munro (ed.), Mammalian protein metabolism, vol. 3. Academic Press, New York, N.Y. [Google Scholar]
- 28.Kaplan, H., and E. Greenberg. 1985. Diffusion of autoinducer is involved in regulation of the Vibrio fischeri luminescence system. J. Bacteriol. 163:1210-1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Karner, M., and J. A. Fuhrman. 1997. Determination of active bacterioplankton: a comparison of universal 16S rRNA probes, autoradiography, and nucleoid staining. Appl. Environ. Microbiol. 63:1208-1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Klee, A. J. 1993. A computer program for the determination of most probable number and its confidence limits. J. Microbiol. Methods 18:91-98. [Google Scholar]
- 31.Kogure, K., U. Simidu, and N. Taga. 1979. A tentative direct microscopic method for counting living marine bacteria. Can. J. Microbiol. 25:415-420. [DOI] [PubMed] [Google Scholar]
- 32.Kumar, S., K. Tamura, I. B. Jakobsen, and M. Nei. 2001. MEGA2: molecular evolutionary genetics analysis software. Bioinformatics 17:1244-1245. [DOI] [PubMed] [Google Scholar]
- 33.Lange, R., and R. Hengge-Aronis. 1991. Identification of a central regulator of stationary-phase gene expression in Escherichia coli. Mol. Microbiol. 5:49-59. [DOI] [PubMed] [Google Scholar]
- 34.Lengeler, J. W., G. Drews, and H. G. Schlegel. 1999. Biology of prokaryotes. Georg Thieme Verlag, Stuttgart, Germany.
- 35.Matin, A., and M. K. Matin. 1982. Cellular levels, excretion, and synthesis rates of cyclic AMP in Escherichia coli grown in continuous culture. J. Bacteriol. 149:801-807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Messing, J. 1983. New M13 vectors for cloning. Methods Enzymol. 101:20-78. [DOI] [PubMed] [Google Scholar]
- 37.Muyzer, G., E. C. De Waal, and A. G. Uitterlinden. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59:695-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Muyzer, G., S. Hottenträger, A. Teske, and C. Waver. 1995. Denaturing gradient gel electrophoresis of PCR-amplified 16S rDNA—a new molecular approach to analyse the genetic diversity of mixed microbial communities, p. 3.4.4.1-3.4.4.22. In A. D. L. Akkermans, J. D. Van Elsas, and F. J. De Bruijn (ed.), Molecular microbial ecology manual. Kluwer, Dordrecht, The Netherlands.
- 39.Okada, T., K. Ueyama, S. Niya, H. Kanazawa, M. Futai, and T. Tsuchiya. 1981. Role of inducer exclusion in preferential utilization of glucose over melibiose in diauxic growth of Escherichia coli. J. Bacteriol. 146:1030-1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Overmann, J., M. J. L. Coolen, and C. Tuschak. 1999. Specific detection of different phylogenetic groups of chemocline bacteria based on PCR and denaturing gradient gel electrophoresis of 16S rRNA gene fragments. Arch. Microbiol. 172:83-94. [DOI] [PubMed] [Google Scholar]
- 41.Rehnstam, A.-S., S. Bäckman, D. C. Smith, F. Azam, and A. Hagström. 1993. Blooms of sequence-specific culturable bacteria in the sea. FEMS Microbiol. Ecol. 102:161-166. [Google Scholar]
- 42.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 43.Schultz, J. E., G. I. Latter, and A. Matin. 1988. Differential regulation by cyclic AMP of starvation protein synthesis in Escherichia coli. J. Bacteriol. 170:3903-3909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Straub, K. L., and B. E. E. Buchholz-Cleven. 1998. Enumeration and detection of anaerobic ferrous iron-oxidizing, nitrate-reducing bacteria from diverse European sediments. Appl. Environ. Microbiol. 64:4846-4856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Suzuki, M., M. S. Rappé, Z. W. Haimberger, H. Winfield, N. Adair, J. Ströbel, and S. J. Giovannoni. 1997. Bacterial diversity among small-subunit rRNA gene clones and cellular isolates from the same seawater sample. Appl. Environ. Microbiol. 63:983-989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Swift, S., M. J. Lynch, L. Fish, D. F. Kirke, J. M. Tomás, G. S. A. B. Stewart, and P. Williams. 1999. Quorum sensing-dependent regulation and blockade of exoprotease production in Aeromonas hydrophila. Infect. Immun. 67:5192-5199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Taddei, F., I. Matic, and M. Radman. 1995. cAMP-dependent SOS induction and mutagenesis in resting bacterial populations. Proc. Natl. Acad. Sci. USA 92:11736-11740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Teske, A., T. Brinkhoff, G. Muyzer, D. P. Moser, J. Rethmeier, and H. W. Jannasch. 2000. Diversity of thiosulfate-oxidizing bacteria from marine sediments and hydrothermal vents. Appl. Environ. Microbiol. 66:3125-3133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 24:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Torsvik, V., J. Goksøyr, F. L. Daae, R. Sørheim, J. Michalsen, and K. Salte. 1993. Diversity of microbial communities determined by DNA reassociation technique, p. 375-378. In R. G. C. Pedrós-Alió (ed.), Trends in microbial ecology. Spanish Society for Microbiology, Barcelona, Spain.
- 51.Walker, G. C. 1996. The SOS response of Escherichia coli, p. 1400-1416. In F. C. Neidhardt (ed.), Escherichia coli and Salmonella: cellular and molecular biology: 2nd ed., vol. 1. ASM Press, Washington, D.C. [Google Scholar]
- 52.Widdel, F., and F. Bak. 1992. Gram-negative mesophilic sulphate reducing bacteria, p. 3353-3378. In T. H. G. Balows, A. M. Dworkin, W. Harder, and K. H. Schleifer (ed.), The prokaryotes. Springer, New York, N.Y.