Abstract
Quantifying the rate at which bacteria colonize aggregates is a key to understanding microbial turnover of aggregates. We used encounter models based on random walk and advection-diffusion considerations to predict colonization rates from the bacteria's motility patterns (swimming speed, tumbling frequency, and turn angles) and the hydrodynamic environment (stationary versus sinking aggregates). We then experimentally tested the models with 10 strains of bacteria isolated from marine particles: two strains were nonmotile; the rest were swimming at 20 to 60 μm s−1 with different tumbling frequency (0 to 2 s−1). The rates at which these bacteria colonized artificial aggregates (stationary and sinking) largely agreed with model predictions. We report several findings. (i) Motile bacteria rapidly colonize aggregates, whereas nonmotile bacteria do not. (ii) Flow enhances colonization rates. (iii) Tumbling strains colonize aggregates enriched with organic substrates faster than unenriched aggregates, while a nontumbling strain did not. (iv) Once on the aggregates, the bacteria may detach and typical residence time is about 3 h. Thus, there is a rapid exchange between attached and free bacteria. (v) With the motility patterns observed, freely swimming bacteria will encounter an aggregate in <1 day at typical upper-ocean aggregate concentrations. This is faster than even starving bacteria burn up their reserves, and bacteria may therefore rely solely on aggregates for food. (vi) The net result of colonization and detachment leads to a predicted equilibrium abundance of attached bacteria as a function of aggregate size, which is markedly different from field observations. This discrepancy suggests that inter- and intraspecific interactions among bacteria and between bacteria and their predators may be more important than colonization in governing the population dynamics of bacteria on natural aggregates.
Bacteria colonize marine aggregates where they grow and are preyed upon. Bacteria typically occur on aggregates in concentrations that are orders of magnitude higher than in the ambient water (10, 11, 40). They cause aggregates to solubilize and remineralize, apparently at high rates (1, 36, 37). Even if microbial processes on aggregates account for only a small fraction of the total microbial activity in the water column (4, 40), seepage of dissolved organic matter from degrading aggregates can affect microbial processes even outside the aggregates. For example, the few available estimates of leakage rates from aggregates suggest that aggregates may be quantitatively significant dissolved organic matter sources for free bacteria and that the solute plumes trailing behind sinking aggregates may be important sites of bacterial growth (19, 23, 39). Thus, marine aggregates are not only sinking vehicles but also important components of the pelagic food webs. Bacterial activities on aggregates are key to understanding this role.
In resolving the population dynamics of bacteria on aggregates one needs to consider the rate at which bacteria colonize the aggregates. Several studies have described the succession of microbes on aggregates (19, 26, 36, 41, 43), but few have examined the colonization process at a relevant—i.e., short (minutes)—time scale (15), and none have adequately considered the effect of the flow environment on colonization. A significant fraction of pelagic bacteria are motile (15, 18, 30), and simulation models have shown that motility and chemosensing capability are important for colonization, even of sinking aggregate (21, 23). Here we develop encounter models to predict colonization rates based on observed motility patterns of the bacteria and the hydrodynamic environments. We then compare model predictions with empirical measurements by using artificial aggregates.
MATERIALS AND METHODS
Models of colonization.
The change in the number of bacteria on an aggregate due to colonization, dN/dt, equals the transport of bacteria toward the aggregate, F, which in turn depends on the ambient concentration of bacteria, C,
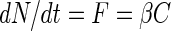 |
(1) |
where β is the “encounter rate kernel,” i.e., the imaginary volume of water from which the aggregate “collects” bacteria per unit time. The colonization of aggregates by bacteria is a result of the motility of the bacteria and/or the scavenging of bacteria by the sinking aggregate. Specific encounter rate kernels can describe each of these processes for our artificial model aggregate, which is spherical and impermeable.
(i) Steady state.
The encounter rate kernel for scavenging, assuming a low Reynolds number flow (25), is:
 |
(2a) |
where U is the aggregate sinking velocity and r is the radius of the bacteria. The kernel for motility colonization depends on the motility pattern. We consider two extremes, viz., swimming along a straight line and random walk. The two kernels are:
 |
(2b) |
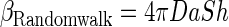 |
(2c) |
where u is the swimming velocity of the bacteria, a is the aggregate radius, D is the diffusivity of randomly moving bacteria (random walk), and Sh is the Sherwood number. For an organism with a run-and-tumble swimming behavior and an exponential distribution of run lengths (“Gaussian random walk”), the diffusivity can be approximated by using the following equation (8)
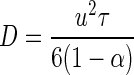 |
(3) |
where τ is the average run length (time) and α the mean value of the cosine of the angle between successive runs. The Sherwood number is the ratio between total (diffusive plus advective) transport of bacteria toward the aggregate and the transport due to diffusion alone (random walk motility). Sh is “1” in the absence of advection (i.e., stationary aggregates). For sinking aggregates, Sh can be approximated as follows (24):
 |
(4) |
where Reynolds number Re = Ua/D and ν is the kinematic viscosity (∼10−2 cm2 s−1). The Sh of typical marine snow aggregates in the size range of 0.1- to 1-cm radius varies from ca. 5 to 20 (24). A comparison of β-values for the scavenging and motility colonization mechanisms demonstrates that colonization is dominated by bacterial motility, and we can thus ignore scavenging when considering motile bacteria.
Our experiments revealed that bacteria that have colonized an aggregate might detach (see Results). If we assume that a constant fraction per unit time (δ) of the bacteria that have colonized the aggregate leave it again, then equation 1 can be modified to:
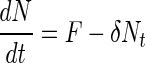 |
(5) |
The cumulated number of bacteria that have colonized the aggregate as a function of time is then:
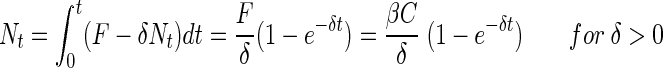 |
(6a) |
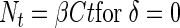 |
(6b) |
(ii) Non-steady state.
For a sinking aggregate, transport of random walk bacteria toward the aggregate reaches steady state almost immediately (24). For bacteria swimming along straight lines, steady state is instantaneous. However, for a nonsinking aggregate colonized by random walk bacteria, steady state is approached slowly. The time-dependent encounter rate kernel for randomly walking bacteria is (33)
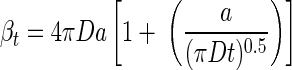 |
(7) |
From equation 7, it follows that it takes ca. 1 to 10 days to reach within 10% of the steady-state transport for diffusivity coefficients (D ∼ 10−6 to 10−5 cm2 s−1) and sphere sizes (a ∼ 0.2 cm) typical of our experiments. Obviously, steady state is not a valid assumption in still-water experiments. The cumulated number of bacteria that have colonized the particle at time t (Nt) under non-steady-state conditions is then
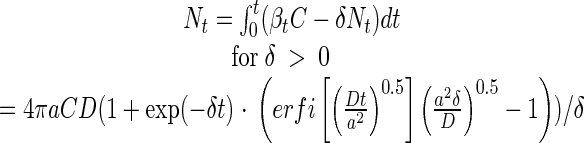 |
(8a) |
where erfi is the imaginary error function, and
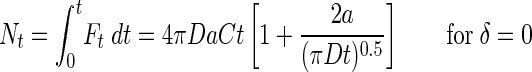 |
(8b) |
Model predictions.
We use equation 8a to describe the colonization process by randomly walking bacteria in still water where a steady state cannot be assumed. Equation 8a predicts that accumulation of bacteria on aggregate increases with the size of the aggregate (a) and diffusion coefficient of the bacteria (D), which in turn is a function of their swimming speed, run lengths, and turn angles (equation 3).
In all other cases (linear swimming, flow), we use equation 6a to describe the colonization process. It follows from equation 6a that the accumulation of bacteria on aggregate increases with the encounter rate kernel (β), which is related to bacterium size, bacteria's motility patterns, aggregate sizes, and ambient flow rate (equations 2a, 2b, and 2c).
To test model predictions, we observed the swimming behavior of 10 bacterial strains (Table 1) and experimentally measured their rates of colonizing artificial aggregates (stationary and sinking). All strains were isolated from marine aggregates or diatoms collected in the Øresund, Denmark, and identified by means of 16S ribosomal DNA sequencing (H.-P. Grossart, unpublished data). We maintained bacterial cultures at exponential growth in marine broth (MB [MB2216; Difco]; 15 g liter−1). In most experiments, we aimed at an ambient concentration of 106 ml−1.
TABLE 1.
Bacterial strains used in the experiments and the number and types of experiments conducted
| Strain | Identification (by GenBank alignment) | % Homology to GenBank sequence | Size
|
No. of expt
|
||
|---|---|---|---|---|---|---|
| Width by length (μm) | Vol (μm3) | Still | Flow | |||
| HP1 | AJ002006, Curacaobacter baltica | 98 | 1 by 2.5 | 2.0 | 2 | |
| HP4 | AJ295154, Oleiphilus messinensis | 93 | 1 by 5 | 3.9 | 1 | |
| HP5 | AF321022, Frigobacterium sp. strain GOB | 98 | 1 by 3 | 2.4 | 1 | |
| HP11 | M58792, Microscilla furvescens | 90 | 1 by 3.5 | 2.7 | 11 | 9 |
| HP15 | AJ000647, Marinobacter strain PCOB-2 | 98 | 1 by 3.5 | 2.7 | 1 | |
| HP25 | AF277514, Cellulophaga strain SIC.834 (uncultured) | 98 | 0.5 by 2 | 0.4 | 3 | 2 |
| HP33 | AF345550, Rhizobium sp. strain SDWo52 | 99 | 0.8 by 2 | 1.0 | 4 | |
| HP39 | AF025321, α-Proteobacterium KAT8 | 99 | 1 by 4 | 3.1 | 6 | 2 |
| HP43 | D90916, Synechocystis sp. | 100 | 1 by 4.5 | 3.5 | 1 | |
| HP46 | AY028198, Marine bacterium TW-3 | 99 | 0.8 by 2 | 1.0 | 1 | |
Swimming behavior observations.
We observed the swimming patterns of the bacteria by means of dark-field microscopy. A small observation chamber was made by gluing a 0.5-cm high, 1.6-cm diameter PVC ring onto a microscopic slide. The chamber was closed by a coverslip. The swimming pattern of each strain was videotaped by using a 10× objective lens yielding a focal depth of ca. 50 μm and a viewing field of ca. 400 by 500 μm. Two-dimensional (2-D) projections of swimming tracks were digitized by LabTrack software (DiMedia) by using a time resolution of 0.04 or 0.16 s. We digitized between 100 and 1,000 tracks per strain. Assuming isotropic distribution of swimming velocities, the mean square velocity in 3-D is 3/2 times the mean square velocity in 2-D, and the average 3-D speed was therefore estimated as 3/2 times the average of the 2-D speed estimate. Swimming speeds were in addition estimated as the maximum speeds that were sustained for long time periods (ensuring that the selected bacteria were swimming in the plane). Tumbling frequencies and turn angles were estimated from visual examination of the 30 to 50 longest swimming tracks per bacterial strain. Since we used 2-D projections of 3-D swimming paths, there is a slight bias in the estimates of turn angles. Measured swimming speeds, run lengths, and turn angles were used to compute the diffusion coefficients of the bacteria (equation 3).
Colonization experiments.
Bacterial colonization was examined by monitoring the accumulation of bacteria on artificial aggregates that were incubated in still or flowing seawater containing bacteria. Model aggregates were made from 2% agar as described by Cronenberg (12). Warm agar was dripped from a pipette into sterile seawater covered by a thin layer of paraffin oil. Almost-perfect agar spheres formed in the oil and subsequently sank into the seawater. Traces of paraffin oil on the spheres were rinsed off with sterilized seawater. The aggregate size (0.12- to 0.28-cm radius) was controlled by the width of the pipette mouth. In our standard experiments, we used spheres with a radius of 0.2 cm. In some experiments we added MB (15 g liter−1) or dimethylsulfoniopropionate (DMSP; 1 mM) to the agar. DMSP is a natural organic substrate leaking from aging algal cells and aggregates (27). In a few experiments we used diamond-cut glass pearls as model aggregates (0.15- to 0.38-cm radius).
A typical experiment consisted of simultaneously placing 24 aggregates in a bacterial suspension. Agar-aggregates were fixed on thin glass needles and suspended in the bacterial suspensions in either still water (2 liters) or in a flume (ca. 10 liters; flow velocities 0.1 to 0.3 cm s−1). Glass pearls were suspended on thin wires of stainless steel. Triplicate aggregates were collected typically at 0, 5, 10, 15, 20, 40, 80, and 160 min. Sampled aggregates were placed in a counting chamber made of an O-ring glued onto a microscopic slide, and a drop of formaldehyde and a drop of DAPI (4′,6′-diamidino-2-phenylindole) were added. The chamber was then closed by a coverslip, and the bacteria were counted at ×1,000 magnification under epifluorescence. Some bacteria may be lost when collecting aggregates and, hence, all abundance estimates are conservative. However, in one experiment we examined the effect of retrieving aggregates and found no measurable effect. Ambient concentrations of bacteria were measured at the onset and termination of each incubation experiment by filtering 1-ml suspensions onto 0.2-μm (pore-size) filters, staining with DAPI, and counting under epifluorescence.
We conducted 44 experiments in which we compared the different strains and examined the effects of flow, aggregate size, bacterial concentration, and the presence of chemical signals (MB and DMSP) in the aggregates on colonization pattern and rate (Table 1).
Observed colonization rates were fitted to either equation 8a (random walk bacteria in still water) or equation 6a (all other cases) by nonlinear regression routines to derive estimates of δ and D (equation 8a) or δ and β (equation 6a). Unfortunately, the two parameters (δ and D or δ and β) are strongly correlated, which results in large confidence limits. From the estimates of β, we finally estimated D · Sh (equation 2c) or bacterial swimming speed, u (equation 2b). These parameters were compared against the u and D values derived from swimming behavior observations as independent validation of the models.
RESULTS
Motility patterns.
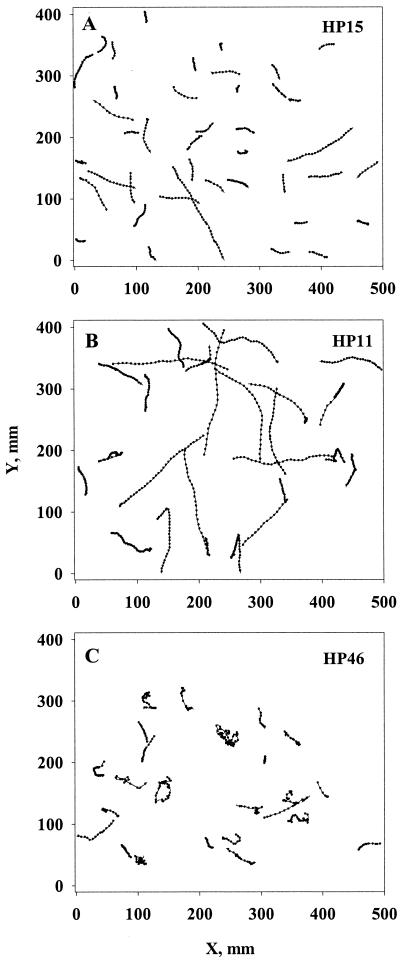
Among the 10 bacterial strains examined, two were nonmotile (strains HP25 and HP43) and the rest were motile. Swimming speeds, tumbling frequencies, and turn angles differed among the motile strains (Table 2), resulting in different swimming paths (Fig. 1). Swimming speed varied between ca. 20 and 60 μm s−1. The motile strains may be categorized into three groups based on their motility patterns (Fig. 1). Group 1 (strains HP4 and HP15) showed no distinct tumbles but was swimming along roughly straight paths. The paths were, however, somewhat wiggly presumably due to Brownian rotation. Group 2 (HP1, HP5, and HP11) tumbled at low frequencies, resulting in run lengths of ca. 10-s duration (range, 6 to 15 s) and somewhat convoluted swimming tracks. Finally, group 3 (HP33, HP39, and HP46) tumbled at high frequencies with run lengths of 0.4 to 3 s, and its swimming paths were very convoluted.
TABLE 2.
Diffusion coefficients estimated from analyses of swimming tracks in 10 strains of bacteriaa
| Strain | No. of tumbles s−1 | τ (s) | α | uavg (μm s−1) | umax (μm s−1) | Davg (cm2 s−1) | Dmax (cm2 s−1) |
|---|---|---|---|---|---|---|---|
| HP1 | 0.16 | 6.2 | −0.67 | 21 | 30 | 2.6 × 10−6 | 5.7 × 10−6 |
| HP4 | 0 | 22 | 26 | ||||
| HP5 | 0.096 | 10.5 | −0.67 | 26 | 34 | 6.8 × 10−6 | 12 × 10−6 |
| HP11 | 0.076 | 14.9 | −0.67 | 32 | 51 | 16 × 10−6 | 38 × 10−6 |
| HP15 | 0 | 29 | 37 | ||||
| HP25 | Nonmotile | ||||||
| HP33 | 0.48 | 2.07 | −0.67 | 22 | 30 | 1.0 × 10−6 | 1.9 × 10−6 |
| HP39 | 0.33 | 3.05 | −0.67 | 22 | 28 | 1.4 × 10−6 | 2.4 × 10−6 |
| HP43 | Nonmotile | ||||||
| HP46 | 2.06 | 0.4 | 0.48 | 28 | 56 | 0.4 × 10−6 | 1.4 × 10−6 |
Tumbling frequencies, average run length (τ), turn angles (as α), and swimming velocities for 10 bacterial strains from analyses of swimming tracks. Two estimates of swimming velocity are provided, uavg, which is the overall average 2-D swimming velocity multiplied by 1.22, and umax, which is the average velocity that is sustained for long periods. A diffusion coefficient is computed for each of these velocities (Davg and Dmax, respectively).
FIG. 1.
2-D projections of swimming tracks of three different bacterial strains representative of the three motility types described in the text: nontumblers (strain HP15), rare tumblers (strain HP11), and frequent tumblers (strain HP46). The dots represent positions at 0.16-s (A and B) or 0.08-s (C) time intervals.
For all of the tumbling strains, average turn angles exceeded 90° and were thus biased backward. For most strains, turn angle distributions did not differ significantly, and these were pooled to yield an average turn angle of ∼140° (α = −0.67). HP46 differed significantly from the rest, with a lower average turn angle (ca. 125°; α = −0.48).
From the motility patterns we estimated the diffusion coefficients for the three motile groups (Table 2). Strains with long run lengths had diffusion coefficients on the order of 10−5 cm2 s−1, whereas strains that tumble frequently had diffusivities about an order of magnitude lower (ca. 10−6 cm2 s−1). No diffusion coefficient could be assigned the nontumbling species on the basis of equation 3.
Colonization pattern.
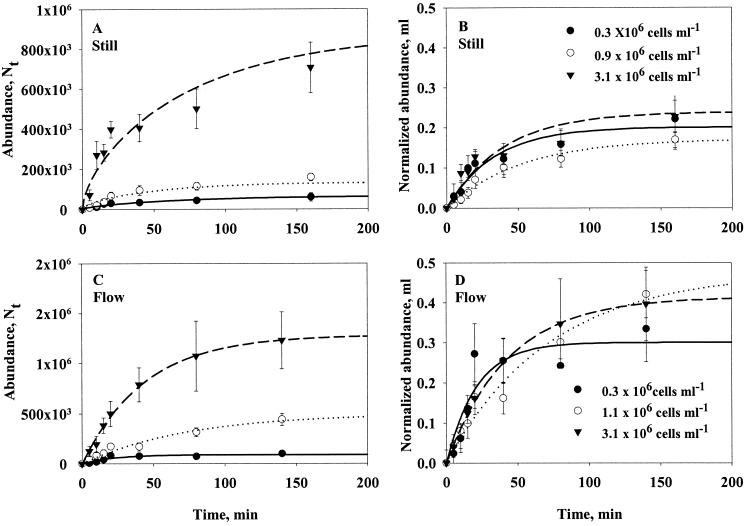
All experiments showed the same colonization pattern, i.e., a decelerating accumulation rate of bacteria (Fig. 2). We first interpreted this as a result of the non-steady-state condition in still water, but obviously equation 8b fits the observations very poorly since the accumulation of bacteria levels off much faster than this model can account for (Fig. 2A). In experiments where the aggregates were suspended in a flow we also found a saturating response (Fig. 2B). Because steady state is almost instantaneous in a flow regime, we had expected a linear increase here. Non-steady state, thus, cannot account for the saturating response in flow experiments either. Microscopic observations showed that bacteria that had attached to an aggregate might detach. We therefore conducted an experiment in which we incubated agar spheres for 60 min in a suspension of bacteria and then moved the spheres to sterile filtered seawater and followed the decrease of bacteria on the aggregate (Fig. 3). From the exponential decline we conclude that bacteria detach at a constant fraction per time; thus, encounter models need to incorporate the detachment rate (δ in equation 6a and equation 8a; Fig. 2A).
FIG. 2.
Colonization of 0.2-cm radius agar spheres in still (A) and flowing (B) water by strain HP11 bacteria. Observations (solid symbols) are described by three different models: non-steady-state diffusion (equation 8b [solid line]), steady state with detachment (equation 6a [dotted line]), and non-steady-state diffusion with detachment (equation 8a [dashed line]). Error bars indicate the standard deviations of 10 measurements.
FIG. 3.
Colonization and detachment of bacteria (strain HP11) on 0.2-cm-radius agar spheres in still water. During the first 60 min, spheres were incubated at a bacterial concentration of 2.7 × 105 ml−1. At 60 min the spheres were transferred to sterile filtered seawater. Equation 8a was fitted to the data taken between 0 and 60 min, and an exponential model was fitted to the data taken from 60 min onward. Error bars indicate the standard deviations of 10 measurements.
Ambient bacterial concentration.
Our models predict that the initial accumulation rate and equilibrium abundance of bacteria on aggregates should increase in proportion to the ambient bacterial concentration, which is confirmed by observations (Fig. 4A). Hence, in order to account for small variations in ambient bacterial concentration, we normalize bacterial abundances on aggregates with ambient concentration in all subsequent presentations (Fig. 4B). The observed dependency of ambient bacterial concentration also implies that colonization rate and equilibrium abundance are not governed by density-dependent mechanisms on the aggregate, at least not at the bacterial concentrations used here.
FIG. 4.
Effect of ambient bacterial concentration on accumulation rate on 0.2-cm agar spheres of strain HP11 bacteria in still (A and B) and flowing water (0.33 cm s−1) (C and D). Panels A and C show absolute abundances of bacteria per aggregate, while panels B and D show abundances normalized with ambient concentration. Equation 8a has been fitted to the data (lines). Error bars indicate the standard deviations of 10 measurements.
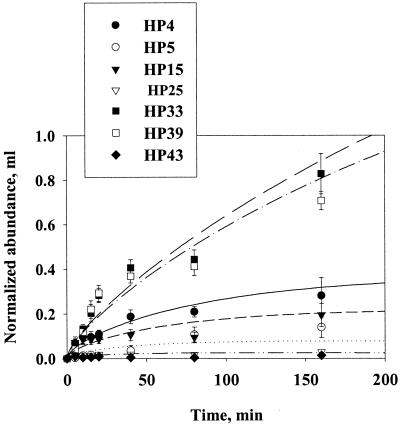
Differences among strains.
All of the motile strains showed colonization patterns similar to that illustrated in Fig. 5. Still-water estimates of D for the different strains varied within 1 order of magnitude, between 0.3 × 10−5 and 2.5 × 10−5 cm2 s−1 (Fig. 6 and Table 3). The estimates of fractional detachment rates varied less between strains than the diffusion coefficients, between 0.7 × 10−4 and 1.7 × 10−4 s−1 (Fig. 6 and Table 3). Nonmotile strains (HP25 and HP43) colonized agar spheres at an insignificant rate in still water, as expected (Fig. 5).
FIG. 5.
Accumulation of different strains of bacteria on 0.2-cm agar spheres in still water. Equation 8a has been fitted to the data (lines). Error bars indicate the standard deviations of 10 measurements.
FIG. 6.
Average (± the standard error) diffusivities and detachment rates estimated from colonization rates of eight motile bacterial strains on 0.2-cm agar unenriched spheres in still water by fitting equation 8a to the observed accumulation of bacteria. The numbers within the columns indicate the numbers of experiments.
TABLE 3.
Swimming velocities (u), diffusivities (D), and fractional detachment rates (δ) estimated from still-water colonization rates of 10 bacterial strainsa
| Strain | U (μm s−1) ± SE | D (cm2 s−1) ± SE | δ (s−1) ± SE | No. of expt |
|---|---|---|---|---|
| HP1 | (6.1 ± 0.3) × 10−6 | (1.6 ± 0.1) × 10−4 | 2 | |
| HP4 | 11 ± 2 | 15 × 10−6 | 1.2 × 10−4 | 1 |
| HP5 | 3.3 × 10−6 | 1.7 × 10−4 | 1 | |
| HP11 | (13 ± 8) × 10−6 | (1.3 ± 0.4) × 10−4 | 10 | |
| HP15 | 15 ± 6 | 9.7 × 10−6 | 1.4 × 10−4 | 1 |
| HP25 | 3 | |||
| HP33 | (21 ± 14) × 10−6 | (0.9 ± 0.7) × 10−4 | 2 | |
| HP39 | (25 ± 5.5) × 10−6 | (0.7 ± 0.3) × 10−4 | 4 | |
| HP43 | 1 | |||
| HP46 | 14 × 10−6 | 1.1 × 10−4 | 1 |
Only experiments with unenriched spheres.
For the bacterial strains that do not tumble (HP4 and HP15), the colonization process can be described by the “linear-swimming” kernel, and their swimming velocities estimated from β (equation 2b) were 11 and 15 μm s−1 (Table 3).
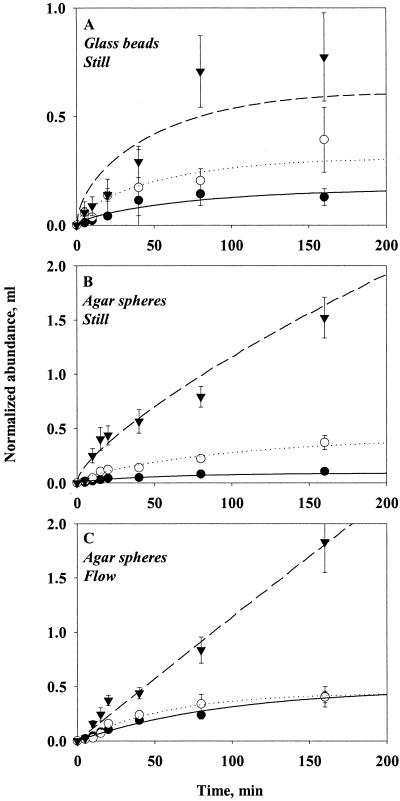
Effect of aggregate size.
Our models predict that the total transport toward a spherical aggregate of bacteria with a random walk motility increases with aggregate radius, both in stationary and in sinking aggregates (cf. equations 2c and 7). At steady state, transport scales with aggregate radius (for constant Sh). At non-steady state, transport increases with radius to a power of 1 to 2 (equation 7). However, D and presumably δ should be independent of aggregate size. These predictions are largely confirmed by observations (Fig. 7). Experiments with glass beads in still water followed the prediction most closely, with estimates of D and δ varying by <10% for bead radii from 0.15 to 0.375 cm (Table 4). D · Sh for agar spheres in flow also varied very little, whereas experiments with agar spheres in still water yielded more variable estimates of D and δ.
FIG. 7.
Effect of sphere size on the accumulation of strain HP11 bacteria in still and flowing water. Glass beads in still water (A), agar spheres in still water (B), and agar spheres in flowing water (0.27 cm s−1) (C). Equation 8a (still water) or equation 6a (flowing water) has been fitted to the data (lines). Error bars indicate the standard deviations of 10 measurements. Symbols: •, small; ○, medium; ▾, large.
TABLE 4.
Estimates of D or D · Sh and δ (in parentheses) for experiments with various sphere sizesa
| Sphere size |
D (δ)
|
Agar spheres [flow ∼ 0.3 cm s−1D·Sh (δ)] | |
|---|---|---|---|
| Glass spheres stagnant | Agar spheres stagnant | ||
| Small | 1.1 × 10−5 (1.4 × 10−4) | 0.9 × 10−5 (1.5 × 10−4) | 5.0 × 10−5 (1.7 × 10−4) |
| Medium | 1.1 × 10−5 (1.5 × 10−4) | 1.8 × 10−5 (1.1 × 10−4) | 5.7 × 10−5 (2.9 × 10−4) |
| Large | 1.2 × 10−5 (1.4 × 10−4) | 3.5 × 10−5 (1.9 × 10−5) | 5.6 × 10−5 (4.3 × 10−6) |
Small, medium, and large spheres measured as 0.15-, 0.25-, and 0.38-cm radii (glass spheres) or 0.12-, 0.17-, and 0.28-cm radii (agar spheres).
Effect of flow.
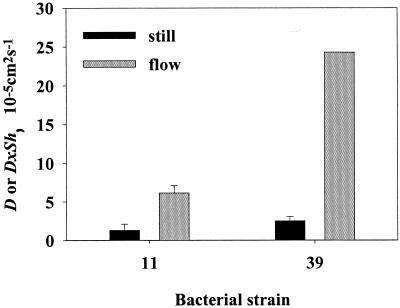
The colonization rate was substantially higher in flowing than in still water, both for motile and nonmotile strains (Fig. 8). In motile strains HP11 and HP39, estimates of D · Sh in flow (∼0.3 cm s−1) were ca. 5 (range, 1 to 9) and 20 times the estimated D in still water (Fig. 9), yielding estimated Sh values of 5 and 20, respectively. The expected Sh value is about 12 for both of these strains (equation 4).
FIG. 8.
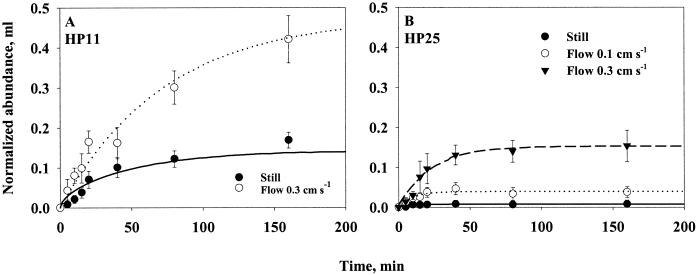
Effect of water flow (0.1 or 0.3 cm s−1) on accumulation of strain HP11 (A) and strain HP25 (B) bacteria on 0.2-cm unenriched agar spheres. Equation 8a (still water) or equation 6a (flowing water) has been fitted to the data (lines). Error bars indicate the standard deviations of 10 measurements.
FIG. 9.
Average (± the standard error) estimates of diffusivity (D) or diffusivity multiplied by the Sherwood number (D · Sh) for strain HP11 and strain HP39 bacteria estimated from accumulation rates on 0.2-cm unenriched agar spheres by fitting equation 8a (still water) or equation 6a (flow) to the observed accumulation of bacteria.
Nonmotile strain HP25 colonized agar spheres at a considerable rate in flow (Fig. 8b). The encounter rate kernels (β) estimated by fitting equation 6a to the data were ca. 3 × 10−5 ml s−1 for a flow of 0.11 cm s−1 and 9 × 10−5 ml s−1 for a flow of 0.32 cm s−1. While the observed threefold increase in β with a threefold increase in flow rate is expected from scavenging of nonmotile cells by a sinking aggregate (equation 2a), the absolute magnitude of β is much higher than expected. With a bacterial radius of ∼1 μm (Table 1), the predicted encounter rate kernels are 0.5 and 1.5 × 10−8 ml s−1 for the low and high flow rates, respectively. The reason for this discrepancy is unclear.
Chemical signals (DMSP and MB).
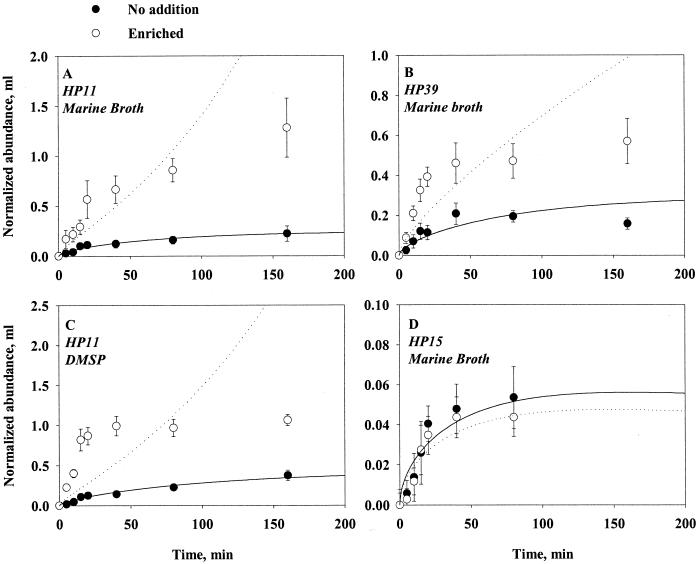
The presence of marine broth in the agar substantially enhanced still-water colonization rates in the tumbling strains (HP33, HP39, and HP11), suggesting a chemosensory response of the bacteria (see Fig. 10A and B for examples). Among these three strains, only HP11 responded to the presence of DMSP in the agar (Fig. 10C). Nontumbling strain HP15 did not respond to the presence of MB in the agar (Fig. 10D)
FIG. 10.
Accumulation of strains HP11, HP39, and HP15 bacteria on enriched (open symbols, dotted lines) and unenriched (closed symbols, solid lines) 0.2-cm agar spheres in still water. Equation 8a has been fitted to the data (lines). In panels A, B, and D the agar spheres were enriched with 15 g of marine broth liter−1; in panel C the agar spheres were enriched with 1 mM DMSP. Error bars indicate the standard deviations of 10 measurements.
The overall colonization pattern of enriched agar spheres was similar to that of unenriched ones. However, equation 8a, which assumes random walk, consistently provides very poor fits to the experiments with tumbling bacteria and enriched spheres. The initial accumulation rate is substantially higher than this model can accommodate and the assumption of random walk is not warranted.
DISCUSSION
Swimming behavior and colonization rates.
Motility enhances the encounter rate between pelagic bacteria and particles or other resource patches, but different motility patterns lead to different encounter rates. The motile strains examined here had swimming velocities of 20 to 60 μm s−1, less than the very high swimming speeds reported for some natural bacterial assemblages (>100 μm s−1 [18, 30]). The present strains differed mainly in their tumbling frequency. A high tumbling frequency and a high turn angle lead to a convoluted swimming path and a lower probability of encountering a particle or a patch. Overall, the observed particle colonization rates were largely consistent with what one would expect from the swimming behavior of the bacteria. A perfect match cannot be expected because not all bacteria encountering a particle will necessarily attach, and therefore observed colonization rates are conservative estimates of encounter rates. However, the diffusivities of motile bacteria estimated by the two independent approaches are all of the same order of magnitude—D ∼ 10−6 to 10−5 cm2 s−1—as are the two estimates of swimming velocities for the nontumbling strains, u ∼ 10 to 30 μm s−1 (Tables 2 and 3). Also, the predicted effects of fluid flow (sinking), ambient bacterial concentration, and aggregate size on colonization are largely consistent with observations for the motile strains.
However, the differences between strains in swimming behavior were only partly reflected in differences in colonization rates. For the group of bacterial strains with long tumbling intervals (HP1, HP5, and HP11), the correspondence between observed colonization rate and swimming behavior was good. However, for strains with short tumbling intervals (HP33, HP39, and HP46), D derived from colonization experiments (Table 3) was ca. 10-fold higher than that predicted from behavior observations and Gaussian random walk assumption (Table 2), i.e., behavioral observations would underestimate their colonization rates. One possible reason for this discrepancy is that Gaussian random walks may not appropriately describe frequently tumbling bacteria. Other types of random walks, Levy walks, provide a more efficient means of encountering food patches or particles and have been suggested as good descriptions of motilities of various organisms, including microbes (28, 42). Levy walks are characterized by bursts of tumbles interrupted by occasional very long runs, not unlike that seen for HP46 (Fig. 1).
The swimming velocities derived from colonization experiments for the nontumbling linear swimmers (strains HP4 and 15; Table 3) are less by a factor of 2 to 3 than those measured directly (Table 2). Thus, behavioral observations would overestimate the colonization rates. However, the efficiency of the linear swimming strategy is constrained by thermally driven Brownian rotation: the cells rotate continuously, causing constant and slight directional shifts (cf. Fig. 1A and B), and at sufficiently large spatial scales, rotational diffusion causes linear swimming to approach a diffusion process. The equivalent diffusion coefficients due to Brownian rotation for strains HP4 and HP15, computed from equations by Berg (8), are ca. 2 × 10−5 cm2 s−1, similar to those derived from the colonization experiments (Fig. 6). Therefore, Brownian rotation can account for the discrepancy between colonization measurements and behavioral observations.
Chemokinetic swimming behavior.
From the discussion above it follows that tumbling reduces the efficiency of encountering resource patches and particles. Why, then, tumble at all? The traditional explanation is that tumbling allows the bacteria to modify their swimming paths in a chemical gradient (8), increasing their chance of locating a food patch. In our experiments, enriching the agar spheres with attractant molecules (MB and DMSP) increased the initial colonization rate by a factor of 5 to 10 among the tumbling strains (Fig. 10A to C), whereas the nontumbling strain was not affected (Fig. 10D). Our observations therefore support the idea that tumbling is a prerequisite of the chemosensory behavior commonly found in marine pelagic bacteria (9, 15, 31).
How long does it take to find an aggregate?
The bacterial communities associated with aggregates are typically different from the bacterial communities in the surrounding water (13, 26, 32). This suggests that some bacteria specialize on colonizing aggregates and that their survival relies on how fast they encounter an aggregate. The rate at which a bacterium encounters aggregates depends on the size distribution and concentration of particles, as well as on the encounter rate kernel of individual bacteria, and can be estimated as
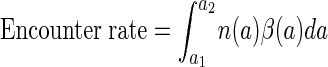 |
(9) |
where
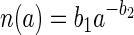 |
(10) |
is the size spectrum of aggregates expressed as a power function of aggregate size, a1 to a2 is the size range of aggregates considered, and β(a) the encounter rate kernel. The encounter rate kernel can also be approximated by a power function of aggregate radius by combining equations 2c and 4, utilizing the fact that the aggregate sinking velocity in situ increases with size as Ucm/s = 0.13acm0.26 (3), and by assuming that D = 10−5 cm2 s−1 and that u = 100 μm s−1:
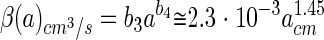 |
(11) |
which is correct to within 1% in a size range (a) of 0.0005 to 1 cm. The encounter rate can now be estimated as follows:
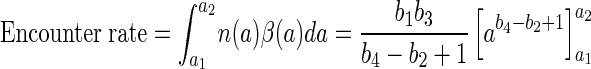 |
(12) |
Based on the aggregate size spectra compiled by Alldredge and Silver (5) and the corresponding b1 and b2 values derived by Kiørboe and Jackson (23), the estimated encounter rates with aggregates (0.0005 to 1 cm) vary between 10−6 and 7 × 10−4 s−1 (median = 3 × 10−5 s−1). This corresponds to an average “searching” time of 0.02 to 12 days (median = 0.4 day). If the average time to encounter an aggregate is 0.4 day and if the aggregate encounter is a Poisson process, then half of the bacteria will encounter an aggregate within 7 h and 92% will do so within 1 day.
Can bacteria swim for long enough to encounter an aggregate? The cost of swimming for bacteria can be computed as the viscous drag (of a sphere) multiplied by the swimming speed (8): 6πηru2, where η is the dynamic viscosity (∼10−2 g cm−1 s−1). Alldredge et al. (2) reported average volumes of 1 μm3 for aggregated-attached bacteria from various environments. For a 0.6-μm radius (1-μm3 volume) bacterium swimming at 100 μm s−1 the power requirement is 10−16 J s−1. A 1-μm3 bacterium contains ca. 10−13g of C (assuming 0.1 g of C cm−3), which by respiratory combustion may release 0.5 × 10−8 J (assuming 50 kJ per g of C). If the efficiency of the propulsion system is 1% and if other metabolic expenses are ignored, then a starving bacterium has energy resources enough for swimming continuously for almost 6 days. Obviously, the metabolic costs of swimming do not limit the chance that a bacterium may encounter an aggregate. This conclusion is independent of the actual swimming velocities of the bacteria because the search time decreases but the swimming cost increases with swimming velocity squared. It follows then that, in most upper-ocean habitats, aggregates are sufficiently abundant so that most bacteria can make it to an aggregate before they starve to death.
Population dynamics of attached bacteria.
Our observations suggest that bacteria that have colonized an aggregate may detach. The estimates of fractional detachment rate (δ) are similar for all of the bacterial strains examined, i.e., ca. 10−4 s−1. Growth of attached bacteria may bias our δ estimates low, but since conceivable magnitudes of specific growth rates are much less than our estimates of specific detachment rates (see below), this is a relatively small error. A detachment rate similar to that found here can be estimated for surface-attached Pseudomonas sp. from data in the study by Baty et al. (7). Such detachment rates imply that the bacteria have an average residence time of ca. 3 h on an aggregate. This is of the same order of magnitude as the residence time in the upper mixed layer of an aggregate sinking at 100 m day−1. Aggregates are risky environments for attached organisms because aggregates are eaten or they sink toward the seafloor. Therefore, to maintain a population in the upper ocean, the bacteria will have to leave the aggregate either by releasing progeny (6) or by detaching. The high detachment rates estimated here suggest that there is a considerable exchange of bacteria between aggregates and the surrounding water (cf. reference 16). The detachment of bacteria is probably only physically possible relatively soon after attachment or cell division because the bacteria may become embedded in the mucus film or matrix that may cover or be part of an aggregate (see reference 34).
The balance between colonization and detachment will lead to an equilibrium abundance of bacteria on an aggregate. From equations 5 and 11, the equilibrium abundance of bacteria on a sinking aggregate is calculated as:
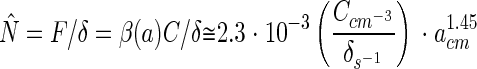 |
(13) |
Thus, provided that bacterial dynamics is governed only by colonization and detachment, the predicted bacterial abundance should scale with aggregate size to the power of ∼1.45 for aggregates with a 0.005- to 1-cm radius. However, this scaling is inconsistent with actual observations: bacterial abundances on natural aggregates scale with aggregate size to the power of 0.25 (Fig. 11; see also reference 4). Bacterial abundances normalized by ambient concentration of bacteria are also much higher on aggregates than predicted. This estimated difference is conservative, since not all ambient bacteria are potential colonizers. There are several possible explanations for these discrepancies.
FIG. 11.
Steady-state abundances of bacteria on aggregates as a function of aggregate size predicted from equation 13 (dashed line) compared to actually observed abundances on field-collected aggregates (symbols and solid line) (taken from the compilation of Kiørboe [22]). The different symbols refer to data from different studies. The predicted relation assumes a bacterial diffusivity of 10−5 cm2 s−1. Abundances have been normalized by ambient concentrations of bacteria. The predicted abundance increases with aggregate size raised to a power of 1.45 within the aggregate size range considered, while the log-log regression for the field-collected aggregates has a slope of 0.25.
First, unlike agar spheres, natural aggregates are fractal with convoluted surfaces that have areas larger by orders of magnitude. However, the fractal nature of a spheroid aggregate has limited influence on colonization rates because the diffusive and advective transport of bacteria toward an aggregate is governed not by its surface area but by the radius of an imaginary sphere circumscribing the aggregate. The enveloping geometry of a natural aggregate may, of course, be different from that of a sphere, and this may influence the colonization rate somewhat. Also, fluid flow through a porous aggregate may enhance colonization rates, but the effect is negligible (24) to small (up to a factor of 4 [38]). Colonization rates and, hence, the equilibrium bacterial abundances on fractal and porous aggregates may therefore be somewhat higher than predicted, but these factors cannot explain the difference in the scaling.
Second, if bacteria grow on the aggregates at a constant rate (μ), the equilibrium abundance of bacteria would be F/(δ − μ) rather than F/δ, i.e., higher than predicted. Reported growth rates of attached bacteria are on the order of 1 day−1 ∼10−5 s−1 (2, 35). Since estimated detachment rates are about 10 times higher, ca. 10−4 s−1, a growth rate of this order only increases the equilibrium abundance by 10%. Some reports suggest that attached bacteria grow much faster than free ones (see, for example, references 16 and 20). However, even a growth rate of 5 day−1 will only double the equilibrium abundance. Thus, growth alone cannot account for the different magnitudes of abundances. Moreover, growth per se does not change the scaling.
Another possible explanation is that the detachment rate declines with time as an increasing fraction of the bacteria become permanently attached. This would lead to higher equilibrium abundances of bacteria. However, it does not account for the difference in scaling, since this phenomenon will probably be more pronounced for the larger and presumably older aggregates.
Finally, the equilibrium bacterial abundances on aggregates may be governed by biological interactions other than colonization and detachment. Bacterial communities on natural aggregates consist of many species, not single-species assemblages as considered in our experiments. While the encounter rate between aggregates and free bacteria is independent of processes on the aggregate, the “decision” to attach or detach may be modified by complex inter- and intraspecific interactions among bacteria, e.g., through the release of signal molecules (“quorum sensing”) and antibiotic substances. For example, swarming behavior in some pathogenic bacteria appears to be regulated through quorum sensing (see, for example, reference 14), and we have evidence that some of the strains isolated from marine particles indeed release signal molecules (17). Likewise, a large fraction of bacterial isolates from marine particles express antagonistic activity (29). Such interactions may change the population dynamics substantially. Marine aggregates also house a rich protozoan fauna that feeds on attached bacteria (10), and predator-prey interactions may render bacterial abundances on aggregates independent of aggregate size (22). Such interactions also depend on the density—not only the abundance—of bacteria and, hence, on the fractal nature of natural aggregates. To more fully understand the population dynamics of bacteria on aggregates, processes of this kind need to be explored.
Acknowledgments
This work was supported by the Danish Natural Science Research Council (9801391), the Carlsberg Foundation (990536/20-950 and 980511/20-513), and the Danish Network for Fisheries and Aquaculture Research, financed by the Danish Ministry for Food, Agriculture, and Fisheries and the Danish Agricultural and Veterinary Research Council.
We thank Uffe H. Thygesen for help with model and software development and Andre Visser, George Jackson, and Josefin Titelman for fruitful discussions.
REFERENCES
- 1.Alldredge, A. L., and Y. Cohen. 1987. Can microscale chemical patches persist in the sea? Microelectrode study of marine snow and fecal pellets. Science 235:689-691. [DOI] [PubMed] [Google Scholar]
- 2.Alldredge, A. L., J. J. Cole, and D. A. Caron. 1986. Production of heterotrophic bacteria inhabiting macroscopic aggregates (marine snow) from surface waters. Limnol. Oceanogr. 31:68-78. [Google Scholar]
- 3.Alldredge, A. L., and C. Gotschalk. 1988. In situ settling behavior of marine snow. Limnol. Oceanogr. 33:339-351. [Google Scholar]
- 4.Alldredge, A. L., and C. Gotschalk. 1990. The relative contribution of marine snow of different origin to biological processes in coastal waters. Cont. Shelf Res. 10:41-58. [Google Scholar]
- 5.Alldredge, A. L., and M. Silver. 1988. Characteristics, dynamics, and significance of marine snow. Prog. Oceanogr. 20:41-58. [Google Scholar]
- 6.Azam, F., and D. C. Smith. 1991. Bacterial influence on the variability in the ocean's biochemical state: a mechanistic view, p. 213-236. In S. Demers (ed.), Particle analysis in oceanography. NATO ASI Series, vol. G27. Springer-Verlag, Berlin, Germany. [Google Scholar]
- 7.Baty, A. M., III, C. C. Eastburn, S. Techkarnjanaruk, A. E. Goodman, and G.-G. Geesey. 2000. Spatial and temporal variations in chitinolytic gene expression and bacterial biomass production during chitin degradation. Appl. Environ. Microbiol. 66:3574-3585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berg, H. C. 1993. Random walks in biology. Princeton University Press, Princeton, N.J.
- 9.Blackburn, N., T. Fenchel, and J. Mitchell. 1998. Microscale nutrient patches in planktonic habitats shown by chemotactic bacteria. Science 282:2254-2256. [DOI] [PubMed] [Google Scholar]
- 10.Caron, D. A. 1987. Grazing of attached bacteria by heterotrophic microflagellates. Microb. Ecol. 13:203-218. [DOI] [PubMed] [Google Scholar]
- 11.Caron, D. A., P. G. Davis, L. P. Madin, and J. M. Sieburth. 1986. Enrichment of microbial populations in macroaggregates (marine snow) from surface waters of the North Atlantic. J. Mar. Res. 44:543-565. [Google Scholar]
- 12.Cronenberg, C. H. C. 1994. Biochemical engineering on a microscale: biofilms investigated with needle-type glucose sensors. Ph.D. thesis. University of Amsterdam, Amsterdam, The Netherlands.
- 13.DeLong, E. F., D. G. Franks, and A. L. Alldredge. 1993. Phylogenetic diversity of aggregate-attached versus free-living marine bacterial assemblages. Limnol. Oceanogr. 38:924-934. [Google Scholar]
- 14.Eberl, L., S. Molin, and M. Givskov. 1999. Surface motility of Serratia liquefaciens MG1. J. Bacteriol. 181:1703-1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fenchel, T. 2001. Eppur si muove: many water column bacteria are motile. Aquat. Microb. Ecol. 24:197-201. [Google Scholar]
- 16.Friedrich, U., M. Schallenberg, and C. Holliger. 1999. Pelagic bacteria-particle interactions and community-specific growth rates in four lakes along a trophic gradient. Microb. Ecol. 37:49-61. [DOI] [PubMed] [Google Scholar]
- 17.Gram, L., H.-P. Grossart, A. Schlingloff, and T. Kiørboe. Quorum sensing in marine snow bacteria? Production of acylated homoserine lactones by Roseobacter strains isolated from marine snow. Appl. Environ. Microbiol., in press. [DOI] [PMC free article] [PubMed]
- 18.Grossart, H.-P., L. Riemann, and F. Azam. 2001. Bacterial motility in the sea and its ecological implications. Aquat. Microb. Ecol. 25:247-258. [Google Scholar]
- 19.Grossart, H.-P., and M. Simon. 1998. Bacterial colonization and microbial decomposition of limnetic organic aggregates (lake snow). Aquat. Microb. Ecol. 15:127-140. [Google Scholar]
- 20.Grossart, H.-P., and H. Ploug. 2001. Microbial degradation of organic carbon and nitrogen on diatom aggregates. Limnol. Oceanogr. 46:267-277. [Google Scholar]
- 21.Jackson, G. A. 1989. Simulation of bacterial attraction and adhesion to falling particles in an aquatic environment. Limnol. Oceanogr. 34:514-530. [Google Scholar]
- 22.Kiørboe, T. 2001. Formation and fate of marine snow. Small-scale processes with large-scale implication. Scientia Marina 65(Suppl. 2):57-71. [Google Scholar]
- 23.Kiørboe, T., and G. A. Jackson. 2001. Marine snow, organic solute plumes, and optimal chemosensory behavior of bacteria. Limnol. Oceanogr. 46:1309-1318. [Google Scholar]
- 24.Kiørboe, T., H. Ploug, and U. H. Thygesen. 2001. Fluid motion and solute distribution around sinking aggregates. I. Small-scale fluxes and heterogeneity of nutrients in the pelagic environment. Mar. Ecol. Prog. Ser. 211:1-13. [Google Scholar]
- 25.Kiørboe, T., and J. Titelman. 1998. Feeding, prey selection and prey encounter mechanisms in the heterotrophic dinoflagellate Noctiluca scintillans. J. Plankton Res. 20:1615-1636. [Google Scholar]
- 26.Knoll, S., W. Zwisler, and M. Simon. 2001. Bacterial colonization of early stages of limnetic diatom microaggregates. Aquat. Microb. Ecol. 25:141-150. [Google Scholar]
- 27.Laroche, D., A. F. Vezina, M. Levasseur, M. Gosselin, J. Stefels, M. D. Keller, P. A. Matrai, and R. L. J. Kwint. 1999. DMSP synthesis and exudation in phytoplankton: a modeling approach. Mar. Ecol. Prog. Ser. 180:37-49. [Google Scholar]
- 28.Levandowsky, M., J. Klafter, and B. S. White. 1988. Swimming behavior and chemosensory responses in the protistan microzooplankton as a function of the hydrodynamic regime. Bull. Mar. Sci. 43:768-773. [Google Scholar]
- 29.Long, R. A., and F. Azam. 2001. Antagonistic interactions among marine pelagic bacteria. Appl. Environ. Microbiol. 67:4975-4983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mitchell, J. G., L. Pearson, A. Bonazinga, S. Dillon, H. Khouri, and R. Paxinos. 1995. Long lag times and high velocities in the motility of natural assemblages of bacteria. Appl. Environ. Microbiol. 61:877-882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mitchell, J. G., L. Pearson, and S. Dillon. 1996. Clustering of marine bacteria in seawater enrichments. Appl. Environ. Microbiol. 62:3716-3721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moeseneder, M. M., C. Winter, and G. J. Herndl. 2001. Horizontal and vertical complexity of attached and free-living bacteria of the eastern Mediterranean Sea, determined by 16S rDNA and 16S rRNA fingerprints. Limnol. Oceanogr. 46:95-107. [Google Scholar]
- 33.Osborn, T. 1996. The role of diffusion for copepods with feeding currents. J. Plankton Res. 18:185-195. [Google Scholar]
- 34.Paerl, H. W. 1975. Microbial attachment to particles in marine and freshwater systems. Microb. Ecol. 2:73-83. [DOI] [PubMed] [Google Scholar]
- 35.Ploug, H., and H.-P. Grossart. 1999. Bacterial production and respiration in suspended aggregates—a matter of the incubation method. Aquat. Microb. Ecol. 20:21-29. [Google Scholar]
- 36.Ploug, H., and H.-P. Grossart. 2000. Bacterial growth and grazing on diatom aggregates: respiratory carbon turnover as a function of aggregate size and sinking velocity. Limnol. Oceanogr. 45:1467-1475. [Google Scholar]
- 37.Ploug, H., H.-P. Grossart, F. Azam, and B. B. Jørgensen. 1999. Photosynthesis, respiration, and carbon turnover in sinking marine snow from surface waters of Southern California Bight: implications for the carbon cycle in the ocean. Mar. Ecol. Prog. Ser. 179:1-11. [Google Scholar]
- 38.Ploug, H., S. Heitanen, and J. Kuparinen. 2002. Diffusion and advection within and around sinking, porous diatom aggregates. Limnol. Oceanogr. 47:1129-1136.
- 39.Smith, D. C., M. Simon, A. L. Alldredge, and F. Azam. 1992. Intensive hydrolytic activity on marine aggregates and implications for rapid particle dissolution. Nature 359:139-141. [Google Scholar]
- 40.Turley, C. M., and P. J. Mackie. 1994. The biogeochemical significance of attached and free-living bacteria and the flux of particles in the deep northeastern Atlantic Ocean. Mar. Ecol. Prog. Ser. 115:191-203. [Google Scholar]
- 41.Unanue, M., I. Azúa, J. M. Arrieta, A. Labirua-Iturburu, L. Egea, and J. Iriberri. 1998. Bacterial colonization and ectoenzymatic activity in phytoplankton-derived model particles: cleavage of peptides and uptake of amino acids. Microb. Ecol. 35:136-146. [DOI] [PubMed] [Google Scholar]
- 42.Viswanathan, G. M., V. Afanasyev, S. V. Buldyrev, E. J. Murphy, P. A. Prince, and H. E. Stanley. 1996. Lévy flight search patterns of wandering albatrosses. Nature 381:413-415. [DOI] [PubMed] [Google Scholar]
- 43.Wörner, U., H. Zimmerman-Timm, and H. Kausch. 2000. The succession of protists on estuarine aggregates. Microb. Ecol. 40:209-222. [DOI] [PubMed] [Google Scholar]