Abstract
From 1975 to 1999, Clostridium perfringens caused 238 food-borne disease outbreaks in Finland, which is 20% of all such reported outbreaks during these years. The fact that C. perfringens is commonly found in human and animal stools and that it is also widespread in the environment is a disadvantage when one is searching for the specific cause of a food-borne infection by traditional methods. In order to strengthen the evidence-based diagnostics of food poisonings suspected to be caused by C. perfringens, we retrospectively investigated 47 C. perfringens isolates by PCR for the cpe gene, which encodes enterotoxin; by reversed passive latex agglutination to detect the enterotoxin production; and by pulsed-field gel electrophoresis (PFGE) to compare their genotypes after restriction of DNA by the enzymes SmaI and ApaI. The strains were isolated during 1984 to 1999 from nine food-borne outbreaks of disease originally reported as having been caused by C. perfringens. In seven of the nine outbreaks our results supported the fact that the cause was C. perfringens. Our findings emphasize the importance of a more detailed characterization of C. perfringens isolates than mere identification to the species level in order to verify the cause of an outbreak. Also, to increase the probability of finding the significant cpe-positive C. perfringens strains, it is very important to isolate and investigate more than one colony from the fecal culture of a patient and screen all these isolates for the presence of the cpe gene before further laboratory work is done.
Clostridium perfringens is an important causative agent of food poisoning. It is also a commonly found member of normal flora of humans, rendering the specific diagnostics difficult in food-borne infections (31). C. perfringens is classified into five types (A to E) according to the exotoxin produced (26, 33). The enterotoxin produced by type A C. perfringens is responsible for acute diarrhea and cramping, which are the predominant symptoms of food poisoning caused by this organism (22, 32, 36). Recent studies indicate that in most C. perfringens isolates from food poisonings, the cpe gene has a chromosomal location, whereas in non-food-borne gastrointestinal illnesses the cpe gene is located on a plasmid (10, 11, 19). Furthermore, the cpe plasmid of at least some non-food-borne gastrointestinal disease isolates can transfer via conjugation (5), and it has been proposed that the chromosomal cpe gene is located on a transposon (4, 6). Thus, both the chromosomal cpe gene and the plasmid cpe gene may be present on mobile genetic elements.
Many genotypic methods, including plasmid analysis (14, 24, 35), ribotyping (15, 34), PCR (7, 12, 20), pulsed-field gel electrophoresis (PFGE) (10, 25, 30), and amplified-fragment length polymorphism (27), have already been used, and their discriminatory powers in the study of C. perfringens isolates have been tested. PFGE has been used to study the location of the cpe gene and the isolates associated with non-food-borne human gastrointestinal diseases (10, 11, 19, 37). However, as far as we know, it has been used as few as three times in order to subtype food-borne C. perfringens strains connected to outbreaks (25, 30, 35).
In Finland, a standardized fecal culture “package”—which includes cultures for Salmonella spp., Shigella spp., Campylobacter spp., Yersinia spp., Staphylococcus aureus, Bacillus cereus, and C. perfringens—has been used to investigate food-borne outbreaks of disease since the 1980s. However, when C. perfringens has been suspected to be the cause of an outbreak, the final diagnosis has mainly been based on the clinical symptoms of the patients and/or findings in foods, and the human fecal C. perfringens isolates have never been studied by genotyping methods.
In this study, a PCR technique was set up for the rapid and specific detection of C. perfringens strains with the cpe gene, which encodes enterotoxin, and used to investigate 47 C. perfringens isolates related to nine food-borne outbreaks of disease suspected of having been caused by C. perfringens. Also, the abilities of these isolates to produce enterotoxin were tested, and they were subtyped by PFGE to give further molecular epidemiological information on the outbreaks.
MATERIALS AND METHODS
Strains.
All available C. perfringens isolates (n = 47) related to nine food-borne outbreaks of disease from 1984 to 1999 were studied (Table 1); 40 isolates were from humans (one pure culture per person), and seven were from foodstuffs. The strains had previously been isolated from primary fecal cultures at the Laboratory of Enteric Pathogens, National Public Health Institute (KTL), or received for verification from other laboratories. The fecal samples from the suspected food poisoning outbreaks were not available for the direct detection of C. perfringens enterotoxin. The identification of the strains was confirmed by standard techniques (1). C. perfringens NCTC 8238 and NCTC 8239 were used as positive controls for cpe, and C. perfringens ATCC 3624 was used as a negative control in PCR. All strains had been stored at −70°C in sterilized skim milk.
TABLE 1.
C. perfringens isolates
| Suspected outbreak cluster | Yr/mo | Origin | Strain | PET-RPLA kit resulta | PCR cpeb | PFGE subtypec | Interpretation |
|---|---|---|---|---|---|---|---|
| I | 1984/8 | Human | AHS 7206 | − | − | NT | Nonoutbreak strain |
| I | 1984/8 | Human | AHS 7207 | − | − | NT | Nonoutbreak strain |
| I | 1984/8 | Human | AHS 7208 | − | − | NT | Nonoutbreak strain |
| I | 1984/8 | Human | AHS 7209 | − | − | Hh | Nonoutbreak strain |
| I | 1984/8 | Human | AHS 7210 | − | − | Ii | Nonoutbreak strain |
| I | 1984/8 | Human | AHS 7211 | − | − | Jj | Nonoutbreak strain |
| II | 1988/11 | Human | AHS 7425 | + | + | Ee | Outbreak strain |
| II | 1988/11 | Human | AHS 7426 | − | − | Kk | Nonoutbreak strain |
| II | 1988/11 | Human | AHS 7427 | −f | + | Ff | Outbreak strain |
| III | 1992/9 | Human | AHS 7563 | + | + | Cc | Outbreak strain |
| III | 1992/9 | Human | AHS 7564 | + | + | Cc | Outbreak strain |
| III | 1992/9 | Human | AHS 7565 | − | − | Gg | Nonoutbreak strain |
| III | 1992/9 | Human | AHS 7566 | + | + | Cc | Outbreak strain |
| III | 1992/9 | Human | AHS 7567 | + | + | Cc | Outbreak strain |
| III | 1992/9 | Human | AHS 7568 | + | + | Cc | Outbreak strain |
| IV | 1993/10 | Human | AHS 7577 | + | + | Dd | Outbreak strain |
| IV | 1993/10 | Human | AHS 7578 | + | + | Dd | Outbreak strain |
| IV | 1993/10 | Human | AHS 7579 | + | + | Dd | Outbreak strain |
| IV | 1993/10 | Human | AHS 7580 | + | + | Dd | Outbreak strain |
| IV | 1993/10 | Human | AHS 7581 | + | + | Dd | Outbreak strain |
| V | 1994/8 | Human | AHS 7616 | − | − | Ll | Nonoutbreak strain |
| V | 1994/8 | Human | AHS 7617 | − | − | Mm | Nonoutbreak strain |
| V | 1994/8 | Human | AHS 7619 | − | − | Gg | Nonoutbreak strain |
| V | 1994/8 | Human | AHS 7620 | − | − | Nn | Nonoutbreak strain |
| VI | 1997/10 | Human | AHS 25154 | + | + | Bb | Outbreak strain |
| VI | 1997/10 | Human | AHS 25155 | + | + | Bb | Outbreak strain |
| VI | 1997/10 | Human | AHS 25156 | + | + | Bb | Outbreak strain |
| VI | 1997/10 | Foodstuffd | AHS 25157 | + | + | Bb | Outbreak strain |
| VI | 1997/10 | Foodstuffd | AHS 25158 | + | + | Bb | Outbreak strain |
| VII | 1998/4 | Human | IH 110562 | + | + | Aa | Outbreak strain |
| VII | 1998/4 | Human | IH 110563 | − | − | Pp | Nonoutbreak strain |
| VIII | 1998/10 | Human | IH 110742 | + | + | Aa | Outbreak strain |
| VIII | 1998/10 | Human | IH 110743 | + | + | Aa | Outbreak strain |
| VIII | 1998/10 | Human | IH 110744 | + | + | Aa | Outbreak strain |
| VIII | 1998/10 | Human | IH 110745 | + | + | Aa | Outbreak strain |
| VIII | 1998/10 | Human | IH 110746 | + | + | Aa | Outbreak strain |
| VIII | 1998/10 | Human | IH 110747 | + | + | Aa | Outbreak strain |
| VIII | 1998/10 | Human | IH 110748 | + | + | Aa | Outbreak strain |
| VIII | 1998/10 | Human | IH 110749 | + | + | Aa | Outbreak strain |
| IX | 1999/3 | Foodstuffe | IH 111211 | + | + | Aa | Outbreak strain |
| IX | 1999/3 | Foodstuffe | IH 111212 | + | + | Aa | Outbreak strain |
| IX | 1999/3 | Foodstuffe | IH 111213 | + | + | Aa | Outbreak strain |
| IX | 1999/3 | Foodstuffe | IH 111214 | + | + | Aa | Outbreak strain |
| IX | 1999/3 | Foodstuffe | IH 111215 | + | + | Aa | Outbreak strain |
| IX | 1999/3 | Human | IH 111218 | − | − | Nonoutbreak strain | |
| IX | 1999/3 | Human | IH 111219 | + | + | Aa | Outbreak strain |
| IX | 1999/3 | Human | IH 111220 | + | + | Aa | Outbreak strain |
+, CPE positive; −, CPE negative.
+, enterotoxin gene positive; −, enterotoxin gene negative.
Uppercase letter indicates the subtype when the strain was digested with SmaI, and lowercase letter indicates the subtype when the strain was digested with ApaI. NT, nontypeable.
Meat casserole.
Minced meat casserole.
Three repetitive determinations for production of enterotoxin gave a negative result.
PCR for cpe gene.
The template DNA for the PCR amplification was obtained from the C. perfringens bacteria by a direct lysis method as described earlier (7). The sequences of the oligonucleotide primers for the cpe enterotoxin gene were selected from the sequence published by Van Damme-Jongsten et al. in 1989, taking into account the differences noted in the publication of Czeczulin et al. in 1993 (7).
The primers were as follows: 5′-TAA CAA TTT AAA TCC AAT GG-3′ and 5′-ATT GAA TAA GGG TAA TTT CC-3′. Two microliters of lysed cells was added into PCR mixture as described earlier (7). PCR was performed in an Eppendorf (Hamburg, Germany) Mastercycler Gradient or Hybaid (Ashford, Middlesex, United Kingdom) PCR Sprint Temperature Cycling apparatus. The following procedure was used: initial denaturation at 95°C for 10 min, followed by 35 cycles consisting of 30 s at 94°C, 30 s at 46°C, and 30 s at 72°C each. The final step was a 10-min incubation at 72°C. The amplification products were analyzed by electrophoresis at 90 V for 1 h and 15 min in a 2% SeaKem ME agarose gel (FMC BioProducts, Rockland, Maine) with a GIBCO BRL Horizon 20.25 system (Life Technologies Inc., Gaithersburg, Md.). The gels were stained with ethidium bromide (0.5 μg/ml) and photographed under UV illumination. pUC Mix Marker 8 (MBI Fermentas Ltd., Vilnius, Lithuania) was used as a molecular weight standard. The size of an amplified fragment was 933 bp.
Assay of enterotoxin production.
The enterotoxin produced by type A C. perfringens strains was detected by reversed passive latex agglutination according to the instructions of the manufacturer (PET-RPLA kit; Oxoid Ltd., Basingstoke, Hampshire, England). Modified Duncan and Strong medium was used for sporulation (13). The growth of strains AHS 7427 and AHS 25157 was monitored by culturing one loopful (10 μl) of cooked meat medium on brucella medium before and after heat treatment at 75°C for 20 min. The sporulation products of these strains after culturing in modified Duncan and Strong medium were examined under the microscope.
PFGE.
C. perfringens strains were grown anaerobically on egg yolk agar overnight at 37°C and then for 5 h at 37°C in Trypticase-glucose-yeast extract broth. Two milliliters of this Trypticase-glucose-yeast extract broth culture was mixed with 5 ml of cold PIV buffer (10 mM Tris, 1 M NaCl). The mix was centrifuged for 15 min at 4°C at 3,000 rpm with a Midispin 2160 LKB (Bromma, Sweden), and the pellet was suspended with 750 μl of cold PIV buffer. This cell suspension was mixed in equal parts with molten 2% low-melting-point agarose (SeaPlaque agarose; FMC BioProducts), and the mixture was pipetted into plug molds. The plugs were incubated overnight at 37°C in EC buffer (6 mM Tris, 1 M NaCl, 100 mM EDTA, 0.5% Brij 58, 0.2% Na-deoxycholate, 0.5% Na-lauroylsarcosine) with 1 mg of lysozyme per ml and were incubated again overnight at 55 to 57°C in ES buffer (0.5 M EDTA, 1% Na-lauroylsarcosine) with 0.3 mg of proteinase K per ml. The washing of the plugs and the conditions for restriction endonuclease digestion and PFGE were as described previously (23). Chromosomal DNA was digested overnight with 15 U of ApaI (MBI Fermentas Ltd.) and SmaI (Boehringer Mannheim GmbH, Mannheim, Germany). A subset of strains was also digested with restriction enzymes SpeI, KspI, AvaI, NaeI, NarI, XhoI, and MluI (all from Boehringer Mannheim GmbH) and AscI (New England BioLabs Inc., Beverly, Mass.). Electrophoresis was performed at 200 V on 1.0% SeaKem ME agarose gel (FMC BioProducts) using the CHEF Mapper 153 or CHEF-DR systems (Bio-Rad Laboratories, Richmond, Calif.). Running conditions for ApaI- and SmaI-digested DNA were 0.5 to 40 s for 20 h, whereas the running conditions for the other restriction enzymes were either 1 to 50 s for 24 h or 5 to 50 s for 24 h. Lambda ladder and/or Low Range PFG markers (New England BioLabs Inc.) were used as molecular weight standards. Any difference between two profiles was considered sufficient to distinguish two different PFGE profiles. PFGE profiles of strains were named with uppercase letters (starting at A) when digested with SmaI and were named with lowercase letters (starting at a) when digested with ApaI.
RESULTS
Thirty-three of the 47 C. perfringens strains related to the outbreaks were positive for cpe in PCR (Table 1). The assay for enterotoxin production was clearly positive (CPE-positive result) for 31 strains. The result of enterotoxin production of one cpe-positive strain (AHS 7427) remained CPE negative even though the whole determination was repeated three times. The growth of the strain on brucella medium before heat treatment was very good, whereas after the heat treatment only a few colonies were found. After being cultured in modified Duncan and Strong medium, the spores were not detected by microscopic examination. The cpe-positive strain (AHS 25157) in cluster VI was tested three separate times: the result remained uncertain twice, and the third time only a CPE-positive result was obtained. In PCR, cpe-positive C. perfringens strains were found in clusters II, III, IV, VI, VII, VIII, and IX. Four of these clusters (II, III, VII, and IX) also included a cpe-negative isolate. All isolates in the suspected clusters I and V were cpe negative.
Ten restriction enzymes were tested for their usefulness in subtyping isolates of C. perfringens by PFGE. Restriction enzymes SmaI and ApaI were chosen for further studies because they produced 7 to 13 well-separated fragments.
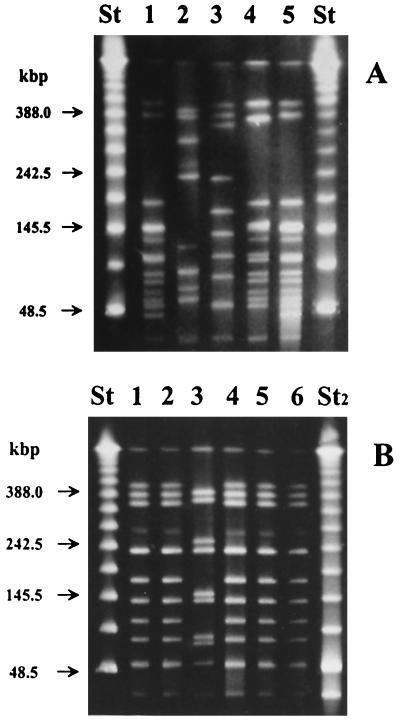
All strains carrying the cpe gene in the five suspected clusters (III, IV, VI, VIII, and IX) had an identical PFGE subtype within each cluster (Cc, Dd, Bb, Aa, and Aa, respectively) when digested with SmaI and ApaI (Table 1 and Fig. 1A). In the suspected cluster II, all three isolates had different PFGE subtypes (Ee, Kk, and Ff). In addition, clusters III, VII, and IX included one strain that belonged to subtype Gg, Pp, and Qq, respectively, whereas all other strains in the same cluster were of the same subtype (Table 1 and Fig. 1B).
FIG. 1.
PFGE banding patterns of C. perfringens isolates obtained when chromosomal DNA was digested with restriction enzyme SmaI. Lanes: St, lambda ladder; St2, Low Range PFG markers (used as molecular size markers). Arrows indicate the positions of the marker DNA fragments. (A) Subtypes of different outbreak clusters. Lane 1, subtype A (cluster VIII); lane 2, subtype B (cluster VI); lane 3, subtype C (cluster III); lane 4, subtype D (cluster IV); lane 5, subtype A (cluster IX). (B) Subtypes of outbreak cluster III. Lane 1, subtype C (AHS 7563); lane 2, subtype C (AHS 7564); lane 3, subtype G (AHS 7565); lane 4, subtype C (AHS 7566); lane 5, subtype C (AHS 7567); lane 6, subtype C (AHS 7568).
Subtype Aa differed from subtype Dd by three PFGE fragments when digested with SmaI (Fig. 1A) and by four when digested with ApaI (data not shown). All other subtypes of the cpe-positive strains differed from one another by more than 10 fragments. The cpe-negative strains AHS 7565 and AHS 7619 in the suspected clusters III and V, respectively, had identical PFGE profiles (Gg), whereas all the other cpe-negative strains had PFGE profiles that differed from each other and from those of the cpe-positive outbreak strains.
All foodstuff isolates that were connected to clusters VI and IX belonged to the same subtype, Bb and Aa, respectively, as did the human isolates of the same cluster (Table 1).
DISCUSSION
The fact that C. perfringens is commonly found in human and animal stools, and that it is also widespread in the environment, is a disadvantage when one is searching for the specific cause of a food-borne outbreak of disease by traditional methods (2, 16, 28, 31, 39). However, demonstrating the presence of C. perfringens enterotoxin in feces of several food poisoning victims is a reliable diagnostic tool for identifying a C. perfringens type A food poisoning outbreak (3). Sporulation is considered necessary for C. perfringens enterotoxin expression, and it is often difficult to achieve for C. perfringens isolates grown in laboratory media (17, 20). Therefore, many laboratories have not performed the detection of enterotoxin type A of C. perfringens routinely. The PCR-based assays for identifying the cpe gene responsible for enterotoxin production have an advantage over serologic assays in that they do not require isolates to sporulate in vitro. On the other hand, the disadvantage of these assays is that they can only identify isolates as potentially enterotoxigenic; i.e., some C. perfringens isolates may carry a silent, unexpressed cpe gene.
From 1975 to 1999, altogether some 58,000 persons in Finland were reported ill as a result of food-borne pathogens during outbreaks of disease; 6,900 cases were reported to be caused by C. perfringens (29). During these 25 years, C. perfringens caused 238 food-borne outbreaks of disease, which is 20% of all outbreaks, thus making C. perfringens one of the most important causes of food-borne infections.
This study aimed to strengthen the evidence-based diagnostics of food poisonings suspected to be caused by C. perfringens. Therefore, we retrospectively studied all C. perfringens isolates that were available in our collections from nine food-borne outbreaks of disease originally reported according to epidemiological data as having been caused by C. perfringens. The isolates were analyzed by PCR for the cpe gene, which encodes enterotoxin and is known to be involved in food poisoning caused by C. perfringens, and also by reversed passive latex agglutination to detect enterotoxin production. Subsequently, the isolates were studied by PFGE to compare their genotypes after restriction of DNA by enzymes SmaI and ApaI.
Two restriction enzymes, SmaI and ApaI, which have also been successfully used in many other studies of C. perfringens (8, 10, 25, 30, 41), were chosen from the set of 10 tested enzymes for further studies. Both enzymes have GC-rich recognition sites and are suitable for generating large DNA fragments in AT-rich genomes of C. perfringens (9).
This study included 40 isolates from humans and seven from foodstuffs. Of the nine infection clusters originally thought to be caused by C. perfringens, in seven (II, III, IV, VI, VII VIII, and IX) our results supported the earlier findings that the cause was C. perfringens. The results also confirmed the earlier reports that the suspected foodstuffs, meat casserole and minced meat casserole, had caused the outbreak in Helsinki in October 1997 and in March 1999, respectively (18, 21). Interestingly, the outbreaks in October 1998 (cluster VIII) and in March 1999 (cluster IX) were caused by the same subtype, Aa. Both outbreaks took place in Helsinki, but there was no other known connection between these outbreaks. Also, in April 1998 in Helsinki (cluster VII), the cpe-positive isolate belonged to this same subtype, Aa, whereas the cpe-negative one belonged to subtype Pp, differing from Aa by more than 10 fragments, and was, therefore, called a nonoutbreak strain according to Tenover et al. (38). When the PFGE profiles of subtypes in outbreak clusters (II, III, IV, VI, VII, VIII, and IX) were compared to each other, they had different origins. Only the strains belonging to subtypes Aa and Dd might be possibly related according to Tenover et al. (38) since they differed by three PFGE fragments when digested with SmaI and by four fragments when digested with ApaI.
In four clusters, II, III, VII, and IX, both the outbreak strains and one nonoutbreak strain were detected. The cpe-negative strains probably were just members of normal flora, as were all the strains in clusters I (six isolates) and V (four isolates). This indicates that C. perfringens was not the cause of the outbreaks in 1984 and 1994. However, the clinical picture of the patients and the fact that, apart from C. perfringens, no food-borne bacterial pathogens (Salmonella spp., Shigella spp., Yersinia spp., Campylobacter spp., S. aureus, or B. cereus) belonging to clusters I and V (data not shown) were found at that time in the fecal samples of these patients supported the hypothesis that C. perfringens was the causative agent. However, the possibility that the symptoms were due to a viral agent cannot completely be excluded. Our findings emphasize the importance of a more detailed characterization of C. perfringens isolates, rather than only identification to the species level, in order to verify the cause of an outbreak.
In cluster II, the cpe-positive strain AHS 7427 remained CPE negative despite three repetitive determinations for the production of enterotoxin. The growth of the strain was clearly lower after the heat treatment than before it. According to the manufacturer of the C. perfringens enterotoxin test kit used (PET-RPLA kit), some enterotoxin-positive strains may actually be killed by heat treatment and will not, therefore, produce enterotoxin in the second medium. Also, no sporulation of the strain was detected. Thus, it may be that the strain was not able to resist the heat treatment enough and therefore did not sporulate and produce a detectable amount of enterotoxin or the gene was a silent, unexpressed cpe gene. In cluster VI, the cpe-positive strain (AHS 25157) was tested three separate times for enterotoxin production: the result remained uncertain twice, and the third time the test only gave a CPE-positive result. Without analyzing the cpe gene encoding enterotoxin and the information on the PFGE types of the other strains in that cluster, the determination of enterotoxin production alone was not adequate enough in investigating this outbreak strain.
One limitation to this study was the availability of only one pure culture per patient, in which a C. perfringens colony of normal flora might have been picked up for further testing instead of enterotoxigenic C. perfringens. Van Damme-Jongsten et al. tested C. perfringens isolates from 186 different food poisoning outbreaks (40). However, they also tested only one or occasionally two strains from each outbreak. Only 60% of their isolates were positive for the cpe gene. It seems that they almost certainly characterized a number of cpe-negative C. perfringens of normal flora—as did we in this study. Thus, it is very important to study more than one isolate from the fecal culture of the patient to ensure that the strain is not part of his or her normal flora.
Maslanka et al. suggested that PFGE provides a reliable method in conjunction with epidemiologic data to diagnose C. perfringens food poisoning outbreaks (25). However, this study clearly indicates that PFGE, in the absence of information regarding whether the isolates are enterotoxigenic or not, can yield misleading or even erroneous conclusions. For example, in this study the investigation of isolates from the suspected outbreaks II and VII could have led to an incorrect conclusion that these outbreaks were not caused by C. perfringens since the isolates within these outbreaks had different PFGE types. The PFGE is very useful in an outbreak situation in showing which isolates have identical PFGE profiles and, therefore, might be a part of the same outbreak. However, this study indicates that before further laboratory work is done, for example, PFGE, all pure C. perfringens isolates should be screened for the presence of the cpe gene.
Only 2 of our 14 cpe-negative strains had identical PFGE profiles. All other profiles were different from each other or from the PFGE profiles of cpe-positive strains. In clusters II, III, VII, and IX, the cpe-negative isolates differed by more than 10 fragments from the cpe-positive outbreak strain of these clusters when digested with SmaI and ApaI. These findings are in contrast to the previous report which stated that in a single outbreak the cpe-positive and -negative isolates have identical or nearly identical PFGE profiles (30). The profiles of the cpe-positive strains within each cluster were identical, except in one cluster, in which two subtypes, Ee and Ff, were detected. However, these subtypes also had a difference of more than 10 fragments. This outbreak might have been caused by two different strains of C. perfringens. Maslanka et al. also obtained four unique PFGE patterns with 18 isolates from one outbreak (25). However, none of those isolates were tested for the ability to produce enterotoxin, so it is unknown whether they were capable of producing symptoms of a food-borne disease or whether some of them were part of the normal flora.
In this study, the true outbreaks in Finland caused by C. perfringens were detected and confirmed by PCR and reversed passive latex agglutination. These results were subsequently supported by the PFGE results. In the future these genotypic or other appropriate methods will be needed to strengthen the evidence-based diagnostics of a food-borne outbreak of disease suspected to be caused by C. perfringens. However, it is recommended that all pure isolates be screened for the presence of the cpe gene before further laboratory work is done.
Acknowledgments
We thank The Food and Environmental Laboratory, Hyvinkää, Finland, and The Laboratory of Environmental Centre, Helsinki, Finland, for foodstuff strains.
The skillful technical assistance of Tarja Heiskanen, Liisa Immonen, and Ritva Taipalinen and the editorial assistance of Maarit Koukkari are gratefully acknowledged.
REFERENCES
- 1.Allen, S. D., C. L. Emery, and J. A. Siders. 1999. Clostridium, p. 654-671. In P. R. Murray, E. J. Baron, M. A. Pfaller, F. C. Tenover, and R. H. Yolken (ed.), Manual of clinical microbiology, 7th ed. American Society for Microbiology, Washington, D.C.
- 2.Bezirtzoglou, E., D. Dimitriou, A. Panagiou, I. Kagalou, and Y. Demoliates. 1994. Distribution of Clostridium perfringens in different aquatic environments in Greece. Microbiol. Res. 149:129-134. [DOI] [PubMed] [Google Scholar]
- 3.Birkhead, G., R. L. Vogt, E. M. Heun, J. T. Snyder, and B. A. McClane. 1988. Characterization of an outbreak of Clostridium perfringens food poisoning by quantitative fecal culture and fecal enterotoxin measurement. J. Clin. Microbiol. 26:471-474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brynestad, S., and P. E. Granum. 1999. Evidence that Tn5565, which includes the enterotoxin gene in Clostridium perfringens, can have a circular form which may be a transposition intermediate. FEMS Microbiol. Lett. 170:281-286. [DOI] [PubMed] [Google Scholar]
- 5.Brynestad, S., M. R. Sarker, B. A. McClane, P. E. Granum, and J. I. Rood. 2001. Enterotoxin plasmid from Clostridium perfringens is conjugative. Infect. Immun. 69:3483-3487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brynestad, S., B. Synstad, and P. E. Granum. 1997. The Clostridium perfringens enterotoxin gene is on a transposable element in type A human food poisoning strains. Microbiology 143:2109-2115. [DOI] [PubMed] [Google Scholar]
- 7.Buogo, C., S. Capaul, H. Häni, J. Frey, and J. Nicolet. 1995. Diagnosis of Clostridium perfringens type C enteritis in pigs using a DNA amplification technique (PCR). J. Vet. Med. B 42:51-58. [DOI] [PubMed] [Google Scholar]
- 8.Canard, B., B. Saint-Joanis, and T. Cole. 1992. Genomic diversity and organization of virulence genes in the pathogenic anaerobe Clostridium perfringens. Mol. Microbiol. 6:1421-1429. [DOI] [PubMed] [Google Scholar]
- 9.Cato, E. P., W. L. George, and S. M. Finegold. 1986. Genus Clostridium Prazmowski 1880, 23AL, p. 1141-1199. In R. G. E. Murray, D. J. Brenner, M. P. Bryant, J. G. Holt, N. R. Krieg, J. W. Moulder, N. Pfenning, P. H. A. Sneath, and J. T. Stanley (ed.), Bergey's manual of systematic bacteriology, vol. 2. Williams & Wilkins, Baltimore, Md. [Google Scholar]
- 10.Collie, R. E., and B. McClane. 1998. Evidence that the enterotoxin gene can be episomal in Clostridium perfringens isolates associated with non-food-borne human gastrointestinal diseases. J. Clin. Microbiol. 36:30-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cornillot, E., B. Saint-Joanis, G. Daube, S. Katayama, P. E. Granum, B. Canard, and S. T. Cole. 1995. The enterotoxin gene (cpe) of Clostridium perfringens can be chromosomal or plasmid-borne. Mol. Microbiol. 15:639-647. [DOI] [PubMed] [Google Scholar]
- 12.Daube, G., B. China, P. Simon, K. Hvala, and J. Mainil. 1994. Typing of Clostridium perfringens by in vitro amplification of toxin genes. J. Appl. Bacteriol. 77:650-655. [DOI] [PubMed] [Google Scholar]
- 13.Duncan, C. L., and D. H. Strong. 1968. Improved medium for sporulation of Clostridium perfringens. Appl. Microbiol. 16:82-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eisgruber, H., M. Wiedmann, and A. Stolle. 1995. Use of plasmid profiling as a typing method for epidemiologically related Clostridium perfringens isolates from food poisoning cases and outbreaks. Lett. Appl. Microbiol. 20:290-294. [DOI] [PubMed] [Google Scholar]
- 15.Forsblom, B., A. Palmu, P. Hirvonen, and H. Jousimies-Somer. 1995. Ribotyping of Clostridium perfringens isolates. Clin. Infect. Dis. 20(Suppl. 2):S323-S324. [DOI] [PubMed] [Google Scholar]
- 16.Gyobu, Y. 1978. Distribution of Clostridium perfringens in the environment and the enterotoxin production by isolated strains. J. Food Hyg. Soc. Jpn. 19:236-241. [Google Scholar]
- 17.Harmon, S. M., and D. A. Kautter. 1986. Improved media for sporulation and enterotoxin production by Clostridium perfringens. J. Food Prot. 49:706-711. [DOI] [PubMed] [Google Scholar]
- 18.Hatakka, M., and H. Halonen. 2000. Food-borne and waterborne outbreaks in Finland in 1999. National Food Administration research notes, vol. 7. National Food Administration, Helsinki, Finland.
- 19.Katayama, S., B. Dupuy, G. Daube, B. China, and S. T. Cole. 1996. Genome mapping of Clostridium perfringens strains with I-CeuI shows many virulence genes to be plasmid-borne. Mol. Gen. Genet. 251:720-726. [DOI] [PubMed] [Google Scholar]
- 20.Kokai-Kun, J. F., J. G. Songer, J. R. Czeczulin, F. Chen, and B. A. McClane. 1994. Comparison of Western immunoblots and gene detection assays for identification of potentially enterotoxigenic isolates of Clostridium perfringens. J. Clin. Microbiol. 32:2533-2539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kukkula, M. 1998. Food-borne outbreaks in Finland in 1997. National Food Administration research notes, vol. 3. National Food Administration, Helsinki, Finland.
- 22.Larsson, H. E., and S. P. Borriello. 1998. Infectious diarrhea due to Clostridium perfringens. J. Infect. Dis. 157:390-391. [DOI] [PubMed] [Google Scholar]
- 23.Lukinmaa, S., R. Schildt, T. Rinttila, and A. Siitonen. 1999. Salmonella Enteritidis phage types 1 and 4: pheno- and genotypic epidemiology of recent outbreaks in Finland. J. Clin. Microbiol. 37:2176-2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mahony, D. E., R. Ahmed, and S. G. Jackson. 1992. Multiple typing techniques applied to a Clostridium perfringens food poisoning outbreak. J. Appl. Bacteriol. 72:309-314. [DOI] [PubMed] [Google Scholar]
- 25.Maslanka, S. E., J. G. Kerr, G. Williams, J. M. Barbaree, L. A. Carson, J. M. Miller, and B. Swaminathan. 1999. Molecular subtyping of Clostridium perfringens by pulsed-field gel electrophoresis to facilitate food-borne-disease outbreak investigations. J. Clin. Microbiol. 37:2209-2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McDonel, J. L. 1986. Toxins of Clostridium perfringens types A, B, C, D, and E, p. 477-517. In F. Dorner and H. Drews (ed.), Pharmacology of bacterial toxins. Pergamon Press, Oxford, United Kingdom.
- 27.McLauchlin, J., G. Ripabelli, M. M. Brett, and E. J. Threlfall. 2000. Amplified fragment length polymorphism (AFLP) analysis of Clostridium perfringens for epidemiological typing. Int. J. Food Microbiol. 56:21-28. [DOI] [PubMed] [Google Scholar]
- 28.Miwa, N., T. Nishina, S. Kubo, and H. Honda. 1997. Most probable numbers of enterotoxigenic Clostridium perfringens in intestinal contents of domestic livestock detected by nested PCR. J. Vet. Med. Sci. 59:557-560. [DOI] [PubMed] [Google Scholar]
- 29.Rahkio, M., H. Korkeala, A. Siitonen, M. Hatakka, V.-M. Niemi, and P. Pakkala (ed.). 2000. Guidebook to unravel food-borne outbreaks, p. 16-21. Elintarvike ja Terveys-lehti, Vammalan Kirjapaino, Finland. (In Finnish.)
- 30.Ridell, J., J. Björkroth, H. Eisgrüber, B. Schalch, A. Stolle, and H. Korkeala. 1998. Prevalence of the enterotoxin gene and clonality of Clostridium perfringens strains associated with food-poisoning outbreaks. J. Food Prot. 61:240-243. [DOI] [PubMed] [Google Scholar]
- 31.Saito, M. 1990. Production of enterotoxin by Clostridium perfringens derived from humans, animals, foods, and the natural environment in Japan. J. Food Prot. 53:115-118. [DOI] [PubMed] [Google Scholar]
- 32.Sarker, M. R., R. J. Carman, and B. A. McClane. 1999. Inactivation of the gene (cpe) encoding Clostridium perfringens enterotoxin eliminates the ability of two cpe-positive C. perfringens type A human gastrointestinal disease isolates to affect rabbit ileal loops. Mol. Microbiol. 33:946-958. [DOI] [PubMed] [Google Scholar]
- 33.Sarker, M. R., U. Singh, and B. A. McClane. 2000. An update on Clostridium perfringens enterotoxin. J. Nat. Toxins 9:251-266. [PubMed] [Google Scholar]
- 34.Schalch, B., J. Björkroth, H. Eisgruber, H. Korkeala, and A. Stolle. 1997. Ribotyping for strain characterization of Clostridium perfringens isolates from food poisoning cases and outbreaks. Appl. Environ. Microbiol. 63:3992-3994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schalch, B., B. Sperner, H. Eisgruber, and A. Stolle. 1999. Molecular methods for the analysis of Clostridium perfringens relevant to food hygiene. FEMS Immunol. Med. Microbiol. 24:281-286. [DOI] [PubMed] [Google Scholar]
- 36.Skjelkvale, R., and T. Uemura. 1977. Experimental diarrhoea in human volunteers following oral administration of Clostridium perfringens enterotoxin. J. Appl. Bacteriol. 43:281-286. [DOI] [PubMed] [Google Scholar]
- 37.Sparks, S. G., R. J. Carman, M. R. Sarker, and B. A. McClane. 2001. Genotyping of enterotoxigenic Clostridium perfringens fecal isolates associated with antibiotic-associated diarrhea and food poisoning in North America. J. Clin. Microbiol. 39:883-888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tschirdewahn, B., S. Notermans, K. Wernars, and F. Untermann. 1991. The presence of enterotoxigenic Clostridium perfringens strains in faeces of various animals. Int. J. Food Microbiol. 14:175-178. [DOI] [PubMed] [Google Scholar]
- 40.Van Damme-Jongsten, M., J. Rodhouse, R. J. Gilbert, and S. Notermans. 1990. Synthetic DNA probes for detection of enterotoxigenic Clostridium perfringens strains isolated from outbreaks of food poisoning. J. Clin. Microbiol. 28:131-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wada, A., Y. Masuda, M. Fukayama, T. Hatakeyama, Y. Yanagawa, H. Watanabe, and T. Inamatsu. 1996. Nosocomial diarrhoea in the elderly due to enterotoxigenic Clostridium perfringens. Microbiol. Immunol. 40:767-771. [DOI] [PubMed] [Google Scholar]