Abstract
We studied the hypersensitivity of clpP and clpB mutants of Escherichia coli to sodium dodecyl sulfate (SDS). Both wild-type E. coli MC4100 and lon mutants grew in the presence of 10% SDS, whereas isogenic clpP and clpB single mutants could not grow above 0.5% SDS and clpA and clpX single mutants could not grow above 5.0% SDS. For wild-type E. coli, cellular ClpP levels as determined by Western immunoblot analysis increased ca. sixfold as the levels of added SDS increased from 0 to 2%. Capsular colanic acid, measured as uronic acid, increased ca. sixfold as the levels of added SDS increased from 2 to 10%. Based on these findings, 3 of the 19 previously identified SDS shock proteins (M. Adamowicz, P. M. Kelley, and K. W. Nickerson, J. Bacteriol. 173:229-233, 1991) are tentatively identified as ClpP, ClpX, and ClpB.
Enteric bacteria such as Escherichia coli and Enterobacter cloacae have evolved to survive the conditions present within mammalian intestinal tracts, where they encounter detergents such as the bile salts, fatty acids, and lysophospholipids. E. coli is also adapted to aquatic environments outside the enteric system (1, 19), where it might also face the challenge of detergents, i.e., from wastewater treatment plants (26). We are interested in the mechanisms of detergent resistance in enteric bacteria and have used sodium dodecyl sulfate (SDS) resistance as our model system. Following our initial discovery that E. cloacae could grow in the presence of 25% SDS (12), we learned the following. (i) Bacteria tolerate SDS rather than metabolize it or modify it (12). (ii) SDS resistance is a common feature among the Enterobacteriaceae, in that 200 of 208 strains grew well in the presence of 5% SDS (14). (iii) This detergent tolerance is for neutral and anionic detergents only; all of the 208 strains studied were highly sensitive to three cationic detergents (14). (iv) SDS stress is accompanied by the synthesis of at least 19 unique or elevated SDS-induced proteins (2). (v) The outer membrane, while necessary for SDS resistance, is not entirely impervious to SDS. For E. cloacae growing in 5% SDS, measurements with 35S-SDS detected ca. 0.15 to 0.6% SDS in the periplasm (19). (vi) SDS resistance is energy dependent. The SDS-grown cells underwent rapid lysis when they ran out of energy (12) or following the addition of sodium azide or dinitrophenol (3, 4). (vii) This energy dependence reflects a requirement for ATP rather than for a proton gradient or a membrane potential (4). In this regard, one of the sites or processes where continuous ATP expenditure could be required for SDS resistance is an ATP-dependent protease (7). The present paper focuses on the role of the ClpP ATP-dependent protease in SDS resistance in E. coli.
Most intracellular proteolysis in E. coli is initiated by energy-dependent proteases. These include the serine proteases ClpAP, ClpXP, and Lon and the zinc metalloprotease HflB. HflB is the only energy-dependent protease that is essential in E. coli (7). Most eubacteria, including E. coli, have only a single clpP gene (7). As the clpP genes from other prokaryotes were identified, it became apparent that the ClpP protease plays more important and more diverse roles in other bacteria than it does in E. coli (22). Apart from the inability of clpP mutants to carry out programmed cell death (6), few phenotypes from the inactivation of ClpP in E. coli have yet been reported (22) and there is no clear answer to the question of what Clp proteases really do in E. coli (25). However, the presence of ca. 0.3% SDS in the periplasm of E. coli (19) suggests that there should also be a significant level of SDS in the cytoplasm, which could in turn contribute to a population of misfolded or denatured cytoplasmic proteins which would be toxic to the cell (25). Thus, their removal by intracellular, ATP-dependent proteolysis would be critical for cell survival. The present paper provides evidence that (i) ClpP has an essential function in E. coli's SDS tolerance, (ii) sensitivity to SDS is a physiologically relevant phenotype resulting from the loss of ClpP, and (iii) there is a significant overlap between the SDS shock proteins (2) and the heat shock proteins (16) in that both SDS and high temperatures induce ClpP, ClpX, and ClpB.
The strains used in this study are listed in Table 1. P1 bacteriophage was obtained from Paul Blum. Isogenic mutants were constructed by P1 transduction using standard techniques (18). Luria-Bertani medium (LB) supplemented with 0.1% glucose was used for studies in the rich medium for E. coli. When required, chloramphenicol (34 μg/ml) and/or kanamycin (50 μg/ml) was added to the medium. All studies were conducted at 37°C unless otherwise specified. Anti-ClpP antibody was a gift from Michael Maurizi (17). For demonstrating the levels of ClpP protein at different SDS concentrations, Western blot analyses were done with cells grown in LB with 0.1% glucose. After 5 h, the optical density at 600 nm of the cultures were determined and cell densities were normalized to an optical density at 600 nm of 1. The cells were then harvested in a Mistral 2000E tabletop centrifuge at 6,000 rpm for 15 min. The cell pellets were washed twice with sterile distilled water and resuspended in 0.5 ml of cold sterile distilled water. Total cell protein (membrane and cytoplasmic) was then extracted by sonication using a Sonifier cell disruptor (model W185). Diluted sonicate (20 μl, 1:10) was then mixed with sample buffer in each case. Polyacrylamide (15%) gel analysis with SDS was carried out according to Laemmli (15). Development of Western blots and detection of ClpP used a ClpP-specific primary antibody and an anti-rabbit immunoglobulin G secondary antibody conjugated with two near-infrared fluorescent dyes (10). Fluorescence emission was determined simultaneously at 720 and 820 nm using an Odyssey infrared imaging system (LI-COR Biosciences, Lincoln, Nebr.).
TABLE 1.
Bacterial strains
| Strain | Relevant genotype or description | Sourcea |
|---|---|---|
| SG20250 | MC4100 F−araD139Δ(argF-lac) U169flbB5301 deoC1 rpsL150 relA1 | S. Gottesman |
| JT4000 | MC4100 Δlon-100 | S. Gottesman |
| SG22098 | MC4100 clpP::cat | S. Gottesman |
| SG12045 | C600 clpA319::kan | S. Gottesman |
| SG12058 | C600 clpX::kan | S. Gottesman |
| JGT3 | MC4100 Δ clpB::kan | F. Baneyx |
| SG22178 | MC4100 clpA::kan, clpX::kan, Δlon | T. Baker from S. Gottesman |
| SG22091 | MC4100 clpB::kan, clpX::kan, ΔclpA | A. L. Goldberg (from S. Gottesman) |
| SG22097 | MC4100 clpP::cat, clpX::kan | S. Gottesman |
| EL99/07 | MC4100 clpA::kan, clpP::cat | A. Taylor |
| EL99/08 | MC4100 clpB::kan, clpP::cat | A. Taylor |
| KNSR-1 | P1 (SG22098) × MC4100 | This study; Camr |
| KNSR-2 | P1 (SG12045) × MC4100 | This study; Kanr |
| KNSR-3 | P1 (SG12058) × MC4100 | This study; Kanr |
| KNSR-4 | P1 (SG22097) × MC4100 | This study; Camr, Kanr |
| KNSR-5 | P1 (EL 99/07) × MC4100 | This study; Camr, Kanr |
| KNSR-6 | P1 (EL 99/08) × MC4100 | This study; Camr, Kanr |
| KNSR-8 | P1 (JGT3) × MC4100 | This study; Kanr |
For strains derived in this study, either chloramphenicol (34 μg/ml) or kanamycin (50 μg/ml) was used to select transductants. The selection of double mutants was carried out in sequence by selecting for chloramphenicol resistance and then using Camr clones to select for kanamycin resistance.
Role of ClpP protease and related proteins in SDS resistance.
Levels of growth achieved by a clpP mutant of E. coli were compared with its parent, MC4100, in the presence and absence of SDS (Table 2). The mutant KNSR-1 has an insertion in clpP and does not contain detectable ClpP protein, as determined by Western blotting with anti-ClpP antibodies. The two strains grew equally well in LB broth without SDS, but the ClpP-defective strain could not grow in the presence of 10% SDS and achieved a maximum turbidity of only 23 Klett units in LB plus 0.5% SDS. clpA clpP, clpX clpP, and clpB clpP double mutants were also sensitive to 0.5% SDS (Table 2). In contrast, the parent MC4100 grew well in 10% SDS. The inability of clpP E. coli to grow in LB plus 10% SDS was partially rescued by reducing the temperature. Maximum turbidity levels of 135, 140, 7, and 1 Klett units were observed at 30, 34, 37, and 42°C, respectively. The inability of clpP E. coli to grow in M63 plus 5% SDS was not rescued by exogenous proline (1 mM), betaine (1 mM), or potassium chloride (10 mM). These additions were tried because 5 to 10% SDS entails both a detergent burden and an osmotic burden (3).
TABLE 2.
Growth of E. coli strains in LB and M63 with added SDSa
| Strain | Protein(s) with mutation | Growth (Klett units) with SDS at:
|
||
|---|---|---|---|---|
| 10% | 5% | 0.5% | ||
| SG20250 | None (parent, MC4100) | 105 ± 3 | 130 ± 3 | 215 ± 0 |
| JT4000 | Lon | 107 ± 5 | 142 ± 2 | 232 ± 1 |
| KNSR-1 | ClpP | 7 ± 5 | 1 ± 0 | 23 ± 5 |
| KNSR-2 | ClpA | 4 ± 0 | 42 ± 9 | 198 ± 5 |
| KNSR-3 | ClpX | 2 ± 0 | 38 ± 5 | 143 ± 7 |
| KNSR-8 | ClpB | 2 ± 0 | 5 ± 0 | 11 ± 0 |
| SG22178 | ClpA and ClpX | 30 ± 6 | 42 ± 5 | 198 ± 10 |
| SG22091 | ClpA, ClpB, and ClpX | 1 ± 0 | 2 ± 0 | 5 ± 0 |
| KNSR-4 | ClpX ClpP | 7 ± 2 | 9 ± 0 | 15 ± 4 |
| KNSR-5 | ClpA ClpP | 2 ± 0 | 8 ± 0 | 19 ± 2 |
| KNSR-6 | ClpB ClpP | 1 ± 0 | 5 ± 5 | 16 ± 9 |
Cultures were grown at 37°C in LB with 0.1% glucose, the appropriate antibiotic, and the indicated concentration of SDS in 250-ml Nephalo flasks with 14-mm-diameter side arms. Growth levels were recorded on a Klett colorimeter with a red 660-nm filter. Values are averages of triplicate experiments with standard errors. For the clpP mutants, the absence of growth was confirmed by plate counts on LB agar. Cellular responses to SDS were the same for growth in LB (rich) and defined M63 (18) media except for the clpA clpX double mutant, which exhibited some growth in LB plus 10% SDS but no growth in M63 plus 10% SDS.
The specificity of clpP sensitivity to SDS was shown by the fact that it achieved the same levels of growth as MC4100 in both 10% polyethylene glycol and 0.2% deoxycholic acid (data not shown). The specificity of the requirement for a functional ClpP protease was also shown by examining a series of isogenic strains defective in related genes (Table 2). The Lon− strain JT4000 still grew well in 10% SDS (Table 2) and the mutants with single defects in ClpA or ClpX grew in 5% SDS, as did the double mutant with defects in both ClpA and ClpX (Table 2). However, the clpB mutant and the triple mutant with defects in ClpA, ClpB, and ClpX were unable to grow in 5% SDS and barely grew in 0.5% SDS (Table 2).
The two-component Clp proteases, such as ClpAP and ClpXP, are elaborate complexes that couple the ATPase and protein unfolding activity of a molecular chaperone with the degradative activity of an endopeptidase (29). ClpP provides the protease site, while ClpX or ClpA acts as an ATPase and in doing so targets the proteins to be degraded. Presumably, ClpX and ClpA target different sets of proteins in vivo (8). However, in vitro experiments can often use ClpA and ClpX interchangeably (9). Our data (Table 2) show that resistance to ≥0.5% SDS requires ClpP but not ClpA or ClpX: both the clpA and clpX single mutants and the clpAX double mutant were able to grow in 5% SDS. Gottesman (7) concluded that a requirement for ClpP but neither ClpA nor ClpX suggests the existence of yet another targeting subunit for ClpP. Based on the SDS sensitivity of the clpB and the clpA clpB clpX mutants (Table 2), the needed targeting subunit could be ClpB (25) but certainly does not have to be ClpB.
ClpP and colanic acid levels in wild-type E. coli.
The importance of ClpP in bacterial SDS resistance was confirmed by measuring ClpP levels in wild-type E. coli MC4100 grown at 37°C in LB plus 0 to 4% SDS. In a typical Western blot experiment (Fig. 1), ClpP levels increased 3-, 7-, and 5-fold for cells grown in 1, 2, and 4% SDS, respectively. Further experiments measuring ClpP levels at increasing concentrations of SDS showed that the ClpP levels reached a plateau for cells grown in 2 to 7% SDS (Fig. 2) and actually declined in cells grown in 10% SDS (Fig. 2).
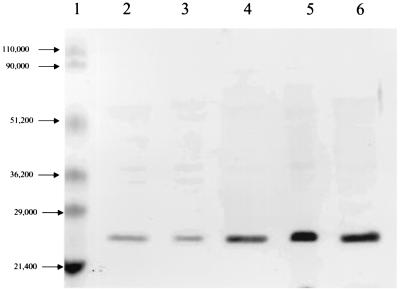
FIG. 1.
Western blot for ClpP levels in wild-type (MC4100) E. coli grown in the presence of SDS. Cells were grown at 37°C in LB with 0, 0.5, 1.0, 2.0, and 4.0% SDS. Cellular proteins were detected with anti-ClpP antibody (17). Lane 1, prestained molecular weight markers. The ClpP band intensities (pixels/mm2) from left to right were as follows: in lane 2, 20,450 (0% SDS); in lane 3, 14,660 (0.5% SDS); in lane 4, 58,190 (1% SDS); in lane 5, 144,550 (2% SDS); and in lane 6, 92,030 (4% SDS). The latter four values correspond to ClpP increases of 0.7-, 3-, 7-, and 5-fold, respectively.
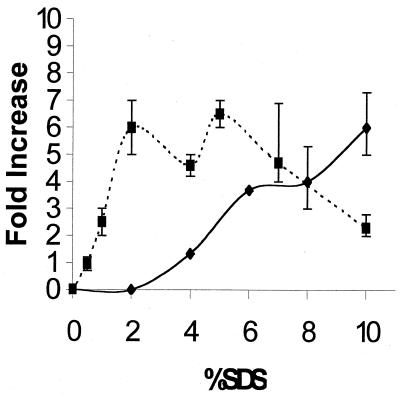
FIG. 2.
Increase in ClpP and capsular material for wild-type (MC4100) E. coli grown at 37°C in the presence of 0 to 10% SDS. ▪, ClpP levels determined by intensity of Western blots; ♦, capsule levels determined by spectrophotometric assay for the uronic acids in colanic acid (5). To make sure that increasing levels of SDS did not interfere with the colanic acid assays, SDS was removed prior to the assay by the addition of KCl to a final concentration of 5% (13). Also, controls with 0 to 10% SDS but no colanic acid gave no color development. All points represent averages of three or more measurements. The data shown in Fig. 1 are one set of the ClpP measurements used here.
Thus, our ClpP results indicated that an alternate mechanism for SDS resistance comes into play at high SDS concentrations. Accordingly, we decided to measure the capsular colanic acid levels for E. coli MC4100 grown in LB plus 0 to 10% SDS (Fig. 2). This choice was made for three reasons. First, capsule synthesis is known to be induced by osmotic shock (24). Second, clpP mutants of E. coli regained the ability to grow in 10% SDS as the temperature was lowered from 37 to 30°C, perhaps because the synthesis of colanic acid is turned on at 30°C (28). Third, lon mutants of E. coli, which grew well in 10% SDS (Table 2), are known to overproduce capsular colanic acid (27).
Total capsule size, as examined by negative-stain microscopy, did not change dramatically. However, capsule measurements based on the uronic acid content of colanic acid (5) increased significantly when cells were grown in ≥ 4% SDS, i.e., levels of SDS at which the ClpP concentration had already plateaued. Above 7% SDS the colanic acid levels continued to increase even as the levels of ClpP decreased. Uronic acid in the capsule is negatively charged (23) and would be expected to repel anionic detergents such as SDS. This electrostatic repulsion provides a rationale for increased colanic acid production in the presence of increasing SDS. We are observing two very different bacterial responses to SDS. From 0 to 2% SDS the cell elevates ClpP, while from 2 to 10% SDS it elevates the colanic acid content of its capsule. Normally ClpP is present as ca. 0.05% of cellular protein (11) so after a sixfold increase it would comprise 0.30% of cellular protein. Perhaps a further increase in ClpP would be unable to cope with >2% SDS, or perhaps colanic acid production provides a more cost-effective solution.
New phenotype for ClpP protease.
ClpP mutants of E. coli are unable to tolerate ≥0.5% SDS. This SDS hypersensitivity could result either from a yet-uncharacterized regulatory circuit being perturbed by the lack of ClpP activity or, more directly, from the need to remove misfolded or denatured proteins. In either case, the involvement of both ClpP and colanic acid in SDS resistance shows that the cellular response to SDS occurs in at least five different locations. Moving progressively inwards, these would be (i) a negatively charged capsule for electrostatic repulsion of SDS, (ii) the outer membrane as a necessary barrier (30), (iii) the periplasm (19), (iv) the cytoplasmic membrane as the site of numerous efflux pumps (20) able to extrude SDS, and (v) the cytoplasm itself in the form of the ClpP and ClpB proteases. Thus, it appears that SDS resistance is a cooperative effort, with relative contributions by five different compartments of the cell. Having five locations participate in a cumulative resistance to SDS provides a ready explanation for any strain-specific differences in SDS resistance observed for E. coli.
A functional ClpP protease is needed for SDS resistance. (i) clpP mutants are inhibited by 0.5 to 10% SDS but not by 10% polyethylene glycol. Showing that the clpP mutant's sensitivity is to anionic detergents is important because Aspedon and Nickerson (3) showed that many of the physiological changes evident in bacteria growing in 10% SDS were due to the osmotic effects of 10% SDS, not the detergent effects. (ii) clpP mutants are inhibited by 10% SDS but lon mutants are not. This distinction occurs despite the fact that Clp and Lon are both intracellular ATP-dependent proteases and that Lon is probably the primary protease degrading abnormally folded proteins in E. coli (7). (iii) SDS sensitivity is due to the absence of ClpP (confirmed by Western blots) rather than the presence of the cat insertion, which inactivated the clpP gene. (iv) The SDS sensitivity of the clpAP and clpXP double mutants (Table 2) shows that SDS sensitivity is due to the absence of ClpP rather than to the accumulation of ClpA or ClpX as “proteins without partners.”
Overlap with SDS shock proteins and heat shock proteins.
Recognition of the importance of Clp proteases in SDS resistance also impacts two of our previous observations. First, Adamowicz et al. (2) used two-dimensional gel electrophoresis to identify at least 19 unique and elevated SDS-induced proteins. These proteins were identified as spots A through S because at that time only their positions on the two-dimensional gels were known. It later became apparent that one of them—spot S, at 12.8 kDa and pI 5.35—was likely the universal stress protein identified by Nyström and Neidhardt (21). It now seems likely that spot O, at 23.0 kDa and pI 5.89, is ClpP, spot G, at 45.3 kDa and pI 5.51, is ClpX, and spot B, at 98.0 kDa and pI 5.43, is ClpB. Adamowicz et al. (2) showed that spots O, G, and B increased 4.8-, 6.8-, and 11.8-fold, respectively, in E. coli grown in the presence of 5% SDS, and these values agree well with the 6-fold increase in ClpP detected by Western blots of E. coli grown in 5% SDS (Fig. 2). It is reasonable that ClpP and ClpX are both induced by SDS (2), because they are cotranscribed in E. coli (7).
Second, Kramer and Nickerson (13) found that growth of E. cloacae in the presence of 10% SDS was accompanied by an energy burden in the form of a 20% decreased cell yield, a 30% higher rate of glucose utilization, and a 70% increased rate of oxygen consumption. One explanation for the SDS hypersensitivity of clpP is that E. coli has to cope with protein denaturation in the cytoplasm during growth in SDS and ClpP plays a major role in getting rid of these denatured or misfolded proteins. Growth in SDS would then include a futile cycle in which ATP was expended both to synthesize proteins and to degrade denatured proteins. Consistent with this theme is the fact that the clpB and clpP clpX operons both have two promoters. The first is a basal promoter, while the second is recognized by σ32 (RpoH) and acts as a heat shock promoter (7, 17), even though ethanol (4%) treatment also induces the heat shock response quite specifically in E. coli (16). As a partial answer to the question posed by LaRossa and Van Dyk (16), whether the nature of the bacterial response allows us to define the nature of the initial insult, it makes sense that there should be some overlap between heat, ethanol, and detergent shock proteins if they all respond to a signal of abnormal or unfolded proteins. Finally, unlike the other Clp ATPases, ClpB has not yet been implicated in the energy-dependent degradation of denatured proteins (7, 25, 31). It has, however, been shown to cooperate with other heat shock proteins in suppressing and reversing protein aggregation (31). Thus, the SDS hypersensitivity of both clpP and clpB mutants of E. coli suggests that SDS stress involves formation of both denatured and aggregated proteins in the bacterial cytoplasm.
Acknowledgments
This research was supported by grants to K.W.N. from the Nebraska Corn Board.
We thank Michael Maurizi for providing the anti-ClpP antiserum and Susan Gottesman for providing many of the Clp-related mutants listed in Table 1 and valuable suggestions on the manuscript. We thank Li-Cor Inc. for providing the Western blot infrared imaging system, the Center for Biotechnology for their facilities, Nancy E. Caceres for assistance with Western blot analysis, and Brian Stalling for technical assistance.
REFERENCES
- 1.Adamowicz, M., T. Conway, and K. W. Nickerson. 1991. Nutritional complementation of oxidative glucose metabolism in Escherichia coli via pyrroloquinoline quinone-dependent glucose dehydrogenase and the Entner-Doudoroff pathway. Appl. Environ. Microbiol. 57:2012-2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adamowicz, M., P. M. Kelley, and K. W. Nickerson. 1991. Detergent (sodium dodecyl sulfate) shock proteins in Escherichia coli. J. Bacteriol. 173:229-233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aspedon, A., and K. W. Nickerson. 1993. A two-part energy burden imposed by growth of Enterobacter cloacae and Escherichia coli in sodium dodecyl sulfate. Can. J. Microbiol. 39:555-561. [DOI] [PubMed] [Google Scholar]
- 4.Aspedon, A., and K. W. Nickerson. 1994. The energy dependence of detergent resistance in Enterobacter cloacae: a likely requirement for ATP rather than a proton gradient or a membrane potential. Can. J. Microbiol. 40:184-191. [DOI] [PubMed] [Google Scholar]
- 5.Blumenkrantz, N., and G. Hansen-Asboe. 1973. New method for quantitative determination of uronic acids. Anal. Biochem. 54:484-489. [DOI] [PubMed] [Google Scholar]
- 6.Engelberg-Kulka, H., M. Reches, S. Narasimhan, R. Schoulaker-Schwarz, Y. Klemes, E. Aizenman, and G. Glaser. 1998. rexB of bacteriophage λ is an anti-cell death gene. Proc. Natl. Acad. Sci. USA 95:15481-15486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gottesman, S. 1996. Proteases and their targets in Escherichia coli. Annu. Rev. Genet. 30:465-506. [DOI] [PubMed] [Google Scholar]
- 8.Gottesman, S., M. R. Maurizi, and S. Wickner. 1997. Regulatory subunits of energy-dependent proteases. Cell 91:435-438. [DOI] [PubMed] [Google Scholar]
- 9.Gottesman, S., E. Roche, Y. N. Zhou, and R. T. Sauer. 1998. The ClpXP and ClpAP proteases degrade proteins with carboxy-terminal peptide tails added by the SsrA-tagging system. Genes Dev. 12:1338-1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang, J. N., I. Park, E. Ellingson, L. E. Littlepage, and D. Pellman. 2001. Activity of the APC Cdh1 form of the anaphase-promoting complex persists until S phase and prevents the premature expression of Cdc 20p. J. Cell Biol. 154:85-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Katayama, Y., S. Gottesman, J. Pumphrey, S. Rudikoff, W. P. Clark, and M. R. Maurizi. 1988. The two-component, ATP-dependent Clp protease of Escherichia coli. J. Biol. Chem. 263:15226-15236. [PubMed] [Google Scholar]
- 12.Kramer, V. C., D. M. Calabrese, and K. W. Nickerson. 1980. Growth of Enterobacter cloacae in the presence of 25% sodium dodecyl sulfate. Appl. Environ. Microbiol. 40:973-976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kramer, V. C., and K. W. Nickerson. 1984. A transport-dependent energy burden imposed by growth of Enterobacter cloacae in the presence of 10% sodium dodecyl sulfate. Can. J. Microbiol. 30:699-702. [DOI] [PubMed] [Google Scholar]
- 14.Kramer, V. C., K. W. Nickerson, N. V. Hamlett, and C. O'Hara. 1984. Prevalence of extreme detergent resistance among the Enterobacteriaceae. Can. J. Microbiol. 30:711-713. [DOI] [PubMed] [Google Scholar]
- 15.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 16.LaRossa, R. A., and T. K. Van Dyk. 2000. Applications of stress responses for environmental monitoring and molecular toxicology, p. 453-468. In G. Storz and R. Hengge-Aronis (ed.), Bacterial stress responses. ASM Press, Washington, D.C.
- 17.Maurizi, M. R., W. P. Clark, Y. Katayama, S. Rudikoff, J. Pumphrey, B. Bowers, and S. Gottesman. 1990. Sequence and structure of ClpP, the proteolytic component of the ATP-dependent Clp protease of Escherichia coli. J. Biol. Chem. 265:12536-12545. [PubMed] [Google Scholar]
- 18.Miller, J. H. 1992. A short course in bacterial genetics. A laboratory manual and handbook for Escherichia coli and related bacteria. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 19.Nickerson, K. W., and A. Aspedon. 1992. Detergent-shock response in enteric bacteria. Mol. Microbiol. 6:957-961. [DOI] [PubMed] [Google Scholar]
- 20.Nikaido, H. 1996. Multidrug efflux pumps of gram-negative bacteria. J. Bacteriol. 178:5853-5859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nyström, T., and F. C. Neidhardt. 1992. Cloning, mapping, and nucleotide sequence of a gene encoding a universal stress protein in Escherichia coli. Mol. Microbiol. 6:3187-3198. [DOI] [PubMed] [Google Scholar]
- 22.Porankiewicz, J., J. Wang, and A. K. Clarke. 1999. New insights into the ATP-dependent Clp protease: Escherichia coli and beyond. Mol. Microbiol. 32:449-458. [DOI] [PubMed] [Google Scholar]
- 23.Rick, P. D., and R. P. Silver. 1996. Enterobacterial common antigen and capsular polysaccharides, p. 104-122. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella, 2nd ed. ASM Press, Washington, D.C.
- 24.Sledjeski, D. D., and S. Gottesman. 1996. Osmotic shock induction of capsule synthesis in Escherichia coli K-12. J. Bacteriol. 178:1204-1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Squires, C., and C. L. Squires. 1992. The Clp proteins: proteolysis regulators or molecular chaperones? J. Bacteriol. 174:1081-1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Swisher, R. D. 1987. Surfactant biodegradation, 2nd ed. Marcel Dekker, Inc., New York, N.Y.
- 27.Torres-Cabassa, A. S., and S. Gottesman. 1987. Capsule synthesis in Escherichia coli K-12 is regulated by proteolysis. J. Bacteriol. 169:981-989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Whitfield, C., and I. S. Roberts. 1999. Structure, assembly and regulation of expression of capsules in Escherichia coli. Mol. Microbiol. 31:1307-1319. [DOI] [PubMed] [Google Scholar]
- 29.Wickner, S., and M. R. Maurizi. 1999. Here's the hook: similar substrate binding sites in the chaperone domains of Clp and Lon. Proc. Natl. Acad. Sci. USA 96:8318-8320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yethon, J. A., D. E. Heinrichs, M. A. Monteiro, M. B. Perry, and C. Whitfield. 1998. Involvement of waaY, waaQ, and waaP in the modification of Escherichia coli lipopolysaccharide and their role in the formation of a stable outer membrane. J. Biol. Chem. 273:26310-26316. [DOI] [PubMed] [Google Scholar]
- 31.Zolkiewski, M. 1999. ClpB cooperates with DnaK, DnaJ, and GrpE in suppressing protein aggregation. J. Biol. Chem. 274:28083-28086. [DOI] [PubMed] [Google Scholar]