Abstract
Microbial diversity studies based on the cloning and sequencing of DNA from nature support the conclusion that only a fraction of the microbial diversity is currently represented in culture collections. Out of over 40 known prokaryotic phyla, only half have cultured representatives. In an effort to culture the uncultured phylotypes from oligotrophic marine ecosystems, we developed high-throughput culturing procedures that utilize the concept of extinction culturing to isolate cultures in small volumes of low-nutrient media. In these experiments, marine bacteria were isolated and cultivated at in situ substrate concentrations—typically 3 orders of magnitude less than common laboratory media. Microtiter plates and a newly developed procedure for making cell arrays were employed to raise the throughput rate and lower detection sensitivity, permitting cell enumeration from 200-μl aliquots of cultures with densities as low as 103 cells/ml. Approximately 2,500 extinction cultures from 11 separate samplings of marine bacterioplankton were screened over the course of 3 years. Up to 14% of the cells collected from coastal seawater were cultured by this method, which was 14- to 1,400-fold higher than the numbers obtained by traditional microbiological culturing techniques. Among the microorganisms cultured were four unique cell lineages that belong to previously uncultured or undescribed marine Proteobacteria clades known from environmental gene cloning studies. These cultures are related to the clades SAR11 (α subclass), OM43 (β subclass), SAR92 (γ subclass), and OM60/OM241 (γ subclass). This method proved successful for the cultivation of previously uncultured marine bacterioplankton that have consistently been found in marine clone libraries.
The term “the great plate count anomaly” was coined by Staley and Konopka in 1985 (31) to describe the difference in orders of magnitude between the numbers of cells from natural environments that form colonies on agar media and the numbers countable by microscopic examination (18). Marine ecosystems are a well-studied example of this phenomenon: only 0.01 to 0.1% of oceanic marine bacterial cells produce colonies by standard plating techniques (19). There are numerous explanations for this anomaly. For example, species that would otherwise be “culturable” may fail to grow because their growth state in nature, such as dormancy, prevents adjustment to conditions found in the medium used for the plate counts (13). This hypothesis does not explain the substantial discrepancy between 16S rRNA genes recovered from seawater directly by cloning and those of the readily cultured marine taxa (22, 34). Another explanation for the “great plate count anomaly” is that many of the microbial species that dominate in natural settings are not adapted for growth in media containing high concentrations of complex organic carbon. Many microorganisms may need oligotrophic or other fastidious conditions to be successfully cultured. There are many examples of microbial strains that are common in nature, but can only be cultivated by specialized techniques (3, 7, 9, 12, 15, 20, 25, 26, 29, 30, 37, 40).
Button and colleagues pioneered an approach that has been successful in isolating novel oligotrophic, heterotrophic cells from marine ecosystems (10). This method uses unamended environmental water as the medium and is often referred to as “extinction culturing” to distinguish it from dilution culturing, which also uses natural water, but involves complex microbial communities (1, 11, 23). Their approach was to dilute natural communities of microorganisms to a known number, ranging from 1 to 10 cells per tube, and then examine these potential cultures for microbial growth by flow cytometry, which is effective for counting very dilute populations of cells. By this method, bacterioplankton culturability from 2 to 60% was reported for marine waters around Alaska and The Netherlands (10). This work resulted in the description of two new oligotrophic bacterioplankton, Sphingomonas alaskensis and “Cycloclasticus oligotrophus” (9, 29, 37, 39). However, this extinction culturing method is relatively laborious. The isolates that have been obtained by this method are of considerable scientific interest, but they are few in number.
The goal of this study was to develop high-throughput culturing (HTC) methods that would enable a large number of extinction cultures to be identified so that the efficacy of this approach could be assessed with a larger sampling of isolates. Over the course of 3 years and 11 separate samplings of marine bacterioplankton, 2,484 extinction wells were examined for growth. The results indicate that these newly developed HTC techniques yield isolates of many novel microbial strains, including members of previously uncultured groups that are believed to be abundant in coastal seawater.
MATERIALS AND METHODS
HTC technique.
A series of protocols and techniques were developed to allow the efficient screening of a large number of extinction culture attempts for growth and subsequent identification (Fig. 1). Slight variations of the method were performed during the development of these HTC techniques over the course of 3 years, but the overall approach remained constant. Microtiter plates were used to culture cells, and cell arrays were made to allow efficient screening of the plates for growth. The cultures acquired were designated with HTC collection (HTCC) numbers.
FIG. 1.
Flow chart of HTC procedures. DMSO, dimethyl sulfoxide.
Preparation of media.
Water for media was collected on the south side of the southern jetty in Newport, Oreg., at high tide with a bucket on 19 March 1998 8 km (44°39.1N, 124°10.6W) offshore from the mouth of Yaquina Bay, Oreg., with a Niskin bottle deployed at 5 m on 7 June 2000. On the day water was collected, it was filtered through a 0.2-μm-pore-diameter Supor membrane and immediately autoclaved. In order to restore the bicarbonate buffer lost during autoclaving, the seawater was sparged with sterile CO2 for at least 6 h, followed by sterile air for at least 12 h. Acid-washed polycarbonate containers were used for media and live samples whenever possible. Dissolved organic carbon concentrations of the seawater media were 107.1 μM (standard deviation [SD], 1.1) for the 19 March 1998 collection, determined with a Shimadzu TOC-500, and 91.6 μM (SD, 1.6) for the 7 June 2000 collection, determined with a Shimadzu TOC-5000A (Shimadzu Co., Kyoto, Japan). Before each use, the liquid media were checked for sterility by directly counting cells stained with 4′,6-diamidino-2-phenylindole (DAPI) as described by Turley (36), except that 1% formaldehyde was used.
Inoculum collection, dilution, and incubation.
Water samples for inocula were collected on the south side of the southern jetty in Newport, Oreg., at high tide with a bucket and at 8 km (44°39.1N, 124°10.6W) and 25 km (44°39.1N, 124°24.7W) offshore from the mouth of Yaquina Bay, Oreg., with a Niskin bottle deployed at 5 m. The water was held in darkness at ambient sea surface temperatures until the processing of samples began, within 1 to 4 h after collection from the jetty and within 9 h after collection off the boat to avoid bottle effects (14). To determine the bacterioplankton cell densities of the inocula, direct cell counts were done by DAPI staining, where at least 300 cells were counted per filter on triplicate filters. To determine viable cell counts (i.e., culturability) by traditional methods, inocula of 50 or 100 μl of seawater were applied to spread plates of MA2216 (Difco Laboratories, Detroit, Mich.), Marine R2A (R2A) (34), and a 1/10 dilution of Marine R2A (1/10R2A). Inoculum samples were diluted into the prepared seawater medium and distributed as 1-ml aliquots into 48-well non-tissue-culture treated Polystyrene plates (Becton Dickinson, Franklin Lakes, N.J.) to a final average inoculum ranging from 1.1 to 5.0 cells per well. At least one control plate was made for each sample collection by distribution of 1-ml aliquots of uninoculated medium. The 48-well plates and agar plates were incubated in the dark at 16°C. The extinction cultures were incubated for 3 weeks, and the agar cultures were incubated until colonies were large enough to count, about 1 week for MA2216 and up to 8 weeks for 1/10R2A.
Detection of growth by using cell arrays.
A cell array was made from each 48-well plate to examine wells for growth. Two hundred microliters from each well in the plate was filtered into the corresponding chamber of a 48-array filter manifold of custom design manufactured by HyTek Plastics, Corvallis, Oreg. Cells were then DAPI stained and vacuum filtered onto a 48-by-60-mm 0.2-μm-pore-diameter white polycarbonate membrane (cut from 8-by-10-in. sheets; Whatman Nuclepore, Newton, Mass.). The membrane was laid on an oiled 75-by-50-mm slide (Corning Glass Works, Corning, N.Y.) and covered with a 48-by-60-mm coverglass (Erie Scientific, Portsmouth, N.H.). The diameter of each sector of the array was 2 mm, which enabled the detection of a culture with a cell titer as low as 1.3 × 103 cells/ml when 200 μl of sample was filtered. The array was then scored for growth by fluorescence microscopy. Cell titers were estimated by counting five random fields within each positive sector.
Culturability statistics.
Percent culturability was determined by the equation for estimation of culturability, V = −ln(1 − p)/X, and the theoretical number of pure cultures was estimated by the equation u = −n(1 − p) ln(1 − p) described by Button and colleagues (10), where u is an estimation of the expected number of pure cultures, n is the number of inoculated wells, V is estimated culturability, p is the proportion of wells positive for growth (wells positive for growth/total inoculated wells), and X is the initial inoculum of cells added per well. To calculate the error, first, the exact lower and upper 95% confidence limits for the binomial proportion (p) were determined by using the SAS package version 6.12 (SAS Institute Inc.). Next, these exact limits were put into the culturability equation and pure culture equation in place of the term p to give the exact lower and upper 95% confidence limits for percent culturability and the theoretical number of pure cultures.
RFLP analysis and sequencing of HTCC isolates.
A subset of 56 HTCC isolates were identified by restriction fragment length polymorphism (RFLP) and rRNA gene sequencing methods. One hundred or 200 μl of culture was put through two cycles of freezing and thawing to promote cell lysis and concentrated in a 10,000-molecular-weight Vivaspin concentrator (Vivascience, Stonehouse, United Kingdom). Some samples were also treated with 150 μl of GES lysis buffer (5 M guanidine thiocyanate, 100 mM EDTA, 0.5% Sarkosyl) while in the concentrator. The lysates were then rinsed three times with 200 μl of Ultrapure water (Specialty Media, Phillipsburg, N.J.) to remove medium salts and lysis buffer. The final volumes of the concentrated samples ranged from 10 to 30 μl. Two to three negative controls (the same procedure with no added culture) were run with each set of concentrated samples.
16S rRNA genes were amplified by nested PCR. Two to 5 μl of each concentrated sample was added to the first PCR, which had a 20-μl reaction volume, and 2 to 5 μl of the first PCR was added to the second PCR, which had a 60- to 100-μl reaction volume. Twenty-five to 33 cycles were used for each PCR, for a total of 50 to 66 cycles of amplification. The PCR cocktail for both reactions contained 0.025 U of Taq per μl (Promega, Madison, Wis., or MBI Fermentas, Hanover, Md.), 5% acetamide, 1.5 mM Mg2+, 200 nM each primer, 220 μM deoxynucleoside triphosphates (dNTP), and 1× PCR buffer (Promega or MBI Fermentas). The PCR cocktail was treated with UV irradiation to reduce the contamination levels present in the reagents (6, 24). The length of UV treatment needed was empirically determined by amplifying a set of negative and positive controls. The amplification conditions for both PCRs were 94°C denaturation for 30 s, 50 to 55°C (depending on primers used) annealing for 1 min, and 72°C extension for 2 min. The second PCR primer set had at least one primer that amplified from a position internal to the set of primers used in the first PCR. The primers used were 8F (5′-AGR GTT TGA TCM TGG CTC AG-3′), 519F (5′-CAG CMG CCG CGG TAA TWC-3′), 1395R (5′-ACG GGC GGT GTG TRC-3′), 1492R (5′-GGT TAC CTT GTT ACG ACT T-3′), and 1522R (5′-AAG GAG GTG ATC CAN CCR CA-3′), which are variations of commonly used primers that target bacteria or prokaryotes (21). The nested set of primers most frequently used was made up of 519F/1492R and 519F/1395R, but other variations of the listed primers were also used. Three negative controls and positive controls with 108, 2,000, 200, and 20 copies of the 16S rRNA gene from the clone SAR242 were run in each PCR set. All primers used have no mismatches to the SAR242 sequence, except for 1492R, which does not match the first and third bases on the 5′ end (nonpriming end). The concentration of the positive control DNA was measured in a Shimadzu UV160U spectrophotometer (Shimadzu Co., Kyoto, Japan). The 20-copy-positive control could be routinely amplified with a total of 50 to 66 cycles of nested PCR.
RFLP of the PCR product was done with the restriction enzymes MboI and HaeIII (MBI Fermentas) (38). HTCC isolates were determined to be a mix of more than one species if RFLP bands from each digest added up to two or more times the length of the expected PCR product. The cultures with fragments that added up to the expected PCR product length were grouped based on matching RFLP patterns, and at least one culture from each RFLP group was sequenced and phylogenetically analyzed.
Before sequencing, the PCR products were purified with the Qiaquick PCR purification kit (Qiagen, Valencia, Calif.). The concentration of the purified product was measured in a Shimadzu UV160U or BioSpec-1601 spectrophotometer (Shimadzu Co., Kyoto, Japan). The purified PCR product was then sequenced by an ABI 373A or 377 automated sequencer (Applied Biosystems, Foster City, Calif.).
Phylogenetic analysis.
HTCC sequences were aligned and masked in ARB (32). Phylogenetic analyses were performed with ARB and PAUP∗ (35). Phylogenetic trees were inferred by neighbor joining with the Jukes and Cantor model to estimate evolutionary distances. Bootstrap values were obtained in PAUP∗ from a consensus of 1,000 neighbor-joining trees. Short sequences of HTCC isolates were added to the tree by using the parsimony insertion tool in ARB. The percent similarity of sequences was determined with the distance matrix tool in ARB; ambiguous bases were not included.
Recovery of HTCC isolates from frozen storage.
The probability of recovering HTCC isolates from frozen storage has not been systematically investigated, and not all cultures were saved for further study. However, isolates from three of the four significant phylogenetic clades in this study, HTCC202 (OM43 clade), HTCC230 and HTCC234 (SAR92 clade), and HTCC223 and HTC227 (OM60/OM241 clade), have been successfully transferred from the initial well, propagated, and stored. Cells were stored in 7% dimethyl sulfoxide and/or 10% glycerol.
DAPI-stained cell images.
Images were obtained with a Hamamatsu ORCA-ER cooled interline charge-coupled device camera (5 Mz) mounted on a Leica DMRB microscope. IPLab Spectrum 3.5 image analysis software was used to acquire images.
Nucleotide sequence accession numbers.
The sequences of the HTCC isolates used in the phylogenetic analyses have been deposited in GenBank under accession no. AY102012 to AY102033.
RESULTS
HTC.
Our general approach to HTC is outlined (Fig. 1). This method, which allows a large number of culture attempts to be efficiently screened for growth and identified, was successful in bringing four major uncultured or undescribed groups of bacterioplankton into culture. These four groups include SAR11 (α subclass) (16), OM43 (β subclass) (27), SAR92 (γ subclass) (8), and OM60/OM241 (γ subclass) (27).
Culturability statistics.
Two hundred fifty-three extinction culture wells were scored positive for growth out of 2,484 wells screened for 3 years and 11 sample collections. A culturability range of 0.4 to 14.3% was calculated for the different sample collections (Table 1). The average culturability for the six samples collected between late May and mid-July was 8.8%, and the average culturability for the five samples collected between early October and early April was 1.2%. Comparisons of culturability were made between the HTC method and traditional plating on nutrient-rich agar media; the culturability ranged from 1.4 to 120 times higher by HTC methods (Table 1). In addition, MA2216 and R2A agar plates were spotted with the first 143 cultures grown from water collected during the summer of 1998 to determine if they had the ability to grow on these media. Only three grew on MA2216, and a fourth grew on R2A; none of these four cultures grew on both agar media (data not shown).
TABLE 1.
Extinction culturability statistics compared to traditional culturability counts
| Date (mo-day-yr) and location of inoculation samplea | Inoculum sample (cells/ml) | Avg no. of cells/well | Total no. of wells inoculated | No. of positive wellsb | Culture designations | % Culturabilityc | % Culturability on nutrient-rich agard
|
||
|---|---|---|---|---|---|---|---|---|---|
| 1/10R2A | R2A | MA2216 | |||||||
| 5-21-98, J | 1.1 × 106 | 1.1 | 144 | 7 | HTCC1-7 | 4.5 (1.8, 9.3) | — | — | — |
| 6-5-98, J | 1.5 × 106 | 1.5 | 192 | 37 | HTCC8-44 | 14.3 (10.0, 19.7) | — | — | — |
| 7-6-98, 8 km | 3.7 × 106 | 3.7 | 192 | 62 | HTCC45-106 | 10.5 (8.0, 13.5) | — | — | — |
| 7-6-98, 25 km | 1.5 × 106 | 1.5 | 192 | 37 | HTCC107-143 | 14.3 (10.0, 19.7) | — | — | — |
| 6-17-99, J | 5.6 × 106 | 3.0 | 192 | 21 | HTCC144-164 | 3.9 (2.4, 5.9) | — | — | — |
| 10-29-99, J | 1.9 × 106 | 3.0 | 192 | 10 | HTCC165-174 | 1.8 (0.9, 3.3) | — | — | — |
| 12-21-99, J | 8.1 × 105 | 5.0 | 384 | 10 | HTCC175-184 | 0.5 (0.3, 1.0) | — | — | — |
| 1-26-00, J | 1.1 × 106 | 5.0 | 192 | 11 | HTCC185-191, 193-196 | 1.2 (0.6, 2.1) | 0.01 | 0.01 | 0.02 |
| 4-5-00, J | 9.0 × 105 | 5.0 | 192 | 20 | HTCC197-216 | 2.2 (1.3, 3.4) | — | 0.15 | 0.12 |
| 7-12-00, J | 1.9 × 106 | 3.0 | 228 | 33 | HTCC217-233, 236-251 | 5.2 (3.6, 7.3) | 0.98 | 0.15 | 0.12 |
| 10-9-00, 8 km | 1.3 × 106 | 3.0 | 384 | 5 | HTCC252-256 | 0.4 (0.1, 1.0) | 0.29 | 0.09 | 0.02 |
Samples were collected on the date indicated from the jetty (J) or 8 or 25 km out from the mouth of Yaquina Bay, Oreg.
Wells were scored for growth after 3 weeks of incubation at 16°C.
Ninety-five percent confidence intervals are shown in parentheses.
Inoculum was the same as that used for the microtiter plates. —, not determined.
Detection of growth and cell densities.
The cell densities of the HTCC cultures ranged from 1.3 × 103 to 1.6 × 106 cells per ml, with a mean of 1.1 × 105 cells per ml and a median of 3.0 × 104 cells per ml. The minimum density for a culture to be detectable was 1.3 × 103 cells per ml. This range of cell densities is the result of as few as 10.0 to as many as 23.3 doublings during the 3-week incubation period, assuming only one cell from the initial inoculum grew in the well (Table 2). The 253 wells that showed cell growth fall into four categories of cell density (Table 2). The maximum cell concentration attained (1.6 × 106 cells/ml) is similar to the natural bacterial numbers in seawater, which ranged from 8.1 × 105 to 5.6 × 106 cells per ml for the 11 inoculum samples collected.
TABLE 2.
Cell densities and inferred doublings attained after 3 weeks of incubation
| Final no. of cells/ml | No. of culturesa | No. of inferred doublingsb |
|---|---|---|
| 1.0 × 103-9.9 × 103 | 66 | 10.0-13.3 |
| 1.0 × 104-9.9 × 104 | 120 | 13.3-16.6 |
| 1.0 × 105-9.9 × 105 | 62 | 16.6-19.9 |
| 1.0 × 106-9.9 × 106 | 5 | 19.9-23.3 |
Out of 253 cultures.
This inference is based on the assumption that only one inoculated cell in each well grew.
Imaging of the DAPI-stained isolates revealed unicellular organisms that were generally of small size. The SAR11 clade isolate HTCC150 was a small, curved rod (ca. 1 to 0.8 μm by 0.3 to 0.2 μm). The OM43 clade isolates HTCC163 and HTCC175 were short rods (ca. 0.8 to 0.5 μm by 0.5 μm). The SAR92 clade isolates HTCC148, HTCC151, and HTCC154 were short rods (ca. 1 to 0.7 μm by 0.7 to 0.5 μm). OM60/OM241 clade isolate HTCC160 was an irregularly shaped coccus that occasionally formed doublets and more rarely chains of three (ca. 0.7 by 0.7 μm). These measurements are subject to sizeable error, since these small cells are at or approach the resolution of visible light microscopes. The cells have been stained with a DNA staining dye and have been fixed with formaldehyde. The images shown are from the original extinction dilutions that yielded the four previously uncultured and undescribed groups (Fig. 2).
FIG. 2.
Fluorescence microscopy images of several of the novel isolates. The cells were stained with DAPI. Size bars, 1 μm.
Phylogenetic analysis and culture identification.
Uncultured or undescribed groups SAR11, OM43, SAR92, and OM60/OM241 accounted for the majority of cultures that were identified out of a subset of 56 cultures (Table 3). All cultured cells from 13 48-well plates (56 cultures) were chosen to represent 5 different sampling months to minimize biases that might emerge as a result of seasonal variation in bacterioplankton abundance. Forty-seven of the 56 cultures were identified; of the 9 cultures that were not identified, 7 were found to be unknown mixtures of several cell types based on RFLP analysis, and 2 did not amplify under the conditions used. There were a total of eight mixed cultures; HTCC149 was found to be a mix of cells from the SAR11 clade and unknown cells. The failure of two cultures to amplify is probably attributable to problems with the DNA extractions and/or low cell densities in the cultures. A considerable effort was made to ensure that these lineages did not fail to amplify because of mismatches to amplification primers. The theoretical statistical estimation for the number of pure cultures versus mixed cultures that should be acquired was consistent with the RFLP analysis (Table 3).
TABLE 3.
Phylogenetic identification and pure culture statistics for 56 cultures
| Inoculation date (mo-day-yr) | No. of wells screened | No. of cultures detected | Theoretical no. of pure culturesa | No. with culture identification
|
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| SAR11 | OM43 | SAR92 | OM60/OM241 | Otherc | Mixed culture | Not identified | ||||
| 5-21-98 | 96 | 7 | 6.7 (2.8, 12.8) | 5 | 2 | |||||
| 6-17-98 | 96 | 11 | 10.3 (5.5, 16.8) | 2 | 1 | 4 | 2 | 3b | ||
| 10-29-99 | 96 | 10 | 9.5 (4.8, 15.9) | 8 | 1 | 1 | ||||
| 1-26-00 | 192 | 11 | 10.7 (5.5, 18.2) | 7 | 2 | 1 | 1 | |||
| 7-12-00 | 144 | 17 | 16.0 (9.8, 23.7) | 3 | 3 | 8 | 1 | 2 | ||
| Total | 624 | 56 | 53.4 (41.2, 67.4) | 2 | 16 | 15 | 3 | 11 | 8 | 2 |
Statistical estimation of the theoretical number of pure cultures acquired with 95% confidence interval. The total 53.4 was determined independently with 624 wells and 56 cultures in the pure culture equation.
One SAR11 culture was mixed with an unknown cell type (RFLP analysis) and is also included under the heading “Mixed culture.”
“Other” indicates cultures that fall into previously cultured groups.
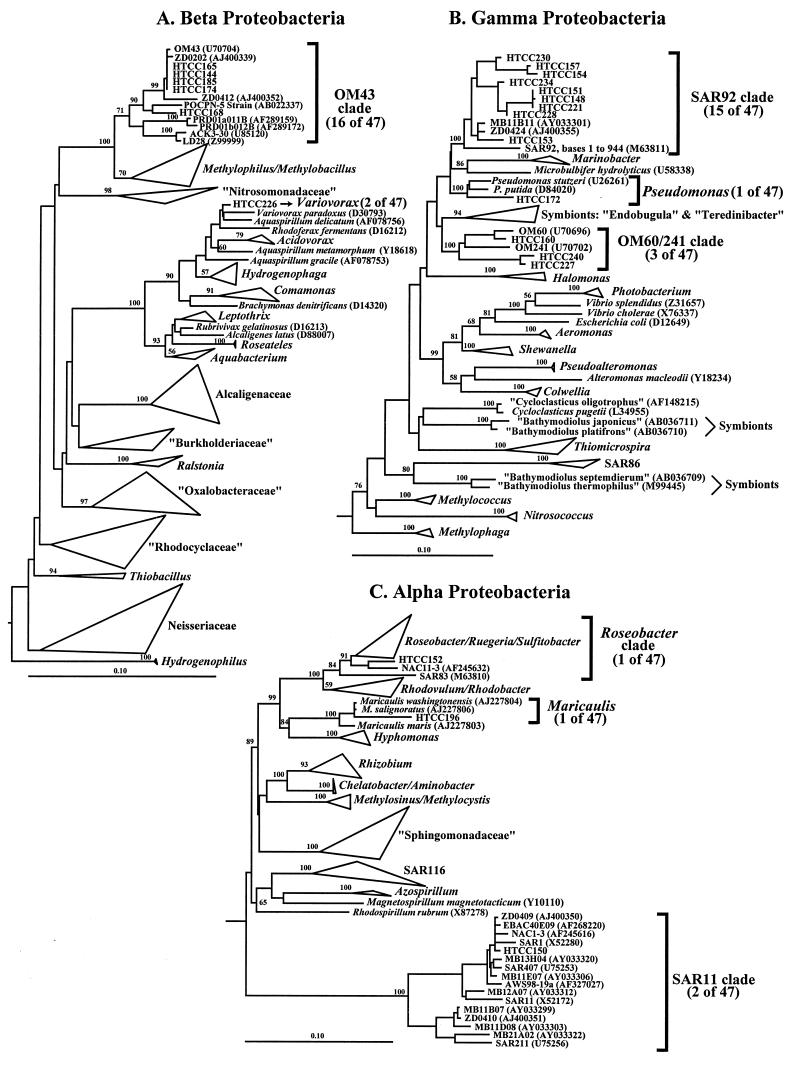
Of the 47 identified cultures, 4 were α-Proteobacteria (Fig. 3C). Two belonged to the SAR11 clade, and one each was from the genus Maricaulis and the Roseobacter clade. Eighteen isolates were identified as β-Proteobacteria (Fig. 3A). These included members of two clades, 16 isolates from the OM43 clade, and two related to the genus Variovorax. Nineteen cultures were γ-Proteobacteria (Fig. 3B). These included three subgroups: the SAR92 clade (15 isolates), the OM60/OM241 clade (3 isolates), and one group from the genus Pseudomonas. Six isolates were members of the phylum Bacteroidetes.
FIG.3.
Neighbor-joining trees showing phylogenetic relationships among the 16S rRNA genes of HTCC isolates compared to those of representative species and environmental clones. Scale bars indicate 0.1 change per nucleotide. Bootstrap values below 50 are not shown. Short sequences (approximately 600 bp) of HTCC isolates were added to the trees by using the parsimony insertion tool in ARB. HTCC230 and HTCC234 are close to full length and were put in the original tree. In parentheses next to HTCC isolates is the number of total cultures from the subset of 47 identified cultures that are included in that clade. However, not all of the HTCC sequences used in the tree are part of the subset of 47 identified cultures. (A) α-Proteobacteria phylogenetic tree. β- and γ-Proteobacteria were used to root the tree; 1,051 characters were used to infer the tree. (B) β-Proteobacteria phylogenetic tree. γ-Proteobacteria isolates were used to root the tree; 789 characters were used to infer the tree. (C) γ-Proteobacteria phylogenetic tree. β-Proteobacteria isolates were used to root the tree; 1,042 characters were used to infer the tree.
The 16S rRNA sequence for the SAR92 clone (M63811) was found to be a chimera. From sequence positions 1 to 944, SAR92 is a member of the γ-Proteobacteria; from positions 1120 to 1354, it is a member of the α-Proteobacteria. The identity of the sequence from 945 to 1119 is ambiguous. The γ portion of the SAR92 clone sequence represents a previously uncultured phylogenetic clade, we have termed the SAR92 clade.
Percent similarities of the sequenced HTCC cultures from the four previously uncultured or undescribed phylogenetic clades were determined, in which several of the sequences were close matches to clones in GenBank (Table 4) (2, 5, 33). Sequences of oligotrophic isolates from the OM43 and OM60/OM241 clades were recently deposited into GenBank as strain POCPN-5 (AB022337) and KI89C (AB022713), respectively, by N. Katanozaka and I. Yoshinaga (unpublished data). HTCC isolates from the four distinct phylogenetic clades SAR11, OM43, SAR92, and OM60/OM241 are more similar to cloned sequences from these clades than to those from previously cultured species, with the exception of HTCC168, which is 99.8% similar to the isolate POCPN-5.
TABLE 4.
16S rRNA sequence similarities of HTCC isolates from previously uncultivated clades to the nearest neighbors in GenBank
| Clade | HTCC isolate | E. coli positiona | Clone | % Similarity |
|---|---|---|---|---|
| SAR11 | 150 | 524-1362 | ZD0409 | 99.8 |
| OM43 | 144 | 712-1386 | OM43 | 100 |
| 165 | 710-1386 | OM43 | 100 | |
| 168 | 711-1371 | POCPN-5b | 99.8 | |
| 174 | 713-1362 | OM43 | 100 | |
| 185 | 719-1378 | OM43 | 100 | |
| SAR92 | 148 | 716-1384 | MB11B11 | 96.9 |
| 151 | 716-1383 | MB11B11 | 96.9 | |
| 153 | 713-1360 | MB11B11 | 99.4 | |
| 154 | 716-1360 | Artic97A-6 | 99.7 | |
| 157 | 716-1351 | Artic97A-6 | 99.5 | |
| 221 | 716-1346 | MB11B11 | 96.7 | |
| 228 | 707-1266 | SAR92 | 96.5 | |
| 230 | 28-1537 | MB11B11 | 97.0 | |
| 234 | 28-1537 | MB11B11 | 97.0 | |
| OM60/OM241 | 160 | 713-1383 | OM60 | 100 |
| 227 | 705-1373 | MERTZ-2CM-38 | 97.0 | |
| 240 | 716-1360 | MERTZ-2CM-38 | 96.9 |
Position of the bases used to determine percent similarity.
POCPN-5 is a cultured strain, not a clone.
DISCUSSION
Culturability statistics and detection of growth.
The goal of this study was to evaluate a culturing format for the high-throughput isolation of uncultured strains of bacterioplankton that are commonly found in gene clone libraries from marine environments. The use of microtiter dishes and a novel technique for making cell arrays enabled us to achieve a higher throughput rate, shorten incubation times, and raise sensitivity for the detection of cells with low growth rates relative to those in previous studies that employed the concept of extinction culturing in natural media.
The percentage of cells that could be cultured by the HTC approach was several orders of magnitude higher than that obtained by culturing on agar plates. Ferguson and colleagues found that the percentage of microbial cells in seawater that could be cultured on a rich nutrient agar medium (MA2216) increased from <0.1% to 13% after 16 h and to 41% after 32 h of confinement in a 4-liter bottle at ambient sample collection temperature (14). Our results cannot easily be explained by this “bottle effect,” because (i) the cells were diluted into the 48-well plates between 1 and 4 h after collection from the jetty and within 9 h after collection from the boat; (ii) readily culturable genera, such as Pseudomonas and Vibrio, were rarely detected in our cultures; and (iii) four previously undescribed lineages were grown by our culture method.
Culturability was observed to be higher in the summer months (8.8%) than in the winter months (1.2%). There are at least two plausible explanations for this observation. First, bacterioplankton cells may be in a dormant state during the winter and either fail to grow or need longer incubation times for growth to be detected. Alternatively, the predominant strains or species of cells present in the winter could be organisms that are unable to grow under the laboratory conditions we provided, which were more similar to summer environmental conditions. The seawater medium used for these experiments was collected during the spring and summer months, and our incubation temperature of 16°C is closer to the summer temperature range of 10.0 to 14.7°C versus the winter range of 9.5 to 10.7°C for the 11 samples collected. Also, the summer months off the coast of Oregon are dominated by upwelling events that bring cool nutrient-rich water to the surface, which subsequently induces large algal blooms. During the winter, the water off the coast is diluted by the Columbia River water plume, mixed by frequent storm events, and not subject to algal blooms. The bacterioplankton that predominate during the summer may be better adapted to the higher nutrient levels and/or nutrient types provided and therefore more amenable to cultivation by the methods we used.
Based on RFLP analysis, the majority of the cultures identified were pure cultures. Theoretical estimates of the number of pure cultures expected were consistent with the number and proportion of pure cultures observed by RFLP analysis; 8 of 54 cultures studied in this manner were mixed cultures. This would indicate that most cultures were the result of only one of the inoculated cells growing in the well. However, RFLP analysis would miss mixed cultures with differential cell lysis or where the primers used for PCR amplification fail to amplify all cell types in a culture. In addition, a dominant cell type may preferentially amplify and thus appears as a pure culture in an RFLP analysis.
Phylogenetic analysis and culture identification.
Phylogenetic identification of the isolates provided striking evidence that extinction culturing in microtiter dishes, with natural seawater and low thresholds of detection, results in the cultivation of microbial groups that appear in environmental clone libraries, but have not been previously detected in culture. The SAR11 and SAR92 clades, which were isolated in this study (transiently, in the case of SAR11) have previously been detected only by environmental rRNA gene cloning. rRNA gene sequences from isolates of other previously uncultivated clades, OM43 and OM60/OM241 (strains POCPN-5 and KI89C, respectively), were recently deposited in GenBank by other investigators.
Some of the isolates that were cultured belong to phylogenetic clades that are highly abundant in marine clone libraries. Clones in the SAR11 clade are abundant in clone libraries made from surface marine waters around the world (17). The OM43 clade is a sister clade to a group of marine methylotrophs that includes Methylophilus and Methylobacillus and is commonly found in clone libraries from coastal sites, but not the open ocean (28). Methylophilus and Methylobacillus are classified as type I methylotrophs, which use the ribulose monophosphate (RuMP) pathway for carbon assimilation. The OM60/OM241 clades are frequently found in coastal marine clone libraries, and the SAR92 clade is found in open ocean as well as coastal clone libraries. In subsequent work (unpublished results), several strains obtained by these procedures were scaled up to 20-liter volumes for further study.
Several other major uncultured groups that are thought to be abundant in surface seawater, such as the SAR86 and SAR116 clusters, did not appear among the HTCC isolates. Further innovations in the HTC approach will be needed to close the gap between culture collections and the microbial species dominating marine bacterioplankton communities. The approach we describe can be used to target specific bacterial groups for cultivation by screening cultures for the microorganisms of interest by means of fluorescence in situ hybridization, so that uncultured targets can be sought in a deliberate manner. The SAR86 cluster has recently been linked to a bacterial rhodopsin gene that facilitates light-mediated proton translocation (4). Incubation of extinction cultures under various conditions, including different sources of carbon, light, and other matrices of variables, may lead to the culturing of microorganisms that have specialized growth requirements. To examine arrays of variables, it would be necessary to increase the rate at which cultures are examined. Work in progress has partially achieved these goals through the application of automation tools, such as robotic liquid handling and the automated scanning of cell arrays (J.-C. Cho, C. S. Alexander, S. Dunlap, S. A. Connon, M. S. Rappé, and S. J. Giovannoni, unpublished results).
Culturing organisms remains an important step in the process of understanding the biology and ecology of microbial species. Cultures can be used to obtain complete genome sequences and to identify properties of organisms that could not be identified by genome sequence alone. Cultures also provide a means to test hypotheses emerging from genome sequences. Combined with proteomics or microarrays, cultures of environmentally significant organisms can be used to examine the adaptations of organisms to environmental change. For example, cultures of key heterotrophic bacterioplankton may enable oceanographers to study how nutrient limitation and other oceanographically relevant variables affect the growth of individual species and thereby help identify the role dominant species play in geochemical cycles.
Acknowledgments
We are indebted to Don Button and Ikuo Yoshinaga, both of whom provided advice and inspiration for this work. We want to thank Jessina McGregor for technical assistance and Bryan Wright for statistical recommendations. We also thank Carol Dimeo, Mike Rappé, Kevin Vergin, Markus Moeseneder, Jang-Cheon Cho, Kate Field, and Kathleen Page for critical review. We give special thanks to Mike Rappé for indicating that SAR92 is a chimeric sequence.
This work was supported by National Science Foundation grants OIA-9977469 and MCB-9977930 and a grant from the Murdock Charitable Trust. In addition, Tartar research fellowships and general grants and funds were provided by Oregon State University.
REFERENCES
- 1.Ammerman, J. W., J. A. Fuhrman, Å. Hagström, and F. Azam. 1984. Bacterioplankton growth in seawater. I. Growth kinetics and cellular characteristics in seawater cultures. Mar. Ecol. Prog. Ser. 18:31-39. [Google Scholar]
- 2.Bano, N., and J. T. Hollibaugh. 2002. Phylogenetic composition of bacterioplankton assemblages from the Arctic Ocean. Appl. Environ. Microbiol. 68:505-518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baxter, M., and J. M. Sieburth. 1984. Metabolic and ultrastructural response to glucose of two eurytrophic bacteria isolated from seawater at different enriching concentrations. Appl. Environ. Microbiol. 47:31-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Béjà, O., L. Aravind, E. V. Koonin, M. T. Suzuki, A. Hadd, L. P. Nguyen, S. B. Jovanovich, C. M. Gates, R. A. Feldman, J. L. Spudich, E. N. Spudich, and E. F. DeLong. 2000. Bacterial rhodopsin: evidence for a new type of phototrophy in the sea. Science 289:1902-1906. [DOI] [PubMed] [Google Scholar]
- 5.Béjà, O., M. T. Suzuki, E. V. Koonin, L. Aravind, A. Hadd, L. P. Nguyen, R. Villacorta, M. Amjadi, C. Garrigues, S. B. Jovanovich, R. A. Feldman, and E. F. DeLong. 2000. Construction and analysis of bacterial artificial chromosome libraries from a marine microbial assemblage. Environ. Microbiol. 2:516-529. [DOI] [PubMed] [Google Scholar]
- 6.Blitchington, R. B., R. Frothingham, R. C. Greene, D. H. Lee, and K. H. Wilson. 1992. UV absorption complicates PCR decontamination. BioTechniques 13:208-210. [PubMed] [Google Scholar]
- 7.Boogerd, F. C., M. M. Q. van Alphen, W. J. van Anrooij, J. C. DeBruyn, P. Bos, and J. G. Kuenen. 1989. The role of growth and maintenance in the oxidation of pyrite in batch culture by a moderately thermophilic, facultative chemolithoautotroph, p. 735-751. In Proceedings of the 1989 International Symposium on Biohydrometallurgy. Canada Centre for Mineral and Energy Technology, Ontario, Canada.
- 8.Britschgi, T. B., and S. J. Giovannoni. 1991. Phylogenetic analysis of a natural marine bacterioplankton population by rRNA gene cloning and sequencing. Appl. Environ. Microbiol. 57:1707-1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Button, D. K., B. R. Robertson, P. W. Lepp, and T. M. Schmidt. 1998. A small, dilute-cytoplasm, high-affinity, novel bacterium isolated by extinction culture and having kinetic constants compatible with growth at ambient concentrations of dissolved nutrients in seawater. Appl. Environ. Microbiol. 64:4467-4476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Button, D. K., F. Schut, P. Quang, R. Martin, and B. R. Robertson. 1993. Viability and isolation of marine bacteria by dilution culture: theory, procedures, and initial results. Appl. Environ. Microbiol. 59:881-891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carlson, C. A., and H. W. Ducklow. 1996. Growth of bacterioplankton and consumption of dissolved organic carbon in the Sargasso Sea. Aquat. Microb. Ecol. 10:69-85. [Google Scholar]
- 12.DeBruyn, J. C., F. C. Boogerd, P. Bos, and J. G. Kuenen. 1990. Floating filters, a novel technique for isolation and enumeration of fastidious, acidophilic, iron-oxidizing, autotrophic bacteria. Appl. Environ. Microbiol. 56:2891-2894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deming, J. W., and J. A. Baross. 2000. Survival, dormancy, and nonculturable cells in extreme deep-sea environments, p. 147-197. In R. R. Colwell and D. J. Grimes (ed.), Nonculturable microorganisms in the environment. ASM Press, Washington, D.C.
- 14.Ferguson, R. L., E. N. Buckley, and A. V. Palumbo. 1984. Response of marine bacterioplankton to differential filtration and confinement. Appl. Environ. Microbiol. 47:49-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ferris, M. J., A. L. Ruff-Roberts, E. D. Kopczynski, M. M. Bateson, and D. M. Ward. 1996. Enrichment culture and microscopy conceal diverse thermophilic Synechococcus populations in a single hot spring microbial mat habitat. Appl. Environ. Microbiol. 62:1045-1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Giovannoni, S. J., T. B. Britschgi, C. L. Moyer, and K. G. Field. 1990. Genetic diversity in Sargasso Sea bacterioplankton. Nature (London) 344:60-63. [DOI] [PubMed] [Google Scholar]
- 17.Giovannoni, S. J., and M. S. Rappé. 2000. Evolution, diversity, and molecular ecology of marine prokaryotes, p. 47-84. In D. L. Kirchman (ed.), Microbial ecology of the oceans. Wiley-Liss, Inc., New York, N.Y.
- 18.Jannasch, H. W., and G. E. Jones. 1959. Bacterial populations in seawater as determined by different methods of enumeration. Limnol. Oceanogr. 4:128-139. [Google Scholar]
- 19.Kogure, K., U. Simidu, and N. Taga. 1979. A tentative direct microscopic method for counting living marine bacteria. Can. J. Microbiol. 25:415-420. [DOI] [PubMed] [Google Scholar]
- 20.Koops, H.-P., and U. C. Möller. 1992. The lithotrophic ammonia-oxidizing bacteria, p. 2625-2637. In A. Balows, H. G. Trüper, M. Dworkin, W. Harder, and K.-H. Schleifer (ed.), The prokaryotes, 2nd ed., vol. 3. Springer-Verlag, New York, N.Y. [Google Scholar]
- 21.Lane, D. J. 1991. 16S/23S rRNA sequencing, p. 115-147. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. John Wiley & Sons, New York, N.Y.
- 22.Lanoil, B. D., C. A. Carlson, and S. J. Giovannoni. 2000. Bacterial chromosomal painting for in situ monitoring of cultured marine bacteria. Environ. Microbiol. 2:654-665. [DOI] [PubMed] [Google Scholar]
- 23.Li, W. K. W., and P. M. Dickie. 1985. Growth of bacteria in seawater filtered through 0.2 μm nucleopore membranes: implications for dilution experiments. Mar. Ecol. Prog. Ser. 26:245-252. [Google Scholar]
- 24.Moore, J. L., C.-Y. Ou, and G. Schochetman. 1991. Use of UV irradiation to reduce false positivity in polymerase chain reaction. BioTechniques 10:442-446. [PubMed] [Google Scholar]
- 25.Nold, S. C., E. D. Kopczynski, and D. M. Ward. 1996. Cultivation of aerobic chemoorganotrophic proteobacteria and gram-positive bacteria from a hot spring microbial mat. Appl. Environ. Microbiol. 62:3917-3921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Partensky, F., W. R. Hess, and D. Vaulot. 1999. Prochlorococcus, a marine photosynthetic prokaryote of global significance. Microbiol. Mol. Biol. Rev. 63:106-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rappé, M. S., P. F. Kemp, and S. J. Giovannoni. 1997. Phylogenetic diversity of marine coastal picoplankton 16S rRNA genes cloned from the continental shelf off Cape Hatteras, North Carolina. Limnol. Oceanogr. 42:811-826. [Google Scholar]
- 28.Rappé, M. S., K. L. Vergin, and S. J. Giovannoni. 2000. Phylogenetic comparisons of a coastal bacterioplankton community with its counterparts in open ocean and freshwater systems. FEMS Microbiol. Ecol. 33:219-232. [DOI] [PubMed] [Google Scholar]
- 29.Schut, F., E. J. de Vries, J. C. Gottschal, B. R. Robertson, W. Harder, R. A. Prins, and D. K. Button. 1993. Isolation of typical marine bacteria by dilution culture: growth, maintenance, and characteristics of isolates under laboratory conditions. Appl. Environ. Microbiol. 59:2150-2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schut, F., J. C. Gottschal, and P. A. Rudolf. 1997. Isolation and characterisation of the marine ultramicrobacterium Sphingomonas sp. strain RB2256. FEMS Microbiol. Rev. 20:363-369. [Google Scholar]
- 31.Staley, J. T., and A. Konopka. 1985. Measurements of in situ activities of nonphotosynthetic microorganisms in aquatic and terrestrial habitats. Annu. Rev. Microbiol. 39:321-346. [DOI] [PubMed] [Google Scholar]
- 32.Strunk, O., W. Ludwig, O. Gross, B. Reichel, M. May, S. Hermann, N. Stuckmann, B. Nonhoff, M. Lenke, T. Ginhart, A. Vilbig, and R. Westram. 1996. ARB—a software environment for sequence data, 2.5b ed. Technical University of Munich, Munich, Germany.
- 33.Suzuki, M. T., O. Béjà, L. T. Taylor, and E. F. DeLong. 2001. Phylogenetic analysis of ribosomal RNA operons from uncultivated coastal marine bacterioplankton. Environ. Microbiol. 3:323-331. [DOI] [PubMed] [Google Scholar]
- 34.Suzuki, M. T., M. S. Rappé, Z. W. Haimberger, H. Winfield, N. Adair, J. Ströbel, and S. J. Giovannoni. 1997. Bacterial diversity among small-subunit rRNA gene clones and cellular isolates from the same seawater sample. Appl. Environ. Microbiol. 63:983-989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Swofford, D. L. 2001. PAUP*. Phylogenetic analysis using parsimony (∗ and other methods), version 4.0b6. Sinauer Associates, Sunderland, Mass.
- 36.Turley, C. M. 1993. Direct estimates of bacterial numbers in seawater samples without incurring cell loss due to sample storage, p. 143-147. In P. F. Kemp, B. F. Sherr, E. B. Sherr, and J. J. Cole (ed.), Handbook of methods in aquatic microbial ecology. Lewis Publishers, Boca Raton, Fla.
- 37.Vancanneyt, M., F. Schut, C. Snauwaert, J. Goris, J. Swings, and J. C. Gottschal. 2001. Sphingomonas alaskensis sp. nov., a dominant bacterium from a marine oligotrophic environment. Int. J. Syst. E vol. Microbiol. 51:73-79. [DOI] [PubMed] [Google Scholar]
- 38.Vergin, K. L., M. S. Rappé, and S. J. Giovannoni. 2001. Streamlined method to analyze 16S rRNA gene clone libraries. BioTechniques 30:938-944. [DOI] [PubMed] [Google Scholar]
- 39.Wang, Y., P. C. K. Lau, and D. K. Button. 1996. A marine oligobacterium harboring genes known to be part of aromatic hydrocarbon degradation pathways of soil pseudomonads. Appl. Environ. Microbiol. 62:2169-2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wirsen, C. O., S. M. Sievert, C. M. Cavanaugh, S. J. Molyneaux, A. Ahmad, L. T. Taylor, and E. F. DeLong. 2002. Characterization of an autotrophic sulfide-oxidizing marine Arcobacter sp. that produces filamentous sulfur. Appl. Environ. Microbiol. 68:316-325. [DOI] [PMC free article] [PubMed] [Google Scholar]