Abstract
The glycan chains of the surface layer (S-layer) glycoprotein from the gram-positive, thermophilic bacterium Aneurinibacillus (formerly Bacillus) thermoaerophilus strain DSM 10155 are composed of l-rhamnose- and d-glycero-d-manno-heptose-containing disaccharide repeating units which are linked to the S-layer polypeptide via core structures that have variable lengths and novel O-glycosidic linkages. In this work we investigated the enzymes involved in the biosynthesis of thymidine diphospho-l-rhamnose (dTDP-l-rhamnose) and their specific properties. Comparable to lipopolysaccharide O-antigen biosynthesis in gram-negative bacteria, dTDP-l-rhamnose is synthesized in a four-step reaction sequence from dTTP and glucose 1-phosphate by the enzymes glucose-1-phosphate thymidylyltransferase (RmlA), dTDP-d-glucose 4,6-dehydratase (RmlB), dTDP-4-dehydrorhamnose 3,5-epimerase (RmlC), and dTDP-4-dehydrorhamnose reductase (RmlD). The rhamnose biosynthesis operon from A. thermoaerophilus DSM 10155 was sequenced, and the genes were overexpressed in Escherichia coli. Compared to purified enterobacterial Rml enzymes, the enzymes from the gram-positive strain show remarkably increased thermostability, a property which is particularly interesting for high-throughput screening and enzymatic synthesis. The closely related strain A. thermoaerophilus L420-91T produces d-rhamnose- and 3-acetamido-3,6-dideoxy-d-galactose-containing S-layer glycan chains. Comparison of the enzyme activity patterns in A. thermoaerophilus strains DSM 10155 and L420-91T for l-rhamnose and d-rhamnose biosynthesis indicated that the enzymes are differentially expressed during S-layer glycan biosynthesis and that A. thermoaerophilus L420-91T is not able to synthesize dTDP-l-rhamnose. These findings confirm that in each strain the enzymes act specifically on S-layer glycoprotein glycan formation.
Crystalline surface layers (S-layers) represent the outermost cell envelope component of many archaea and bacteria (for reviews see references 41, 45, and 46). Distinct functions of S-layers have been reported only in a few cases (47). For pathogenic bacteria variation of surface antigens is important for evading the immune response of the host. Campylobacter fetus achieves antigenic variation by expression of different S-layer proteins from at least eight homologs of the structural S-layer gene (6). Variations in the expression of S-layer proteins are not limited to pathogenic species and have also been described for nonpathogenic bacteria, such as Bacillus sphaericus (3), Geobacillus (formerly Bacillus) stearothermophilus (24), and Lactobacillus acidophilus (2). These organisms seem to adapt to altered environmental conditions by changing the S-layer protein. Modification of the surface properties is also achieved by posttranslational modification of the S-layer proteins, particularly by glycosylation (for reviews see references 15, 31, 32, 34, 42, and 51). For example, the closely related Aneurinibacillus (formerly Bacillus) thermoaerophilus strains L420-91T (= DSM 10154T) and DSM 10155 synthesize different glycans composed of either d-rhamnose and 3-acetamido-3,6-dideoxy-d-galactose (22) or l-rhamnose and d-glycero-d-manno-heptose (23).
Few analyses have been carried out so far to elucidate the biosynthesis of bacterial S-layer glycoprotein glycans (12, 44). The studies that have been performed confirmed that there are differences in activation of mono- and oligosaccharides for eukaryotic glycoprotein biosynthesis (20) and even for the formation of other bacterial glycoconjugates. Most of the nucleotide-activated monosaccharides used for biosynthesis of S-layer glycoproteins are identical to those used for biosynthesis of other glycoconjugates. However, in Thermoanaerobacter thermosaccharolyticum strain E207-71 N-acetylmannosamine is activated not as the common UDP-N-acetylmannosamine but as GDP-N-acetylmannosamine (44). These monosaccharide precursors are not used for the formation of lipid-activated oligosaccharides but as precursors for the synthesis of nucleotide-activated oligosaccharides.
In our attempts to elucidate the formation of specific sugar components of bacterial S-layer glycans, we focused on the biosynthesis of l-rhamnose. This sugar has often been found in S-layer glycoproteins (for reviews see references 31, 32, 34, and 42), and its biosynthetic pathway in gram-negative organisms has been known since the 1960s (8, 21, 36). The reaction steps and enzymes and the corresponding gene loci are as follows (10, 11, 14, 16, 38): (i) dTTP + d-Glc-1-P → dTDP-d-Glc + PPi (glucose-1-phosphate thymidylyltransferase; EC 2.7.7.24; rmlA); (ii) dTDP-d-Glc → dTDP-6-deoxy-d-xylo-4-hexulose (dTDP-d-glucose 4,6-dehydratase; EC 4.2.1.46; rmlB); (iii) dTDP-6-deoxy-d-xylo-4-hexulose → dTDP-6-deoxy-l-lyxo-4-hexulose (dTDP-4-dehydrorhamnose 3,5-epimerase; EC 5.1.3.13; rmlC); and (iv) dTDP-6-deoxy-l-lyxo-4-hexulose + NADPH → dTDP-l-rhamnose + NADP+ (dTDP-4-dehydrorhamnose reductase; EC 1.1.1.133; rmlD).
All previous biochemical studies of dTDP-l-rhamnose synthesis except those with mycobacteria (50) were related to the formation of lipopolysaccharides and capsules, while rml genes of gram-negative and gram-positive bacteria and archaea have been cloned and sequenced without further functional analyses (14, 35, 48, 53, 54). None of these studies reported involvement of the genes in the biosynthesis of prokaryotic glycoproteins. The dTDP-l-rhamnose biosynthesis pathway has been discussed as a potential therapeutic target (7) since dTDP-l-rhamnose is not synthesized in humans. Recently, a screening test has been described based on RmlB to -D from Mycobacterium tuberculosis, and 8,000 substances have been screened for inhibitory activity. Some of these substances at high concentrations acted against M. tuberculosis (28). A serious problem for high-throughput screening is the instability of the enzymes used, as reported for RmlB from Salmonella enterica LT2 (49).
In this paper we describe preparation and characterization of the Rml proteins from the thermophilic strain A. thermoaerophilus DSM 10155. Comparison of the enzyme activities of A. thermoaerophilus strains DSM 10155 and L420-91T provided evidence that these enzymes are indeed involved in S-layer protein glycosylation.
MATERIALS AND METHODS
Materials.
dTDP-glucose, NADPH, NADH, NADP+, NAD+, glucose 1,6-diphosphate, phosphoglucomutase, and glucose-6-phosphate dehydrogenase were obtained from Sigma (Vienna, Austria). dTDP-6-deoxy-d-xylo-4-hexulose and dTDP-l-rhamnose were synthesized as described previously (10).
Bacterial strains and growth conditions.
A. thermoaerophilus strains L420-91T (= DSM 10154T) and DSM 10155 (13, 23, 30) were obtained from F. Hollaus (Zuckerforschung Tulln GmbH, Tulln, Austria). The bacteria were cultivated in several 15-liter batches in a Braun Biostat C fermentor (B. Braun Biotech, Melsungen, Germany) by using a modified SVIII medium (0.25% peptone, 0.5% yeast extract, 0.13% dipotassium hydrogen phosphate, 0.01% magnesium sulfate) and the following conditions: 55°C; pH 7.0 to 7.5; airflow, 10 liters/min; and 300 rpm. Under these conditions oxygen limitation was reached rapidly (40). Cells were pelleted by centrifugation when the optical density at 600 nm reached levels greater than 3 and were stored at −20°C until they were used. Escherichia coli B strain ATCC 23848 (36) was used as reference strain for dTDP-l-rhamnose synthesis.
Plasmids were maintained in E. coli DH5α [K-12 F− φ80d lacZΔM15 endA1 recA1 hsdR17 (rK− mK−) supE44 thi-1 gyrA96 relA1 Δ(lacZYA-argF)U169] (39). For enzyme overexpression, E. coli BL21(λDE3) [F− ompT hsdSB (rB− mB−) gal dcm (λDE3); Novagen, Madison, Wis.] was used. E. coli strains were grown in Luria-Bertani medium. Media were supplemented, when required, with kanamycin (30 μg/ml). Cultures were grown at 37°C with or without agitation.
DNA manipulation, PCR, and sequencing.
All standard DNA recombinant methods used were methods described by Sambrook et al. (39). PCR was performed by using a PCR sprint thermocycler (Hybaid, Ashford, United Kingdom). Chromosome walking was done as described previously (18). DNA sequencing was performed either by MWG Biotech (Ebersberg, Germany) or by Agowa (Berlin, Germany).
Sequence analyses.
Nucleotide and protein sequences were analyzed by using online analysis tools, including BLAST (Basic local alignment search tool) (1) and Multalin (5).
Construction of plasmids.
DNA sequence analysis identified four open reading frames corresponding to RmlA to -D. The four open reading frames were individually amplified by PCR by using primers designed to introduce a unique NcoI or NdeI site overlapping the initiating ATG codon. A downstream SstI site was introduced to facilitate cloning of the amplified fragment. The following primers were used: RMLA1F (5′-TGAACAGCATATGAAAGGAATTATT-3′; NdeI site underlined); RMLA25R(5′-TTCGAGCTCTTGCACTCTTCTCACTCC-3′; SstI site underlined); RMLB3F (5′-ATACCATGGAAGTTTTAGTTACAGG-3′; NcoI site underlined); RMLB4R (5′-ATTGAGCTCTCATCGCTGAACC-3′; SstI site underlined); RMLC11F (5′-GATCCAACATATGCAAAGAATAGAAACG-3′; NdeI site underlined); RMLC10R (5′-TTTGAGCTCTAAGATCAAGACATCT-3′; SstI site underlined); RMLD3F (5′-GTTCAGCCATGGAAGTTCTTG-3′; NcoI site underlined); and RMLD4R (5′-CTTTCCTTGAGCTCTCATTCC-3′; SstI site underlined).
PCR products were digested with the appropriate restriction endonucleases according to the manufacturer's recommendations and cloned in plasmid pET-28a(+) (rmlA rmlC) or pET-30a(+) (rmlB rmlD) (Novagen) for histidine-tagged proteins and in pET-30a (rmlA) for native expression. Plasmids were electrotransformed into E. coli DH5α and BL21(λDE3).
Protein overexpression and purification.
Expression (10) and purification of histidine-tagged proteins from plasmids pRmlA3, pRmlB2, pRmlC3, and pRmlD2 were done as described previously (17).
RmlA without histidine tag modification was purified by the following method. RmlA was overexpressed by using plasmid pRmlA1 in E. coli BL21(λDE3), and extracts were prepared as described previously (10). The extract was applied to a DEAE-HiPrep column (Amersham Pharmacia Biotech, Uppsala, Sweden) equilibrated with 20 mM Tris-HCl buffer (pH 7.7). RmlA eluted in a linear 0 to 1 M KCl gradient at 0.4 M KCl. Fractions showing activity were pooled, dialyzed, and further purified on a MonoQ HR5/5 column (Amersham Pharmacia Biotech) as described previously (10). RmlA eluted at 0.3 M KCl, and buffer was exchanged against 50 mM piperazine-HCl buffer (pH 5.5). For chromatofocusing, protein was applied to a MonoP HR 5/5 column (Amersham Pharmacia Biotech) and eluted with polybuffer 74 (pH 4.0) at approximately pH 4.2. Final purification was achieved by rechromatography on a MonoQ column after buffer exchange (20 mM Tris-HCl buffer, pH 7.7).
Analytical techniques.
Nucleotides and nucleotide-activated sugars were analyzed on a CarboPac PA-1 column by the method of Palmieri et al. (37). UV spectra were recorded in 0.1 M NaOH with a Beckman DU-65 spectrophotometer. Protein contents were determined by the method of Bradford (4). Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) by the method of Laemmli (26) was performed as described previously (33).
Preparation of polyclonal antibodies against RmlA and immunoblotting.
Immunization of rabbits was done at the Institut für Labortierkunde und Genetik, Universität Wien (Himberg, Austria). Immunization was performed with 200 μg of purified native RmlA on days 0, 28, and 49. A serum sample (5 ml) was obtained on day 56 and used for further analysis. The immunospecificity of the polyclonal antibody was tested by using Western blots with purified RmlA and bacterial extracts. Proteins from SDS-PAGE gels were blotted on polyvinylidene difluoride membranes (ImmobiloP; Millipore, Bedford, Mass.) by using the discontinuous buffer system (25). RmlA was detected by using a polyclonal rabbit anti-RmlA antibody according to the rapid immunodetection method described by the manufacturer of the membranes. Incubation with antisera was performed by using 1:1,000 dilutions of antisera in 1% bovine serum albumin-0.05% Tween 20 in phosphate-buffered saline (10 mM sodium phosphate buffer [pH 7.2] and 0.9% sodium chloride [buffer A]) for 1 h at room temperature. Subsequently, blots were incubated with goat anti-rabbit immunoglobulin G (Sigma) at a 1:2,000 dilution in buffer A for 30 min at room temperature. For detection of bound alkaline phosphatase-conjugated antiserum, 5-bromo-4-chloro-3-indoyl-phosphate (Roche Diagnostics, Vienna, Austria) and 4-nitroblue tetrazolium chloride (Roche) were used as chromogenic substrates.
Immunogold labeling of RmlA on thin sections and electron microscopic examination.
Labeling was accomplished by using cells from the early stationary growth phase which were cultivated in 200-ml flasks. Cell pellets from an overnight culture were fixed in 0.1% glutaraldehyde-4% formaldehyde in 0.1 M sodium cacodylate buffer (pH 7.4). Some of the preparations were also fixed with 1% osmium tetroxide in distilled water before dehydration was performed with a graded series of ethanol solutions that ended in absolute ethanol. The pellets were embedded in Spurr resin and cured for 48 h at 50°C, and thin sections were cut with an Ultracut ultramicrotome (Reichert-Jung AG, Vienna, Austria). The sections were incubated overnight with anti-RmlA antibody by using a 1:5,000 dilution in buffer A, blocked with 0.5% bovine serum albumin in the same buffer, and immunolabeled with anti-rabbit immunoglobulin G-gold conjugate (10-nm gold particles; Sigma) diluted 1:100 in buffer A (9). The labeled thin sections were postlabeled with lead citrate for 5 min (A. Fammy, Proc. 25th Annu. Electron Microsc. Soc. Am. Meet., p. 148, 1967) and examined with a Philips CM 12 electron microscope operated at an acceleration voltage of 80 kV.
Preparation of crude bacterial extracts.
Bacterial cells were washed twice with 20 mM Tris-HCl buffer (pH 7.7) containing 5 mM MgCl2, 0.5 mM EDTA, and 0.5 mM dithiothreitol (buffer B) and resuspended in buffer B (0.1 g [wet weight] of cell pellet per ml). Cells were disrupted by ultrasonication with a Branson model 450 Sonifier (5 min; intensity, 6; 50% duty cycle; cooled in an ice bath); cell fragments were removed by centrifugation at 28,300 × g for 30 min at 4°C, and membranes were removed by centrifugation at 247,000 × g for 60 min. Supernatants were dialyzed against buffer B and stored at −20°C until they were used (29).
To prove that the strains investigated could produce dTDP-l-rhamnose, reactions were performed with crude extracts. Reaction mixtures containing 1 μmol of dTDP-d-glucose, 1 μmol of NAD+, 4.5 mg of crude protein, and an appropriate volume of 20 mM Tris-HCl buffer (pH 7.7) were incubated for 1 h at 50°C (or at 37°C for E. coli B). Then 5 μmol of NADPH was added to each reaction mixture, and the mixture was incubated for an additional 1 h. Proteins were removed by precipitation with ethanol, and the samples were concentrated to a smaller volume and desalted with a Sephadex G-10 column. UV-absorbing fractions were pooled, lyophilized, and stored at −20°C. Reaction products were analyzed by high-performance anion-exchange chromatography with a CarboPac PA-1 column (Dionex, Sunnyvale, Calif.) as described previously (37).
Enzyme assays.
Glucose-1-phosphate thymidylyltransferase (EC 2.7.7.24; RmlA) activity was determined by the assay of Kornfeld and Glaser (21). This assay is based on the reverse reaction and enzymatic determination of the glucose 1-phosphate formed. Alternatively, the forward reaction was assayed by determining the incorporation of glucose 1-phosphate and dTTP and the excess of RmlB, RmlC, and RmlD and monitoring the decrease in NADH. A typical reaction mixture contained 0.36 mM dTTP, 0.36 mM glucose 1-phosphate, and 0.15 mM NADH, as well as 0.2 U of RmlB, 0.2 U of RmlC, and 0.2 U of RmlD from S. enterica LT2, in a final volume of 0.55 ml (10).
dTDP-d-glucose 4,6-dehydratase (EC 4.2.1.46; RmlB) activity was assayed as described by Vara and Hutchinson (55). A typical reaction mixture contained 0.18 mM dTDP-glucose and 0.15 mM NADH, as well as 0.2 U of RmlC and 0.2 U of RmlD from S. enterica LT2 (10).
dTDP-4-dehydrorhamnose 3,5-epimerase (RmlC) and dTDP-4-dehydrorhamnose reductase (RmlD) activities were determined by an assay described recently (10). This assay uses the NADH-dependent conversion of dTDP-6-deoxy-d-xylo-4-hexulose to dTDP-d-rhamnose by RmlC and RmlD. By using an excess of purified RmlC or RmlD from S. enterica LT2, the individual activity of each enzyme can be determined (10).
Mannose-1-phosphate guanylyltransferase activity was detected by the method of Szumilo et al. (52). A typical reaction mixture contained 0.18 mM GDP-mannose, 0.45 mM NADP+, 0.9 mM glucose, 0.18 mM ADP, 2.3 mM inorganic pyrophosphate, 0.5 U of nucleoside diphosphate kinase, 0.5 U of hexokinase, and 0.5 U of glucose-6-phosphate dehydrogenase. Activities were calculated from the linear increase in the absorbance at 340 nm resulting from the NADPH produced.
The combined activity of GDP-d-mannose 4,6-dehydratase and GDP-6-deoxy-d-lyxo-4-hexulose reductase was determined by using the overall reaction starting from GDP-mannose and resulting in oxidation of NADH as described above for RmlC and RmlD.
Characterization of enzymes.
Kinetic constants were determined at 25°C as described previously (10). For RmlA and RmlB the photometric assays described above were used. For determination of Km and kcat values the concentrations were varied as follows: TTP (RmlA), 0.036 to 0.55 mM; glucose 1-phosphate (RmlA), 0.018 to 0.36 mM; dTDP-d-glucose (RmlA), 0.0091 to 0.18 mM; inorganic pyrophosphate (RmlA), 0.045 to 0.91 mM; dTDP-d-glucose (RmlB), 0.0036 to 0.055 mM; dTDP-6-deoxy-d-xylo-4-hexulose (RmlC), 0.018 to 0.27 mM; NADH (RmlD), 0.0036 to 0.073 mM; and NADPH (RmlD), 0.0036 to 0.073 mM.
To probe thermal stability, enzymes were diluted in 50 mM phosphate buffer and incubated at 37 or 55°C. At different times samples were analyzed to determine the remaining activity. From the data obtained the half-life was estimated.
Enzymatic synthesis of dTDP-l-rhamnose.
Enzymatic synthesis of dTDP-l-rhamnose was done essentially as described previously (10). However, instead of enzymes from S. enterica LT2, the counterparts of these enzymes from A. thermoaerophilus DSM 10155 were used. Briefly, 75 μmol of dTDP-d-glucose was converted to dTDP-l-rhamnose by 1 U of RmlB, 1 U of RmlC, and 1 U of RmlD in 2 h at 25°C. NADH was regenerated from NAD+ formed by formate dehydrogenase from Candida boidinii (ASA Spezialenzyme GmbH, Braunschweig, Germany).
NMR spectroscopy.
The anomeric configuration of dTDP-l-rhamnose prepared with enzymes from A. thermoaerophilus DSM 10155, as well as from S. enterica LT2, was determined by nuclear magnetic resonance (NMR) measurements of heteronuclear coupling constants taken from heteronuclear multiple-bond correlation spectroscopy spectra, as described elsewhere (43).
Nucleotide sequence accession number.
The nucleotide sequences of the rhamnose operon and the heptose operon of A. thermoaerophilus DSM 10155 have been deposited in the GenBank/EMBL Data Bank under accession number AF324836.
RESULTS
Detection of enzymes involved in dTDP-l-rhamnose biosynthesis.
In enterobacteria dTDP-l-rhamnose is synthesized in a four-step reaction sequence starting from glucose 1-phosphate and dTTP (7, 8, 21). The four enzymes (RmlA, RmlB, RmlC, RmlD) involved in biosynthesis of this nucleotide-activated precursor were detected in crude extracts of A. thermoaerophilus DSM 10155 by using specific spectrophotometric assays. The activities ranged from 2.5 to 8.6 mU/mg of protein for the enzymes (Table 1). These values correspond well with those determined for E. coli B, the strain used as a reference for dTDP-l-rhamnose synthesis (36). To probe the influence of S-layer glycan composition on the activity of the enzymes involved in dTDP-l-rhamnose biosynthesis, A. thermoaerophilus strain L420-91, the type strain of the genus, was used. The S-layer protein of this strain is modified, with glycans composed of d-rhamnose and 3-acetamido-3,6-dideoxy-d-galactose. d-Rhamnose is activated as GDP-d-rhamnose (17), and it has been proposed that 3-acetamido-3,6-dideoxy-d-galactose is activated as dTDP-3-acetamido-3,6-deoxy-d-galactose (56). GDP-d-rhamnose is produced from mannose 1-phosphate and GTP by the action of mannose-1-phosphate guanylyltransferase, GDP-mannose dehydratase, and GDP-6-deoxy-d-lyxo-4-hexulose reductase. In crude extracts of A. thermoaerophilus L420-91T the activity of mannose-1-phosphate guanylyltransferase was 2.3 mU/mg, and the combined activity of GDP-mannose dehydratase and GDP-6-deoxy-d-lyxo-4-hexulose reductase for the NADH-dependent conversion of GDP-d-mannose to GDP-d-rhamnose was 0.53 mU/mg. In A. thermoaerophilus DSM 10155 the activity of these enzymes was not detectable. On the other hand, in A. thermoaerophilus L420-91T RmlC and RmlD activities were not detected. Interestingly, the RmlA and RmlB activities in strain L420-91T were comparable to those in A. thermoaerophilus DSM 10155 (Table 1).
TABLE 1.
Activities of enzymes involved in dTDP-l-rhamnose biosynthesis with crude extracts from different A. thermoaerophilus strains and E. coli B
| Strain | Enzyme activities (mU/mg of protein)
|
|||
|---|---|---|---|---|
| RmlA | RmlB | RmlC | RmlD | |
| E. coli B | 27.4 | 5.65 | 2.33 | 0.42 |
| A. thermoaerophilus DSM 10155 | 8.6 | 4.76 | 4.95 | 2.54 |
| A. thermoaerophilus L420-91T | 2.0 | 0.61 | NDa | ND |
ND, not detectable.
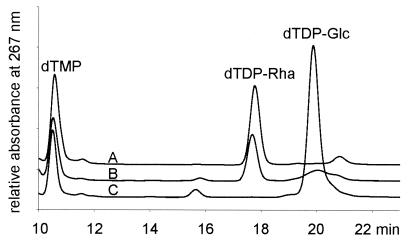
In agreement with the enzymatic assays, when crude extracts from A. thermoaerophilus DSM 10155 were used, dTDP-d-glucose was converted to dTDP-l-rhamnose in the presence of NADPH. The synthesis of dTDP-l-rhamnose was even more efficient than it was in E. coli B. As expected, conversion was not possible with extracts from A. thermoaerophilus L420-91T due to the lack of RmlC and RmlD in this organism (Fig. 1).
FIG. 1.
Synthesis of dTDP-l-rhamnose. Crude extracts from A. thermoaerophilus DSM 10155 (line A), E. coli B (line B), and A. thermoaerophilus L420-91T (line C) were used to synthesize dTDP-l-rhamnose from dTDP-d-glucose in the presence of NADPH.
Cloning and sequencing of the rml locus of A. thermoaerophilus DSM 10155.
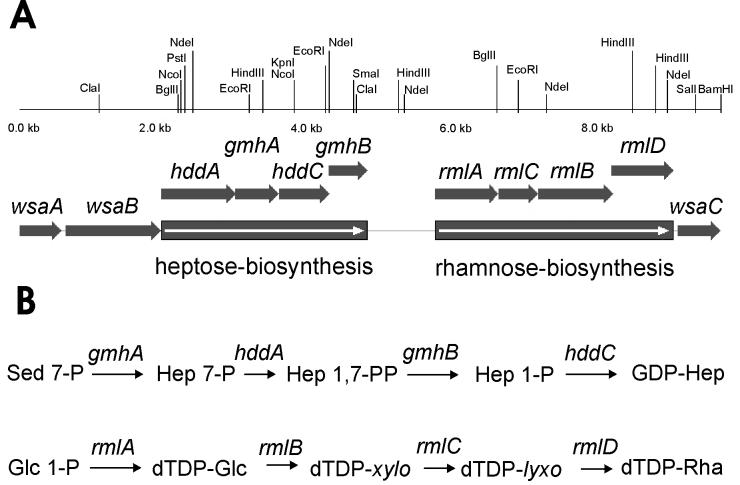
In a previous study a 4.6-kb BglII fragment from the chromosome of A. thermoaerophilus DSM 10155 containing part of the rmlA gene was cloned and sequenced (19). By chromosome walking (18) the downstream region was sequenced. Proteins that were encoded were identified by BLAST analysis and multiple alignment, and the genes were found to be in the order rmlACBD (Fig. 2). Proteins showing the highest levels of similarity to the enzymes are listed in Table 2.
FIG. 2.
Precursor formation in A. thermoaerophilus DSM 10155. (A) Map of the genes involved in formation of GDP-d-glycero-α-d-manno-heptose and dTDP-β-l-rhamnose. (B) Scheme of the biosynthetic pathway of the nucleotide-activated precursors of the S-layer glycoprotein of A. thermoaerophilus DSM 10155.
TABLE 2.
Homologs of the dTDP-l-rhamnose biosynthetic enzymes from A. thermoaerophilus DSM 10155
| Enzyme | Homolog (organism) | Identity/ % similarity | Accession no. |
|---|---|---|---|
| RmlA | RmlA (Enterococcus faecalis) | 74/86 | AAC35920 |
| RmlA (Streptococcus pyogenes) | 73/85 | AAK33848 | |
| RmlA (Lactococcus lactis subsp. lactis) | 72/86 | AAK04292 | |
| RmlB | RmlB (Methanobacterium thermo- autotrophicum) | 68/84 | H69105 |
| RmlB (Leptospira borgpetersenii) | 55/72 | AAD12971 | |
| RmlB (Xanthomonas campestris) | 56/69 | AAK53466 | |
| RmlC | RmlC (Enterococcus faecalis) | 61/71 | AAC35921 |
| RmlC (Shigella flexneri) | 52/66 | P37780 | |
| RmlC (Pyrococcus abyssi) | 53/65 | E75098 | |
| RmlD | RmlD (Methanobacterium thermo- autotrophicum) | 49/66 | D69106 |
| RmlD (Actinobacillus actinomycetem- comitans) | 47/66 | BA94404 | |
| RmlD (Bacillus subtilis) | 42/59 | P39631 |
Southern hybridization experiments with oligonucleotide probes, as well as rmlA-specific DNA probes, yielded specific signals, indicating that only one rmlA gene was present (data not shown).
Expression and purification of the Rml enzymes from A. thermoaerophilus DSM 10155.
For further characterization and for use in enzymatic synthesis, the enzymes responsible for dTDP-l-rhamnose synthesis in A. thermoaerophilus DSM 10155 were overexpressed as His-tagged fusion proteins to facilitate purification. This modification enabled purification to apparent homogeneity in only one chromatographic step. The proteins were desalted, and purity and molecular mass were checked by SDS-PAGE analysis (data not shown).
The stability of RmlB from S. enterica LT2 was shown to be a serious problem for productivity in enzymatic synthesis (49). The stability of the enzymes from A. thermoaerophilus DSM 10155 was tested at 37 and 55°C and compared with the stability of the enzymes from S. enterica LT2 (Table 3). At 37°C RmlB, the most unstable of the enzymes, had a half-life of 11 h. This is 11 times longer than the half-life of RmlB from S. enterica LT2. The other enzymes (RmlA, RmlC, and RmlD) from A. thermoaerophilus DSM 10155 were stable for at least 48 h, while the corresponding enzymes from S. enterica LT2 were stable for 1 to 6 h. At the optimal growth temperature for the thermophilic organism (55°C) the stabilities of the thermophilic enzymes differed considerably. The half-lives ranged from 11 min (RmlB) to 27 h (RmlC). The divalent ion requirements and pH preferences of the mesophilic and thermophilic enzymes are almost identical, making mixtures of individual enzymes for analytical or synthetic purposes possible.
TABLE 3.
Properties of dTDP-l-rhamnose biosynthetic enzymes
| Enzyme | Organism | Stability at 37°C (half-life [h]) | Stability at 55°C (half-life) | Maximum activity pH | Divalent ion required |
|---|---|---|---|---|---|
| RmlA | S. enterica LT2 | 6 | 7.0 | Mg2+ | |
| A. thermoaerophilus DSM 10155 | ≥24 | 15 min | 7.5 | Mg2+ | |
| RmlB | S. enterica LT2 | 1 | 8.0 | Mg2+ | |
| A. thermoaerophilus DSM 10155 | 11 | 11 min | 7.5 | Mg2+ | |
| RmlC | S. enterica LT2 | 3 | 7.5 | ||
| A. thermoaerophilus DSM 10155 | ≥24 | 27 h | 8.0 | ||
| RmlD | S. enterica LT2 | 1.5 | 6.5 | ||
| A. thermoaerophilus DSM 10155 | ≥24 | 2 h | 7.5 |
Kinetic constants for the Rml enzymes from A. thermoaerophilus DSM 10155.
Recently, RmlB, RmlC, and RmlD from M. tuberculosis have been used for screening potential therapeutic agents acting on the basis of TDP-l-rhamnose biosynthesis (28). For high-throughput screening the enzymes from A. thermoaerophilus DSM 10155 could replace the enzymes from mesophilic organisms in order to eliminate effects from protein instability. To prove the applicability of the Rml enzymes from A. thermoaerophilus DSM 10155, kinetic constants were determined and compared with those previously reported for enzymes from S. enterica LT2 (10, 27, 49). No dramatic differences were observed (Table 4). Only the Km of RmlC for dTDP-6-deoxy-d-xylo-4-hexulose was 10-fold reduced for the thermophilic enzyme. Since the enzymes from the thermophilic organism were tested at a temperature that was 30°C below the normal growth temperature of that organism, the kcat values were lower than those of the enzymes from the mesophilic strain (Table 4).
TABLE 4.
Apparent kinetic constants for dTDP-l-rhamnose biosynthetic enzymes
| Enzyme | Substrate | Km (mM) | kcat (s−1) |
|---|---|---|---|
| LT2 RmlAa | dTTP | 0.020 | |
| α-d-Glucose 1-phosphate | 0.11 | 30 | |
| dTDP-d-glucose | 0.083 | ||
| Pyrophosphate | 0.15 | 180 | |
| DSM 10155 RmlA | dTTP | 0.031 ± 0.0040 | 8.9 ± 0.24 |
| α-d-Glucose 1-phosphate | 0.20 ± 0.036 | 13 ± 1.1 | |
| dTDP-d-glucose | 0.13 ± 0.036 | 15 ± 2.0 | |
| Pyrophosphate | 0.055 ± 0.0082 | 10 ± 0.32 | |
| LT2 RmlBb | dTDP-d-glucose | 0.0072 | 3.0 |
| DSM 10155 RmlB | dTDP-d-glucose | 0.012 ± 0.0016 | 1.9 ± 0.079 |
| LT2 RmlCc | dTDP-6-deoxy-d-xylo-4- hexulose | 0.71 ± 0.17 | 39 ± 6.6 |
| DSM 10155 RmlC | dTDP-6-deoxy-d-xylo-4- hexulose | 0.062 ± 0.0063 | 2.0 ± 0.67 |
| LT2 RmlDc | NADH | 0.0063 ± 0.0019 | 39 ± 6.6 |
| NADPH | 0.013 ± 0.0020 | 25 ± 1.1 | |
| DSM 10155 RmlD | NADH | 0.012 ± 0.0017 | 1.5 ± 6.6 |
| NADPH | 0.0076 ± 0.0015 | 2.3 ± 0.14 |
Enzymatic synthesis with Rml enzymes from A. thermoaerophilus DSM 10155.
RmlD from A. thermoaerophilus DSM 10155 showed the same dual-cofactor specificity for NADH and NADPH as RmlD from S. enterica LT2. Thus, this enzyme is suitable for substrate regeneration with NAD+-dependent formate dehydrogenase from C. boidinii. For example, 75 μmol of dTDP-d-glucose was converted to dTDP-l-rhamnose in 2 h with 1 U of RmlB, 1 U of RmlC, and 1 U of RmlD from A. thermoaerophilus DSM 10155. By using high-performance anion-exchange chromatography, the yield was shown to be 100%, and the purity was comparable to the purity of the product synthesized with enzymes from the mesophilic source. By using NMR analysis, dTDP-l-rhamnose was shown to possess a β-configuration with a C,H-coupling constant of 161 Hz. Thus, the anomeric configuration is identical to that of the reaction product obtained with RmlB, RmlC, and RmlD from S. enterica LT2 (1JC,H = 161 Hz).
Cytoplasmic localization of RmlA.
To show the localization of the rhamnose biosynthetic enzymes, a polyclonal antibody was generated against one of the corresponding enzymes. RmlA from A. thermoaerophilus DSM 10155 was overexpressed in E. coli and purified by several chromatographic steps to apparent homogeneity. Rabbits were immunized, and sera were tested for RmlA specificity by Western blot analysis. A single positive signal was obtained with a crude protein preparation from A. thermoaerophilus DSM 10155. Immunolabeling of thin sections for RmlA with the polyclonal antibody (Fig. 3) resulted in weak but specific deposition of colloidal gold particles in the vicinity of the cytoplasmic membrane of A. thermoaerophilus.
FIG. 3.
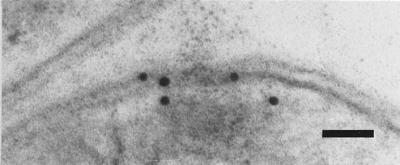
Localization of RmlA in A. thermoaerophilus DSM 10155 cells: electron micrograph of an ultrathin section after immunolabeling with colloidal gold, showing the cytoplasmic localization of the enzyme. Bar = 50 nm.
DISCUSSION
S-layer glycoproteins form the outermost protective coat of several thermophilic members of the Bacillus-Clostridium group (34, 42). Other organisms in this group form S-layers composed of nonglycosylated proteins (46). So far, the biological function of the glycan chains covalently linked to the S-layer glycoproteins is not known (47). Nevertheless, in structure these glycoconjugates resemble the most important molecules in the outer leaflet of the outer membrane of gram-negative bacteria, the lipopolysaccharides (44). They exhibit enormous diversity based on composition and linkages of the glycan chains. Although compared to lipopolysaccharides only a small number of S-layer glycans have been characterized, a similar diversity of monosaccharide constituents and structure has been observed (31, 32). For example, A. thermoaerophilus L420-91T produces a glycan composed of d-rhamnose and 3-acetamido-3,6-dideoxy-d-galactose with the structure {→3)-[α-d-Fucp3NAc-(1→2)]-α-d-Rhap-(1→3)-[α-d-Fucp3NAc-(1→2)]-α-d-Rhap-(1→2)-α-d-Rhap-(1→2)-α-d-Rhap-(1→}n∼15 (22), while the structure of the glycan chain of A. thermoaerophilus DSM 10155 is [→4)-α-l-Rhap-(1→3)-β-d-glycero-d-manno-Hepp-(1→]n∼18 (23, 57).
As with other bacterial glycoconjugates, the differences in S-layer glycan compositions should be the result of different sets of glycosylation enzymes that the strains have acquired during evolution. So far, the glycan biosynthesis clusters of A. thermoaerophilus strains L420-91T and DSM 10155 and G. stearothermophilus NRS 2004/3a have been sequenced partially (unpublished data). Functional analysis allowed workers to establish the biosynthetic pathway of GDP-d-rhamnose in A. thermoaerophilus L420-91T (17) and the biosynthetic pathway of GDP-d-glycero-d-manno-heptose in A. thermoaerophilus DSM 10155 (19). However, due to the lack of methods for transformation and gene knockout for A. thermoaerophilus, no direct evidence for involvement of these genes in glycan formation has been obtained so far. In A. thermoaerophilus DSM 10155 the genes rmlA, rmlB, rmlC, and rmlD map directly downstream of the genes responsible for heptose biosynthesis (Fig. 2). Southern blot analysis with degenerate primers and DNA probes for rmlA yielded a single specific signal. These results indicate that no further copies of this gene are present in the genome of A. thermoaerophilus DSM 10155. The part of the S-layer glycan biosynthesis cluster which has recently been sequenced (GenBank accession number AF324836) encodes the heptose operon and the rhamnose operon and is sufficient to provide the precursor nucleotide-activated monosaccharides for synthesis of the S-layer glycan repeating unit. However, characterization of the glycosyltransferases involved in this glycosylation pathway is not yet complete.
Enzymatic analyses with crude extracts from A. thermoaerophilus L420-91T and DSM 10155 confirmed the genetic data. All genes of the rhamnose operon are transcribed and translated to yield active enzymes and are located in the cytoplasm of A. thermoaerophilus DSM 10155 (Fig. 3). On the other hand, A. thermoaerophilus L420-91T lacks enzymatic activities for RmlC and RmlD, and in A. thermoaerophilus DSM 10155 no activities for GDP-d-rhamnose biosynthesis have been detected. These results show the specificity of precursor formation, tailored for each individual S-layer glycan. The enzymatic activities of RmlA and RmlB in A. thermoaerophilus L420-91T may correspond to the formation of dTDP-3-acetamido-3,6-dideoxy-d-galactose, the precursor of 3-acetamido-3,6-dideoxy-d-galactose (56). Biosynthesis of this intermediate has been proposed to branch off the dTDP-l-rhamnose pathway after the RmlB-catalyzed reaction step.
Enzymes involved in biosynthesis of dTDP-l-rhamnose are targets for antibacterial therapy. In a recent study several thousand substances have been screened in a microtiter plate-based assay for inhibition of RmlB, RmlC, or RmlD from M. tuberculosis (28). Use of Rml enzymes for enzymatic synthesis of dTDP-l-rhamnose, its intermediates, and dTDP-activated monosaccharides has been discussed (10, 49). However, the instability of enzymes from the mesophilic source limits productivity (49). Here we describe a simple method for expression and purification of the Rml enzymes from A. thermoaerophilus, which exhibit at least 11-fold-higher stability at 37°C than the enzymes from S. enterica LT2. The enzymes from the thermophilic source have comparable kinetic parameters. However, the Km of RmlC for dTDP-6-deoxy-d-xylo-4-hexulose is reduced 10-fold. This could be due to the instability of dTDP-6-deoxy-l-lyxo-4-hexulose (10). Since instability is expected to be greater at an elevated temperature, tighter binding of dTDP-6-deoxy-d-xylo-4-hexulose and dTDP-6-deoxy-l-lyxo-4-hexulose might be required to minimize degradation reactions. Due to the mechanistic and kinetic similarity the enzymes from A. thermoaerophilus can be used alone or in combination with other enzymes from mesophilic sources for enzymatic synthesis of dTDP-activated monosaccharides. For example, 75 μmol of dTDP-d-glucose has been converted to dTDP-l-rhamnose with a 100% yield, as previously determined with enzymes from S. enterica LT2. For characterization of the individual enzymes of the entire Rml reaction cascade, spectrophotometric assays are now available which allow analysis of each reaction step, starting from glucose 1-phosphate and dTTP. For screening purposes, these assays are easily adaptable for use in microtiter plates, as shown previously for the cascade from RmlB to -D, starting from dTDP-d-glucose (28). Similar solution requirements (divalent ions and pH value) and kinetic constants make these enzymes powerful alternatives for use in high-throughput screening, as well as for use in enzymatic synthesis.
Acknowledgments
We thank Paul Kosma for the NMR measurements and appreciate the excellent technical assistance of Sonja Zayni.
This work was supported by the Austrian Science Fund (projects P12966-MOB and P14209-MOB to P.M.) and by the Austrian Federal Ministry of Education, Science, and Culture.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boot, H. J., and P. H. Pouwels. 1996. Expression, secretion and antigenic variation of bacterial S-layer proteins. Mol. Microbiol. 21:1117-1123. [DOI] [PubMed] [Google Scholar]
- 3.Bowditch, R. D., P. Baumann, and A. A. Yousten. 1989. Cloning and sequencing of the gene encoding a 125-kilodalton surface-layer protein from Bacillus sphaericus 2362 and of a related cryptic gene. J. Bacteriol. 171:4178-4188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bradford, M. M. 1976. A rapid and sensitive method for quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 5.Corpet, F. 1988. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 16:10881-10890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dworkin, J., and M. J. Blaser. 1996. Generation of Campylobacter fetus S-layer protein diversity utilizes a single promoter on an invertible DNA segment. Mol. Microbiol. 19:1241-1253. [DOI] [PubMed] [Google Scholar]
- 7.Giraud, M. F., and J. H. Naismith. 2000. The rhamnose pathway. Curr. Opin. Struct. Biol. 10:687-696. [DOI] [PubMed] [Google Scholar]
- 8.Glaser, L., and S. Kornfeld. 1961. The enzymatic synthesis of thymidine-linked sugars. II. Thymidine diphosphate l-rhamnose. J. Biol. Chem. 236:1795-1799. [PubMed] [Google Scholar]
- 9.Glauert, A. M. 1974. Fixation, dehydration and embedding of biological specimens, p. 1-198. In A. M. Glauert (ed.), Practical methods in electron microscopy, vol. 3, part I. North-Holland Publishing Co., Amsterdam, The Netherlands. [Google Scholar]
- 10.Graninger, M., B. Nidetzky, D. E. Heinrichs, C. Whitfield, and P. Messner. 1999. Characterization of dTDP-4-dehydrorhamnose 3,5-epimerase and dTDP-4-dehydrorhamnose reductase, required for dTDP-l-rhamnose biosynthesis in Salmonella enterica serovar Typhimurium LT2. J. Biol. Chem. 274:25069-25077. [DOI] [PubMed] [Google Scholar]
- 11.Gronow, S., and H. Brade. 2001. Lipopolysaccharide biosynthesis: which steps do bacteria need to survive? J. Endotoxin Res. 7:3-23. [PubMed] [Google Scholar]
- 12.Hartmann, E., P. Messner, G. Allmeier, and H. König. 1993. Proposed pathway for biosynthesis of the S-layer glycoprotein of Bacillus alvei. J. Bacteriol. 175:4515-4519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heyndricks, M., L. Lebbe, M. Vancanneyt, K. Kersters, P. de Vos, N. A. Logan, G. Forsyth, S. Nazli, N. Ali, and R. C. W. Berkeley. 1997. A polyphasic reassessment of the genus Aneurinibacillus, reclassification of Bacillus thermoaerophilus (Meier-Stauffer et al. 1996) as Aneurinibacillus thermoaerophilus comb. nov., and emended descriptions of A. aneurinilyticus corrig., A. migulanus, and A. thermoaerophilus. Int. J. Syst. Bacteriol. 47:808-817. [Google Scholar]
- 14.Jiang, X.-M., B. Neal, F. Santiago, S. J. Lee, L. K. Romana, and P. R. Reeves. 1991. Structure and sequence of the rfb (O antigen) gene cluster of Salmonella serovar typhimurium (strain LT2). Mol. Microbiol. 5:695-713. [DOI] [PubMed] [Google Scholar]
- 15.Kandler, O. 1993. Archaea (Archaebacteria). Prog. Bot. 54:1-24. [Google Scholar]
- 16.Keenleyside, W. J., and C. Whitfield. 1999. Genetics and biosynthesis of lipopolysaccharide O-antigens, p. 331-358. In H. Brade, D. C. Morrison, S. Vogel, and S. Opal (ed.), Endotoxin in health and disease. Marcel Dekker, Inc., New York, N.Y.
- 17.Kneidinger, B., M. Graninger, G. Adam, M. Puchberger, P. Kosma, S. Zayni, and P. Messner. 2001. Identification of two GDP-6-deoxy-d-lyxo-4-hexulose reductases synthesizing GDP-d-rhamnose in Aneurinibacillus thermoaerophilus L420-91T. J. Biol. Chem. 276:5577-5583. [DOI] [PubMed] [Google Scholar]
- 18.Kneidinger, B., M. Graninger, and P. Messner. 2001. Chromosome walking by cloning of distinct PCR fragments. BioTechniques 30:248-249. [DOI] [PubMed] [Google Scholar]
- 19.Kneidinger, B., M. Graninger, M. Puchberger, P. Kosma, and P. Messner. 2001. Biosynthesis of nucleotide-activated d-glycero-d-manno-heptose. J. Biol. Chem. 276:20935-20944. [DOI] [PubMed] [Google Scholar]
- 20.Kornfeld, R., and S. Kornfeld. 1985. Assembly of asparagine-linked oligosaccharides. Annu. Rev. Biochem. 54:631-664. [DOI] [PubMed] [Google Scholar]
- 21.Kornfeld, S., and L. Glaser. 1961. The enzymatic synthesis of thymidine-linked sugars. I. Thymidine diphosphate glucose. J. Biol. Chem. 236:1791-1794. [PubMed] [Google Scholar]
- 22.Kosma, P., C. Neuninger, R. Christian, G. Schulz, and P. Messner. 1995. Glycan structure of the S-layer glycoprotein of Bacillus sp. L420-91. Glycoconjugate J. 12:99-107. [DOI] [PubMed] [Google Scholar]
- 23.Kosma, P., T. Wugeditsch, R. Christian, S. Zayni, and P. Messner. 1995. Glycan structure of a heptose-containing S-layer glycoprotein of Bacillus thermoaerophilus. Glycobiology 5:791-796. (Erratum, 6:5, 1996.) [DOI] [PubMed] [Google Scholar]
- 24.Kuen, B., A. Koch, E. Asenbauer, M. Sára, and W. Lubitz. 1997. Molecular characterization of the Bacillus stearothermophilus PV72 S-layer gene sbsB induced by oxidative stress. J. Bacteriol. 179:1664-1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kyhse-Anderson, J. 1984. Electroblotting of multiple gels: a simple apparatus without buffer tank for rapid transfer of proteins from polyacrylamide to nitrocellulose. J. Biochem. Biophys. Methods 10:203-209. [DOI] [PubMed] [Google Scholar]
- 26.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 27.Lindqvist, L., R. Kaiser, P. R. Reeves, and A. A. Lindberg. 1993. Purification, characterization and HPLC assay of Salmonella glucose-1-phosphate thymidylyl transferase from the cloned rfbA gene. Eur. J. Biochem. 211:763-770. [DOI] [PubMed] [Google Scholar]
- 28.Ma, Y., R. J. Stern, M. S. Scherman, V. D. Vissa, W. Yan, V. C. Jones, F. Zhang, S. G. Franzblau, W. H. Lewis, and M. R. McNeil. 2001. Drug targeting Mycobacterium tuberculosis cell wall synthesis: genetics of dTDP-rhamnose synthetic enzymes and development of a microtiter plate-based screen for inhibitors of conversion of dTDP-glucose to dTDP-rhamnose. Antimicrob. Agents Chemother. 45:1407-1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marumo, K., L. Lindqvist, N. Verma, A. Weintraub, P. R. Reeves, and A. A. Lindberg. 1992. Enzymatic synthesis and isolation of thymidine diphosphate-6-deoxy-d-xylo-4-hexulose and thymidine diphosphate-l-rhamnose. Production using cloned gene products and separation by HPLC. Eur. J. Biochem. 204:539-545. [DOI] [PubMed] [Google Scholar]
- 30.Meier-Stauffer, K., H.-J. Busse, F. A. Rainey, J. Burghardt, A. Scheberl, F. Hollaus, B. Kuen, A. Makristathis, U. B. Sleytr, and P. Messner. 1996. Description of Bacillus thermoaerophilus sp. nov., to include sugar beet isolates and Bacillus brevis ATCC 12990. Int. J. Syst. Bacteriol. 46:532-541. [Google Scholar]
- 31.Messner, P. 1996. Chemical composition and biosynthesis of S-layers, p. 35-76. In U. B. Sleytr, P. Messner, D. Pum, and M. Sára (ed.), Crystalline bacterial cell surface proteins. R. G. Landes/Academic Press Inc., Austin, Tex.
- 32.Messner, P. 1997. Bacterial glycoproteins. Glycoconjugate J. 14:3-11. [DOI] [PubMed] [Google Scholar]
- 33.Messner, P., F. Hollaus, and U. B. Sleytr. 1984. Paracrystalline cell wall surface layers of different Bacillus stearothermophilus strains. Int. J. Syst. Bacteriol. 34:202-210. [Google Scholar]
- 34.Messner, P., and C. Schäffer. 2000. Surface layer glycoproteins of Bacteria and Archaea, p. 93-125. In R. J. Doyle (ed.), Glycomicrobiology. Kluwer Academic/Plenum Publishers, New York, N.Y.
- 35.Munõz, R., M. Mollerbach, R. López, and E. García. 1997. Molecular organization of the genes required for the synthesis of type 1 capsular polysaccharide of Streptococcus pneumoniae: formation of binary encapsulated pneumococci and identification of cryptic dTDP-rhamnose biosynthesis genes. Mol. Microbiol. 25:79-92. [DOI] [PubMed] [Google Scholar]
- 36.Okazaki, R., T. Okazaki, J. L. Strominger, and A. M. Michelson. 1962. Thymidine diphosphate 4-keto-6-deoxy-d-glucose, an intermediate in thymidine diphosphate l-rhamnose synthesis in Escherichia coli strains. J. Biol. Chem. 237:3014-3026. [PubMed] [Google Scholar]
- 37.Palmieri, M. J., G. T. Berry, D. A. Player, S. Rogers, and S. Segal. 1991. The concentration of red blood cell UDPglucose and UDPgalactose determined by high-performance liquid chromatography. Anal. Biochem. 184:388-393. [DOI] [PubMed] [Google Scholar]
- 38.Reeves, P. R., M. Hobbs, M. A. Valvano, M. Skurnik, C. Whitfield, D. Coplin, N. Kido, J. Klena, D. Maskell, C. R. H. Raetz, and P. D. Rick. 1996. Bacterial polysaccharide synthesis and gene nomenclature. Trends Microbiol. 4:495-503. [DOI] [PubMed] [Google Scholar]
- 39.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 40.Sára, M., and U. B. Sleytr. 1994. Comparative studies of S-layer proteins from Bacillus stearothermophilus strains expressed during growth in continuous culture under oxygen-limited and non-oxygen-limited conditions. J. Bacteriol. 176:7182-7189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sára, M., and U. B. Sleytr. 2000. S-layer proteins. J. Bacteriol. 182:859-868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schäffer, C., and P. Messner. 2001. Glycobiology of surface layer proteins. Biochimie 83:591-599. [DOI] [PubMed] [Google Scholar]
- 43.Schäffer, C., T. Scherf, R. Christian, P. Kosma, S. Zayni, P. Messner, and N. Sharon. 2001. Purification and structure elucidation of the N-acetylbacillosamine-containing polysaccharide from Bacillus licheniformis ATCC 9945. Eur. J. Biochem. 268:857-864. [DOI] [PubMed] [Google Scholar]
- 44.Schäffer, C., T. Wugeditsch, C. Neuninger, and P. Messner. 1996. Are S-layer glycoproteins and lipopolysaccharides related? Microb. Drug Resist. 2:17-23. [DOI] [PubMed] [Google Scholar]
- 45.Sleytr, U. B., and T. J. Beveridge. 1999. Bacterial S-layers. Trends Microbiol. 7:253-260. [DOI] [PubMed] [Google Scholar]
- 46.Sleytr, U. B., P. Messner, D. Pum, and M. Sára (ed.). 1996. Crystalline bacterial cell surface proteins. R. G. Landes/Academic Press, Austin, Tex.
- 47.Sleytr, U. B., P. Messner, D. Pum, and M. Sára. 1999. Crystalline bacterial cell surface layers (S-layers): from supramolecular cell structure to biomimetics and nanotechnology. Angew. Chem. Int. Ed. Engl. 38:1034-1054. [DOI] [PubMed] [Google Scholar]
- 48.Smith, D. R., L. A. Doucette-Stamm, C. Deloughery, H. Lee, J. Dubois, T. Aldredge, R. Bashirzadeh, D. Blakely, R. Cook, K. Gilbert, D. Harrison, L. Hoang, P. Keagle, W. Lumm, B. Pothier, D. Qiu, R. Spadafora, R. Vicaire, Y. Wang, J. Wierzbowski, R. Gibson, N. Jiwani, A. Caruso, D. Bush, H. Safer, D. Patwell, S. Prabhakar, S. McDougall, G. Shimer, A. Goyal, S. Pietrokovski, G. M. Church, C. J. Daniels, J.-I. Mao, P. Rice, J. Nölling, and J. N. Reeve. 1997. Complete genome sequence of Methanobacterium thermoautotrophicum ΔH: functional analysis and comparative genomics. J. Bacteriol. 179:7135-7155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stein, A., M. R. Kula, and L. Elling. 1998. Combined preparative enzymatic synthesis of dTDP-6-deoxy-4-keto-d-glucose from dTDP and sucrose. Glycoconjugate J. 15:139-145. [DOI] [PubMed] [Google Scholar]
- 50.Stern, R. J., T.-Y. Lee, T.-J. Lee, W. Yan, M. S. Scherman, V. D. Vissa, S.-K. Kim, B. L. Wanner, and M. R. McNeil. 1999. Conversion of dTDP-4-keto-6-deoxyglucose to free dTDP-4-keto-rhamnose by the rmlC gene products of Escherichia coli and Mycobacterium tuberculosis. Microbiology 145:663-671. [DOI] [PubMed] [Google Scholar]
- 51.Sumper, M., and F. T. Wieland. 1995. Bacterial glycoproteins, p. 455-473. In J. Montreuil, J. F. G. Vliegenthart, and H. Schachter (ed.), Glycoproteins. Elsevier, Amsterdam, The Netherlands.
- 52.Szumilo, T., R. R. Drake, J. L. York, and A. D. Elbein. 1993. GDP-mannose pyrophosphorylase. Purification to homogeneity, properties, and utilization to prepare photoaffinity analogs. J. Biol. Chem. 268:17943-17950. [PubMed] [Google Scholar]
- 53.Tsukioka, Y., Y. Yamashita, Y. Nakano, T. Oho, and T. Koga. 1997. Identification of a fourth gene involved in dTDP-rhamnose synthesis in Streptococcus mutans. J. Bacteriol. 179:4411-4414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tsukioka, Y., Y. Yamashita, T. Oho, Y. Nakano, and T. Koga. 1997. Biological function of the dTDP-rhamnose synthesis pathway in Streptococcus mutans. J. Bacteriol. 179:1126-1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vara, J. A., and C. R. Hutchinson. 1988. Purification of thymidine-diphospho-d-glucose 4,6-dehydratase from an erythromycin-producing strain of Saccharopolyspora erythrea by high resolution liquid chromatography. J. Biol. Chem. 263:14992-14995. [PubMed] [Google Scholar]
- 56.Volk, W. A., and G. Ashwell. 1963. Enzymatic formation of TDP-3-acetamido-3,6-dideoxy hexose. Biochem. Biophys. Res. Commun. 12:116-120. [DOI] [PubMed] [Google Scholar]
- 57.Wugeditsch, T., N. E. Zachara, M. Puchberger, P. Kosma, A. A. Gooley, and P. Messner. 1999. Structural heterogeneity in the core oligosaccharide of the S-layer glycoprotein from Aneurinibacillus thermoaerophilus DSM 10155. Glycobiology 9:787-795. [DOI] [PubMed] [Google Scholar]