Abstract
The adzuki bean beetle, Callosobruchus chinensis, is infected with three distinct lineages of endosymbiotic bacteria belonging to the genus Wolbachia, which were designated wBruCon, wBruOri, and wBruAus. In an attempt to understand the mechanisms underlying the infection with these three organisms, the spatiotemporal infection dynamics of the three Wolbachia strains was investigated in detail by using a quantitative PCR technique. During the development of C. chinensis, the wBruCon, wBruOri, and wBruAus infection levels consistently increased but the growth patterns were different. The levels of infection plateaued at the pupal stage at approximately 3 × 108, 2 × 108, and 5 × 107 wsp copy equivalents per insect for wBruCon, wBruOri, and wBruAus, respectively. At the whole-insect level, the population densities of the three Wolbachia types did not show remarkable differences between adult males and females. At the tissue level, however, the total densities and relative levels of the three Wolbachia types varied significantly when different tissues and organs were compared and when the same tissues derived from males and females were compared. The histological data obtained by in situ hybridization and electron microscopy were concordant with the results of quantitative PCR analyses. Based on the histological data and the peculiar Wolbachia composition commonly found in nurse tissues and oocytes, we suggest that the Wolbachia strains are vertically transmitted to oocytes not directly, but by way of nurse tissue. On the basis of our results, we discuss interactions among the three coinfecting Wolbachia types, reproductive strategies of Wolbachia, and factors involved in the different cytoplasmic incompatibility phenotypes.
Members of the genus Wolbachia constitute a group of rickettsial endosymbiotic bacteria in the α-subdivision of the Proteobacteria. Infection with Wolbachia is found frequently in insects (39, 40, 41) and less frequently in mites (8), spiders (28), crustaceans (6), and nematodes (2). It has been shown that Wolbachia infection causes reproductive alterations in the arthropod hosts, such as cytoplasmic incompatibility (CI), parthenogenesis, feminization, and male killing (for a review see reference 29). Because Wolbachia is inherited solely through the maternal lineage of the host by transovarial transmission, these reproductive changes can increase the frequency of infected females in host populations, often at the expense of host fitness (for a review see reference 38).
Since Wolbachia strains are transovarially transmitted through the hosts and manipulate host reproduction, they are expected to be associated with germ line tissues of the host organisms. Hence, much of the research on Wolbachia infections has been directed toward host reproductive tissues. In contrast, descriptions of Wolbachia interactions with nonreproductive tissues are primarily fragmentary (3, 25, 32, 42). Recently, however, it has been recognized that Wolbachia infections are also found in a variety of somatic tissues in different insects (13, 14, 23, 26). To understand the relationships between Wolbachia infection and the phenotypic effects of Wolbachia on a host, it is necessary to understand Wolbachia tissue tropism and infection densities in different host tissues. Several studies have suggested that the density of Wolbachia in a host is positively correlated with the intensity of CI expression (7, 9, 10, 27). In the “popcorn” Wolbachia infection in Drosophila melanogaster, extensive infection of somatic tissues of adult insects resulted in a significant reduction in the life span of the insects (26). In these studies, however, the density of Wolbachia was estimated only for the whole body (7, 27), for eggs (9), or for sperm cysts (10) or was assessed qualitatively (26). No study has quantitatively estimated the levels of Wolbachia infection in different tissues and organs of the same host. Throughout growth and development of a host insect, the density and localization of Wolbachia in various tissues and organs may exhibit dynamic and highly controlled changes. However, spatial and temporal infection dynamics of Wolbachia have not been described in any host system.
The adzuki bean beetle, Callosobruchus chinensis (Coleoptera: Bruchidae), is a pest of stored adzuki bean, Vigna angularis. In previous studies (23, 24), it was demonstrated that infection with three phylogenetically distinct strains of Wolbachia, designated wBruCon, wBruOri, and wBruAus, is widespread in Japanese populations of C. chinensis. Interestingly, the three Wolbachia strains caused different levels of CI; wBruCon caused complete CI, wBruOri caused partial CI, and wBruAus caused no or very weak CI. In addition, notably, the densities of the three Wolbachia strains in adult insects were consistently different; the density of wBruOri was 107 to 108 wsp copy equivalents per insect, the density of wBruCon was 107 to 108 wsp copy equivalents per insect, and the density of wBruAus was 106 to 107 wsp copy equivalents per insect. These results suggest that there are potential interactions among the three Wolbachia strains, including competition for space and resources, differential tissue tropism, and various infection dynamics during host development.
To further understand these aspects of Wolbachia superinfection in C. chinensis, we investigated the spatiotemporal infection dynamics of the three Wolbachia types during host development by using a quantitative PCR technique and other molecular and histological methods.
MATERIALS AND METHODS
Materials.
A long-established laboratory strain of C. chinensis, strain jC, was used in this study. The insects were reared in petri dishes filled with adzuki beans at 30°C and a relative humidity of 70% by using a long-day regimen (16 h of light and 8 h of darkness). The materials used in this study are available upon request from the corresponding author.
Definition of developmental stages of C. chinensis.
To collect strictly staged eggs of C. chinensis, mated females were allowed to oviposit on adzuki beans in petri dishes for 1 h. The eggs were harvested 1, 6, 12, 18, 24, 48, 72, and 96 h after the start of oviposition. To collect first-instar larvae, we gently detached and collected eggs from the substratum and allowed them to hatch in a petri dish. To collect older larvae and pupae, infested beans were broken with forceps under a dissecting microscope. Based on size, second-, third-, and fourth-instar larvae were separated. Pupae that were white were considered early pupae, whereas pupae that displayed the coloration of the adult cuticle were considered late pupae. Adult insects were collected within 1 day after emergence.
Preparation of samples for DNA analysis.
To monitor the infection dynamics of the three Wolbachia types during C. chinensis development, strictly staged eggs, larvae, pupae, and adults were collected and immediately preserved in acetone (17). Samples were fixed and dehydrated in acetone, dried in air, weighed individually, and subjected to DNA extraction. To investigate the titers of the three Wolbachia strains in tissues and organs of C. chinensis, fat bodies, Malpighian tubules, guts, nurse tissues, oocytes, and testes were isolated from adult insects. Individual insects were carefully dissected with fine forceps under a dissecting microscope in a petri dish covered with silicon rubber and filled with phosphate-buffered saline containing Tween 20 (PBT) (1.9 mM NaH2PO4, 8.1 mM Na2HPO4, 175 mM NaCl, 0.1% Tween 20 [pH 7.4]). Isolated tissues and organs were individually washed several times with fresh PBT to minimize possible microbial contamination and immediately subjected to DNA extraction.
DNA extraction.
A sample was placed in a 1.5-ml plastic tube, combined with 200 μl of lysis buffer containing sodium dodecyl sulfate and proteinase K, homogenized with a plastic pestle, incubated at 55°C for 3 h or longer, and subjected to DNA extraction with a QIAamp tissue kit (QIAGEN). Total DNA of a sample was eluted with 200 μl of TE buffer (10 mM Tris-HCl [pH 8.0], 0.1 mM EDTA).
PCR detection.
Collective PCR detection of Wolbachia was conducted by using two sets of universal primers for the ftsZ and wsp genes as previously described (24). A 0.75-kb ftsZ segment was amplified by using primers ftsF and ftsR, and a 0.6-kb wsp fragment was amplified by using primers wspF and wspR. Specific PCR detection of wBruCon, wBruOri, and wBruAus was performed by using specific reverse PCR primers wspConR, wspOriR, and wspAusR in combination with universal forward primer wspF, which produced an approximately 0.4-kb wsp gene segment (24). Three plasmids, one containing the wsp gene of wBruCon, one containing the wsp gene of wBruOri, and one containing the wsp gene of wBruAus, were used as negative and positive control samples for specific detection. The PCR products were electrophoresed in agarose gels, stained with ethidium bromide, and observed on a UV transilluminator.
Quantitative PCR.
Real-time fluorescence detection quantitative PCR was performed by using the TaqMan PCR and ABI Prism 7700 sequence detection system (PE Applied Biosystems) as previously described (24). To estimate the titers of the three Wolbachia strains, the copy number of the wsp gene was quantified. A double-fluorescence-labeled probe for detection, TQwspPRB, was targeted to a conserved region of the three types of wsp sequences. The following highly specific amplifying primers were designed for variable flanking regions between the three sequences: TQwspCF and TQwspCR, TQwspOF and TQwspOR, and TQwspAF and TQwspAR (24). The probe and these primers were designed by using the Primer Express 1.0 program package (PE Applied Biosystems). Standard curves were drawn by using standard plasmid samples that contained the different wsp genes at concentrations of 102, 103, 104, 105, 106, and 107 copies/μl. To estimate the amount of a host gene, the mitochondrial cytochrome oxidase II (COII) gene was quantified. Quantification was conducted by using a probe, TQAzkCIIPrb (5′-FAM-ATGCAACCCCGGGTCGTCTTAA-TAMRA-3′), and primers TQAzkCIIF (5′-CTTGAACTATCCCATCTCTAGGAGT-3′) and TQAzkCIIR (5′-AACCAGCTCGATTAATAATAAATCTG-3′).
Whole-mount in situ hybridization.
All incubations were carried out in 1.5-ml plastic tubes on a shaker. Dissected tissues and organs from adult insects were fixed with 4% paraformaldehyde in PBT for 2 h. After samples were washed twice with PBT for 5 min, they were dehydrated with a water-ethanol series (25, 50, 75, and 100% ethanol in PBT, 5 min for each step). Then the samples were rehydrated with an ethanol-water series (100, 75, 50, and 25% ethanol in PBT, 5 min for each step), washed twice with PBT for 5 min, and treated with 6 μg of proteinase K per ml in PBT for 3 min at 37°C to improve the infiltration of probe and reagents. After incubation with 2 mg of glycine per ml in PBT for 3 min to inhibit proteinase activity, the samples were washed with PBT for 5 min, fixed with 0.2% glutaraldehyde-4% paraformaldehyde in PBT for 20 min, and washed twice with PBT for 3 min. Then PBT was replaced with hybridization buffer (HB) (20 mM Tris-HCl [pH 8.0], 0.9% NaCl, 0.01% sodium dodecyl sulfate, 30% formamide) by incubation three times (10 min each). A digoxigenin-labeled oligonucleotide probe, DIG-Wol16S49 (5′-digoxigenin-ACCATAGCAAGCTACAAT-3′), was designed in this study for specific detection of Wolbachia. Note that this probe recognizes wBruCon, wBruOri, and wBruAus. A universal probe for eubacteria, DIG-EUB338 (5′-digoxigenin-GCTGCCTCCCGTAGGAGT-3′) (1), was also used as a positive control probe. Probe DIG-Wol16S49cmp (5′-digoxigenin-ATTGTAGCTTGCTATGGT-3′), which is complementary to the specific probe DIG-Wol16S49, was used for negative control experiments. A reaction with no probe was also conducted as a negative control. The samples were hybridized with 66 pmol of probe per ml in HB overnight. Detection of the bound probe was performed with a DIG nucleic acid detection kit (Boehringer Mannheim).
Electron microscopy.
Nurse tissues and Malpighian tubules were dissected from adult females under a dissecting microscope in 2.5% glutaraldehyde in 0.1 M phosphate buffer (PB) (19 mM NaH2PO4, 81 mM Na2HPO4; pH 7.4), prefixed with the fixative at 4°C overnight, postfixed with 2% osmium tetroxide in 0.1 M PB at 4°C for 60 min, and subjected to block staining with 0.5% uranyl acetate for 1 h. The tissues were dehydrated with an ethanol series and embedded in Epon 812. Ultrathin sections were cut with a ultramicrotome (Leichert-Nissei Urtracut-N), mounted on collodion-coated copper mesh, stained with uranyl acetate and lead citrate, and observed with a transmission electron microscope (Hitachi H-7000) at 75 kV.
RESULTS
Infection dynamics of the three Wolbachia strains during development.
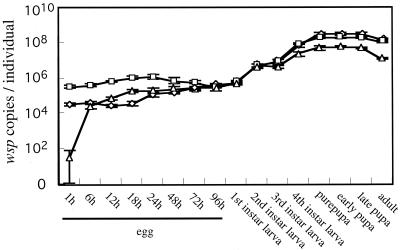
The populations of wBruCon, wBruOri, and wBruAus were monitored throughout development of C. chinensis by using a quantitative PCR technique. Figure 1 shows the infection dynamics in terms of wsp copies per insect. The populations of wBruCon, wBruOri, and wBruAus consistently increased as development of the host proceeded. Notably, however, the three Wolbachia strains exhibited different population growth patterns. In early eggs, wBruOri was the most abundant (around 4 × 105 wsp copy equivalents per insect), the amount of wBruCon was intermediate (around 4 × 104 wsp copy equivalents per insect), and wBruAus was the least abundant (around 1 × 102 wsp copy equivalents per insect). In late eggs and first-instar larvae, the sizes of the populations of wBruCon, wBruOri, and wBruAus were almost the same, around 6 × 105 wsp copy equivalents per insect. At the second-instar and later stages, the sizes of the populations of wBruCon and wBruOri were comparable whereas the populations of wBruAus were consistently and significantly smaller. In prepupae and pupae, the populations of the Wolbachia strains reached plateaus; the concentrations of wBruCon, wBruOri, and wBruAus were around 3 × 108, 2 × 108, and 5 × 107 wsp copy equivalents per insect, respectively.
FIG. 1.
Dynamics of wBruCon, wBruOri, and wBruAus infection during the development of C. chinensis, expressed in terms of number of wsp copies per insect. Symbols: ⋄, wBruCon; □, wBruOri; ▵, wBruAus. The error bars indicate standard deviations (n = 10). The pupae and adults were females. Sex could not be distinguished at the other stages.
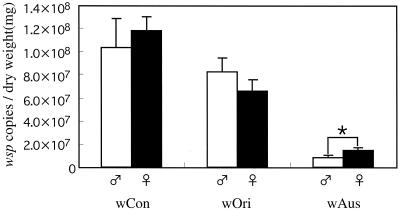
Infection densities of the three Wolbachia strains in adult males and females.
Figure 2 shows a comparison of the infection densities of wBruCon, wBruOri, and wBruAus in adult males and females of C. chinensis. In general, the difference in the densities between the sexes was not remarkable for each of the Wolbachia strains. However, the difference was statistically significant for wBruAus.
FIG. 2.
Comparison of the population densities of wBruCon (wCon), wBruOri (wOri), and wBruAus (wAus) in females and males of newly emerged adults of C. chinensis, expressed in terms of number of wsp copies per milligram (dry weight) of insect. The error bars indicate standard deviations (n = 10). The asterisk indicates a statistically significant difference (Mann-Whitney U test, P < 0.01).
Densities of the three Wolbachia strains in tissues and organs.
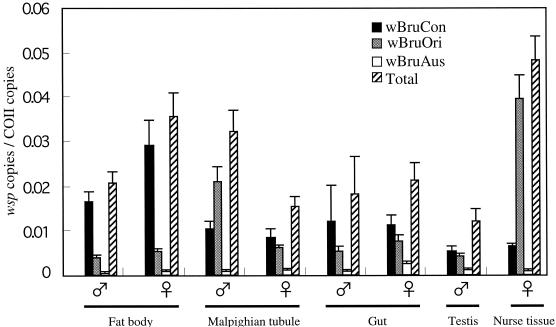
To quantitatively investigate the localization of wBruCon, wBruOri, and wBruAus in C. chinensis tissues, dissected tissues and organs from adult insects were subjected to quantitative PCR analysis. In order to compare different tissues and organs, the wsp gene copy number of the Wolbachia strains was standardized by using the copy number of a host gene, mitochondrial COII. It was found that both the density and the composition of the three Wolbachia strains were specific for tissues and organs of C. chinensis.
The total densities of the three Wolbachia strains differed significantly in different tissues and organs (Fig. 3). For the tissues and organs examined, for example, higher levels of Wolbachia infection were detected in the nurse tissue and fat body than in the gut and testis. The total densities of the Wolbachia strains also showed sex-related differences. For example, in the fat bodies the Wolbachia density was higher in females than in males, while in Malpighian tubules the Wolbachia density was lower in females than in males.
FIG. 3.
Infection densities of wBruCon, wBruOri, and wBruAus in different tissues and organs of C. chinensis expressed in terms of number of wsp copies per COII copy. The mean and standard deviation for five samples are shown for each tissue or organ. Newly emerged adults were subjected to the analysis.
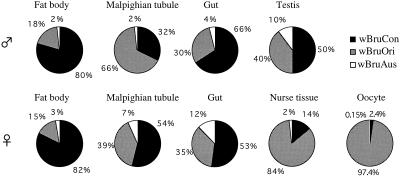
The compositions of the three Wolbachia strains were also significantly different in different tissues and organs (Fig. 4). In fat bodies, wBruCon accounted for around 80% of the total Wolbachia population, wBruOri accounted for about 15 to 18%, and wBruAus accounted for a small fraction. In guts and testes, wBruCon comprised about 50 to 70% of the total Wolbachia population, wBruOri comprised around 30 to 40%, and wBruAus comprised around 10% or less. In Malpighian tubules, a sex-related difference was detected; in males wBruOri was the predominant strain, whereas in females wBruCon was the major component. In nurse tissues and oocytes, strikingly, wBruOri accounted for more than 80% of the total Wolbachia population, whereas wBruCon and wBruAus accounted for only small fractions.
FIG. 4.
Relative amounts of wBruCon, wBruOri, and wBruAus in different tissues and organs of C. chinensis. The mean values in Fig. 3 were used to construct the pie graphs.
In situ hybridization and electron microscopy of Wolbachia.
To directly observe Wolbachia in tissues and organs of C. chinensis, histological analyses were performed by using in situ hybridization and electron microscopy. In these experiments, the three Wolbachia strains were visualized collectively, not differentially.
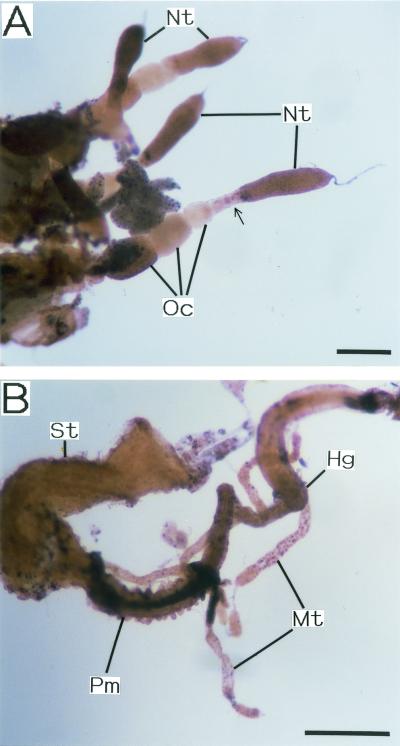
Figure 5 shows the results of whole-mount in situ hybridization of dissected internal organs when an oligonucleotide probe targeted to Wolbachia 16S rRNA was used. Wolbachia signals were detected in all the tissues and organs examined, although the intensity and mode of staining were not uniform. In female ovarioles, nurse tissues and maturing oocytes exhibited particularly dense signals, while very young oocytes were stained only faintly (Fig. 5A). At the junction of nurse tissues and young oocytes, Wolbachia signals were observed frequently, as if the Wolbachia cells were transported between these tissues (Fig. 5A). Malpighian tubules were not uniformly stained but exhibited a dotted staining pattern (Fig. 5B). Although the posterior midgut exhibited particularly deep staining, the signal was due to endogenous alkaline phosphatase activity (Fig. 5B). In negative control experiments (i.e., complementary probe control and control containing no probe), most of these signals were not detected; the only exception was the signal in the posterior midgut (data not shown). In positive control experiments performed with a universal eubacterial probe, similar staining patterns were observed (data not shown).
FIG. 5.
Whole-mount in situ hybridization of dissected tissues and organs of C. chinensis performed with oligonucleotide probe DIG-Wol16S49 targeted to 16S rRNA of Wolbachia. (A) Ovarioles from an adult female. (B) Gut and Malpighian tubules from an adult male. Bars = 0.5 mm. Abbreviations: Hg, hindgut; Mt, Malpighian tubule; Nt, nurse tissue; Oc, oocyte; Pm, posterior midgut; St, stomach. The arrow indicates the signal of Wolbachia presumably in the process of vertical transmission from nurse tissue to oocyte.
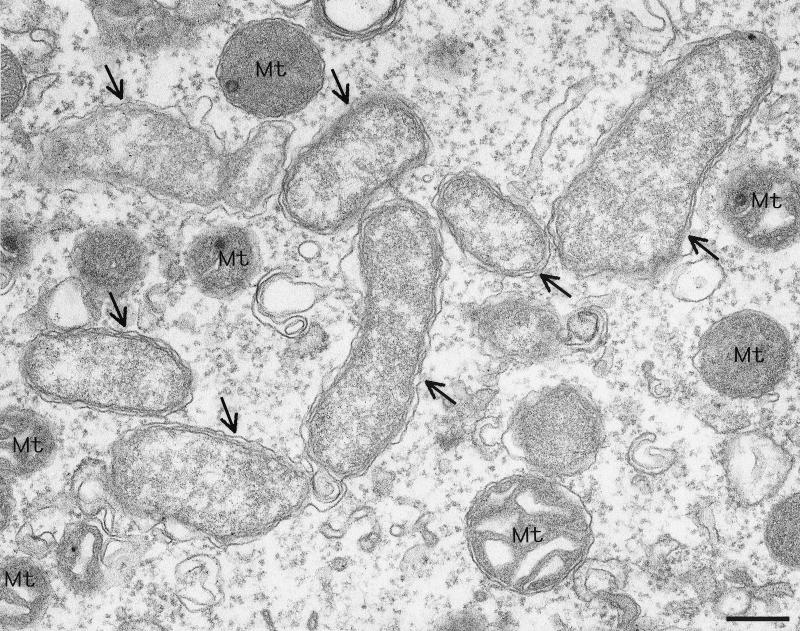
Figure 6 is an electron microscopic image of a nurse tissue cell. A number of bacterial cells whose morphology was typical of Wolbachia cells were observed intracellularly. The same bacteria were also found in the cells of Malpighian tubules (data not shown).
FIG. 6.
Electron micrograph of Wolbachia in nurse tissue of C. chinensis. The arrows indicate Wolbachia cells. Bar = 0.2 μm. Mt, mitochondrion.
DISCUSSION
This study was the first detailed quantitative analysis of the spatiotemporal infection dynamics of Wolbachia endosymbionts during development of an insect host. The real-time fluorescence detection quantitative PCR technique allowed us to perform reliable, robust, and efficient quantification of the Wolbachia strains in DNA samples extracted from individual insects or tissues in terms of wsp gene copy number. The insect which we examined, C. chinensis, was shown to be stably infected with three strains of Wolbachia, wBruCon, wBruOri, and wBruAus, which are phylogenetically distinct from each other and exhibit different CI phenotypes (24). From this study, therefore, it was expected that we would be able to gain insight into interactions among the three Wolbachia strains in the same host body, the reproductive strategies which the three Wolbachia strains adopt for survival and transmission, and factors that are involved in the different CI phenotypes.
Monitoring the populations of wBruCon, wBruOri, and wBruAus throughout the development of C. chinensis showed that the three Wolbachia strains exhibited different population growth patterns (Fig. 1). As a consequence, the relative amounts of the three Wolbachia strains changed as development of the host proceeded. In eggs wBruOri was the predominant strain, whereas in old larvae, pupae, and adults wBruCon constituted the majority. At all stages, the amount of wBruAus was the smallest. These results suggested that the three Wolbachia strains might be subject to differential control of proliferation during development of the host.
By using tissue-specific quantitative PCR analyses, we demonstrated that there are interesting patterns of differential tissue tropism of wBruCon, wBruOri, and wBruAus in the same host (Fig. 3 and 4). Although the mechanism is unknown, there are two possible components that may contribute to the tissue-dependent differences in density and composition of these three Wolbachia strains. One component is tissue tropism inherent in the Wolbachia strains. The other component is interactions between coinfecting Wolbachia strains in the same tissue. These two components could be sorted out by examining the density and localization of the Wolbachia strains in singly infected and doubly infected lines of C. chinensis.
The results of the whole-mount in situ hybridization analysis performed with a probe targeted to 16S rRNA of Wolbachia (Fig. 5) were concordant with the results of the tissue-specific quantitative PCR analyses (Fig. 3 and 4). All the tissues and organs examined were Wolbachia positive, although the intensities of the staining revealed differences among them. Nurse tissue, whose Wolbachia content was estimated to be high, was stained deeply. Malpighian tubules were not uniformly stained but exhibited a dotted staining pattern, which suggests that the Wolbachia strains do not infect all cells uniformly but preferentially infect specific cells that are distributed rather sparsely in this tissue. The cytological identity of the infected cells is unknown. In the in situ hybridization analysis, wBruCon, wBruOri, and wBruAus were collectively detected by using a Wolbachia-specific probe. To investigate the localization of the three Wolbachia strains in greater histological detail, it is necessary to develop highly specific probes for each of them.
At the level of the whole body, there were no remarkable differences in the densities and compositions of the populations of the three Wolbachia strains between males and females (Fig. 2). At the level of tissues and organs, in contrast, significant sex-related differences were identified (Fig. 3 and 4). The densities and compositions of the three Wolbachia strains were markedly different in testes and oocytes (Fig. 4), which might be relevant to the different CI and vertical transmission phenotypes. In several somatic tissues, such as Malpighian tubules and fat bodies, the densities and compositions of the populations of the three Wolbachia strains also were different in the two sexes (Fig. 3 and 4), although the biological significance of the differences is not obvious.
In C. chinensis, the three Wolbachia strains caused different levels of CI; wBruCon caused complete CI, wBruOri caused partial CI, and wBruAus caused no or very weak CI (24). The expression and intensity of Wolbachia-induced CI are thought to be affected by the following factors: (i) the CI-inducing mechanism inherent in the Wolbachia strain (7, 18, 30, 33, 34); (ii) the density of the Wolbachia strain in the host (7, 9, 10, 27, 36); and (iii) the genetic background of the host (5, 7, 31). One of these factors, the effect of the host genetic background, can be disregarded in this study because of the superinfection of the same host used. The effect of Wolbachia density in the host may explain the absence of detectable CI with wBruAus, because the density of wBruAus was consistently and significantly less than that of wBruCon and that of wBruOri. On the other hand, the infection densities of wBruCon and wBruOri were largely comparable throughout the development of the host. Therefore, the differences in CI phenotype between wBruCon and wBruOri cannot simply be attributed to the density effect.
Although molecular mechanisms of CI are not understood, the modification-rescue model has been proposed to explain the phenomena observed in Wolbachia-induced CI (19, 20, 37). According to this model, it is expected that Wolbachia cells infecting testes are involved in the modification process and Wolbachia cells that accumulate in oocytes are responsible for the rescue process. In this study, we quantified the bacterial titer and composition of the CI-inducing Wolbachia strains, wBruCon and wBruOri, in testes and oocytes (Fig. 3 and 4). In testes, the proportions of wBruCon and wBruOri were relatively high, 50 and 40%, suggesting that these two Wolbachia types have equal potential access to modify the sperm during spermatogenesis. In oocytes, interestingly, wBruCon accounted for only 2.4% of the Wolbachia population, whereas wBruOri accounted for 97.4%. The complete CI and rescue caused by wBruCon (24) suggest that the small amount of wBruCon in oocytes can efficiently rescue the modified sperm. In contrast, the complete rescue of wBruOri-modified sperm (24) may be explained by the high infection titer of wBruOri in oocytes. The partial CI caused by wBruOri may be due to the weak modification ability of this strain.
In general, intracellular symbiotic bacteria of arthropods, including Wolbachia, are inherited by transovarial transmission to developing oocytes in the ovarioles in the maternal body. Histologically, it has been observed that such symbionts either actively find their way to eggs or are transferred there by specialized cells or via other structures (11, 21). However, the detailed processes of vertical transmission for Wolbachia have been poorly described. In this study, of the tissues examined, only nurse tissue and oocytes exhibited a peculiar Wolbachia composition in which wBruOri was highly predominant (Fig. 4). In the process of insect egg formation, proteins, lipids, RNAs, and other cytoplasmic components are actively synthesized by nurse cells and are transported to developing oocytes through nutritive cord with trophic flow (4, 16). One plausible explanation for the similar Wolbachia compositions of nurse tissue and oocytes is that the Wolbachia first infect and proliferate in the nurse tissue and are subsequently transported to oocytes by the nutritive cord. This idea was supported by the in situ hybridization results, which showed that Wolbachia cells were present in the nutritive cord, presumably being transported (Fig. 5A). The peculiar Wolbachia composition in nurse tissue and oocytes also suggests that wBruOri may have some adaptation for transmission to and proliferation in the female germ tissues.
In natural populations of C. chinensis, wBruCon, wBruOri, and wBruAus exhibited very high infection frequencies (24). How the three different Wolbachia strains are stably maintained in the host populations is an interesting problem. Considering that they showed remarkable differences in CI expression, tissue tropism, and infection dynamics during host development, the three Wolbachia strains may have adopted different strategies for survival, reproduction, and transmission. Theoretically, the following strategies can favor maintenance of maternally inherited endosymbionts in host populations: (i) efficient vertical transmission, (ii) positive fitness effect on the host, (iii) reproductive manipulation, such as CI, and (iv) mechanism for horizontal transmission (12, 15, 35, 36). Since wBruCon caused complete CI and its bacterial titer in eggs was small, maintenance of wBruCon may principally be realized through an efficient mechanism for CI. In contrast, wBruOri caused partial CI, and its bacterial titer in eggs was very high, suggesting that maintenance of wBruOri may be mainly attributed to a sophisticated mechanism for vertical transmission. How wBruAus, which caused no CI and accounted for a very small fraction of the bacteria in eggs, can be stably maintained in host populations is puzzling. At present, we have no data for fitness effects and horizontal transmission of the three Wolbachia strains. Therefore, future examination of these aspects of the Wolbachia infection in C. chinensis is needed.
Using the quantitative PCR technique, we successfully quantified the three Wolbachia strains in the same DNA samples from individual insects or tissues in terms of wsp gene copy number. Since Wolbachia strains are difficult to culture, gene copy number is a reliable and useful index of bacterial amount. It should be noted, however, that gene copy number is not always in agreement with microbial cell number obtained by standard quantification methods, such as CFU determination or direct cell counting. There may be multiple copies of some target genes in the genome (for example, 16S ribosomal DNA in many bacteria). In Buchnera, an essential endosymbiont of aphids, the number of copies of whole genomic DNA per bacterial cell is amplified more than 100 times (22). When quantitative PCR is used, DNA copies derived from dead Wolbachia cells and cell-free exogenous DNA present in the host are also counted. Considering these factors, we emphasize that the wsp copy number obtained in this study may not strictly reflect the real number of Wolbachia cells. To assess the Wolbachia densities in tissues and organs, the copy number of a mitochondrial gene, the COII gene, was quantified to normalize wsp copy number. However, we recognized the possibility that mitochondrial densities might be different in different tissues and organs and might be affected by the physiological condition of the host.
Acknowledgments
We thank A. Sugimura, S. Kumagai, and K. Sato for technical and secretarial assistance and S. Dobson for reading the manuscript.
This research was supported by the Industrial Science and Technology Frontier program Technological Development of Biological Resources in Bioconsortia of the Ministry of International Trade and Industry of Japan and by the Program for Promotion of Basic Research Activities for Innovation Biosciences (ProBRAIN) of the Bio-Oriented Technology Research Advancement Institution.
REFERENCES
- 1.Amann, R. I., L. Krumholz, and D. A. Stahl. 1990. Fluorescent-oligonucleotide probing of whole cells for determinative, phylogenetic, and environmental studies in microbiology. J. Bacteriol. 172:762-770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bandi, C., T. J. C. Anderson, C. Genchi, and M. L. Blaxter. 1998. Phylogeny of Wolbachia in filarial nematodes. Proc. R. Soc. Lond. Ser. B 265:2407-2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Binnington, K. C., and A. A. Hoffmann. 1989. Wolbachia-like organisms and cytoplasmic incompatibility in Drosophila simulans. J. Invertebr. Pathol. 54:344-352. [Google Scholar]
- 4.Blackman, R. L. 1987. Reproduction, cytogenetics and development, p. 163-195. In A. K. Minks and P. Harrewijn (ed.), Aphids: their biology, natural enemies and control, vol. 2A. Elsevier, Amsterdam, The Netherlands. [Google Scholar]
- 5.Bordenstein, S. R., and J. H. Werren. 1998. Effects of A and B Wolbachia and host genotype on interspecies cytoplasmic incompatibility in Nasonia. Genetics 148:1833-1844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bouchon, D., T. Rigaud, and P. Juchault. 1998. Evidence for widespread Wolbachia infection in isopod crustaceans: molecular identification and host feminization. Proc. R. Soc. Lond. Ser. B 265:1081-1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bourtzis, K., A. Nirgianaki, G. Markakis, and C. Savakis. 1996. Wolbachia infection and cytoplasmic incompatibility in Drosophila species. Genetics 144:1063-1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Breeuwer, J. A. J., and G. Jacobs. 1996. Wolbachia: intracellular manipulators of mite reproduction. Exp. Appl. Acarol. 20:421-434. [DOI] [PubMed] [Google Scholar]
- 9.Breeuwer, J. A. J., and J. H. Werren. 1993. Cytoplasmic incompatibility and bacterial density in Nasonia vitripennis. Genetics 135:565-574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bressac, C., and F. Rousset. 1993. The reproductive incompatibility system in Drosophila simulans: DAPI-staining analysis of the Wolbachia symbionts in sperm cysts. J. Invertebr. Pathol. 61:226-230. [DOI] [PubMed] [Google Scholar]
- 11.Buchner, P. 1965. Endosymbiosis of animals with plant microorganisms. Interscience, New York, N.Y.
- 12.Caspari, E., and G. S. Watson. 1959. On the evolutionary importance of cytoplasmic sterility in mosquitoes. Evolution 13:568-570. [Google Scholar]
- 13.Cheng, Q., T. D. Ruel, W. Zhou, S. K. Moloo, P. Majiwa, S. L. O'Neill, and S. Aksoy. 2000. Tissue distribution and prevalence of Wolbachia infections in tsetse flies, Glossina spp. Med. Vet. Entomol. 14:44-50. [DOI] [PubMed] [Google Scholar]
- 14.Dobson, S. L., K. Bourtzis, H. R. Braig, B. F. Jones, W. Zhou, F. Rousset, and S. L. O'Neill. 1999. Wolbachia infections are distributed throughout insect somatic and germ line tissues. Insect Biochem. Mol. Biol. 29:153-160. [DOI] [PubMed] [Google Scholar]
- 15.Fine, P. E. 1978. On the dynamics of symbiote-dependent cytoplasmic incompatibility in culicine mosquitoes. J. Invertebr. Pathol. 30:10-18. [DOI] [PubMed] [Google Scholar]
- 16.Foldi, I. 1990. Internal anatomy, p. 65-84. In D. Rosen (ed.), Armored scale insects: their biology, natural enemies and control, vol. 4A. Elsevier, Amsterdam, The Netherlands. [Google Scholar]
- 17.Fukatsu, T. 1999. Acetone preservation: a practical technique for molecular analysis. Mol. Ecol. 8:1935-1945. [DOI] [PubMed] [Google Scholar]
- 18.Giordano, R., S. L. O'Neill, and H. M. Robertson. 1995. Wolbachia infections and the expression of cytoplasmic incompatibility in Drosophila sechellia and D. mauritiana. Genetics 140:1307-1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hoffmann, A. A., and M. Turelli. 1997. Cytoplasmic incompatibility in insects, p. 42-80. In S. L. O'Neill, A. A. Hoffman, and J. H. Werren (ed.), Influential passengers: inherited microorganisms and arthropod reproduction. Oxford University Press, Oxford, United Kingdom.
- 20.Hurst, L. D., and G. T. McVean. 1996. Clade selection, reversible evolution and the persistence of selfish elements: the evolutionary dynamics of cytoplasmic incompatibility. Proc. R. Soc. Lond. Ser. B 263:97-104. [Google Scholar]
- 21.Koch, A. 1967. Insects and their endosymbionts, p. 1-106. In S. M. Henry (ed.), Symbiosis, vol. II. Academic Press, New York, N.Y. [Google Scholar]
- 22.Komaki, K., and H. Ishikawa. 1999. Intracellular bacterial symbionts of aphids possess many genomic copies per bacterium. J. Mol. Evol. 48:717-722. [DOI] [PubMed] [Google Scholar]
- 23.Kondo, N., M. Shimada, and T. Fukatsu. 1999. High prevalence of Wolbachia in the azuki bean beetle Callosobruchus chinensis (Coleoptera, Bruchidae). Zool. Sci. 16:955-962. [Google Scholar]
- 24.Kondo, N., N. Ijichi, M. Shimada, and T. Fukatsu. 2002. Prevailing triple infection with Wolbachia in Callosobruchus chinensis (Coleoptera: Bruchidae). Mol. Ecol. 11:167-180. [DOI] [PubMed] [Google Scholar]
- 25.Louis, C., and L. Nigro. 1989. Ultrastructural evidence of Wolbachia rickettsiales in Drosophila simulans and their relationships with unidirectional cross-incompatibility. J. Invertebr. Pathol. 54:399-407. [Google Scholar]
- 26.Min, K. T., and S. Benzer. 1997. Wolbachia, normally a symbiont of Drosophila, can be virulent, causing degeneration and death. Proc. Natl. Acad. Sci. USA 94:10792-10796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Noda, H., Y. Koizumi, Q. Zhang, and K. Deng. 2001. Infection density of Wolbachia and incompatibility level in two planthopper species, Laodelphax striatellus and Sogatella furcifera. Insect Biochem. Mol. Biol. 31:727-737. [DOI] [PubMed] [Google Scholar]
- 28.Oh, H. W., M. G. Kim, S. W. Shin, K. S. Bae, Y. J. Ahn, and H. Y. Park. 2000. Ultrastructural and molecular identification of a Wolbachia endosymbiont in a spider, Nephila clavata. Insect Mol. Biol. 9:539-543. [DOI] [PubMed] [Google Scholar]
- 29.O'Neill, S. L., A. A. Hoffmann, and J. H. Werren (ed.). 1997. Influential passengers: inherited microorganisms and arthropod reproduction. Oxford University Press, Oxford, United Kingdom.
- 30.Perrot-Minnot, M. J., L. R. Guo, and J. H. Werren. 1996. Single and double infections with Wolbachia in the parasitic wasp Nasonia vitripenis: effects on compatibility. Genetics 143:961-972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Poinsot, D., K, Bourtzis, G., Markakis, C., Savakis, and H. Merçot. 1998. Wolbachia transfer from Drosophila melanogaster into D. simulans: host effect and cytoplasmic incompatibility relationships. Genetics 150:227-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rigaud, T., C. Souty-Grosset, R. Raimond, J. Mocquard, and P. Juchault. 1991. Feminizing endocytobiosis in the terrestrial crustacean Armadillidium vulgare Latr. (Isopoda): recent acquisitions. Endocytobiosis Cell Res. 7:259-273. [Google Scholar]
- 33.Rousset, R., H. R. Braig, and S. L. O'Neill. 1999. A stable triple Wolbachia infection in Drosophila with nearly additive incompatibility effects. Heredity 82:620-627. [DOI] [PubMed] [Google Scholar]
- 34.Sinkins, S. P., H. R. Braig, and S. L. O'Neill. 1995. Wolbachia superinfections and the expression of cytoplasmic incompatibility. Proc. R. Soc. Lond. Ser. B 261:325-330. [DOI] [PubMed] [Google Scholar]
- 35.Turelli, M. 1994. Evolution of incompatibility-inducing microbes and their hosts. Evolution 48:1500-1513. [DOI] [PubMed] [Google Scholar]
- 36.Turelli, M., and A. A. Hoffmann. 1995. Cytoplasmic incompatibility in Drosophila simulans: dynamics and parameter estimates from natural populations. Genetics 140:1319-1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Werren, J. H. 1997. Biology of Wolbachia. Annu. Rev. Entomol. 42:587-609. [DOI] [PubMed] [Google Scholar]
- 38.Werren, J. H., and S. L. O'Neill. 1997. The evolution of heritable symbionts, p. 1-41. In S. L. O'Neill, A. A. Hoffmann, and J. H. Werren (ed.), Influential passengers: inherited microorganisms and arthropod reproduction. Oxford University Press, Oxford, United Kingdom.
- 39.Werren, J. H., and D. M. Windsor. 2000. Wolbachia infection frequencies in insects: evidence of a global equilibrium? Proc. R. Soc. Lond. Ser. B 267:1277-1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Werren, J. H., D. Windsor, and L. R. Guo. 1995. Distribution of Wolbachia among neotropical arthropods. Proc. R. Soc. Lond. B Biol. Sci. 262:197-204. [Google Scholar]
- 41.West, S. A., J. M. Cook, J. H. Werren, and H. C. J. Godfray. 1998. Wolbachia in two insect host-parasitoid communities. Mol. Ecol. 7:1457-1465. [DOI] [PubMed] [Google Scholar]
- 42.Yen, J. H. 1975. Transovarial transmission of Rickettsia-like microorganisms in mosquitoes. Ann. N. Y. Acad. Sci. 266:152-161. [DOI] [PubMed] [Google Scholar]