Abstract
Numerous microorganisms, including bacteria, yeasts, and molds, are present in cheeses, forming a complex ecosystem. Among these organisms, bacteria are responsible for most of the physicochemical and aromatic transformations that are intrinsic to the cheesemaking process. Identification of the bacteria that constitute the cheese ecosystem is essential for understanding their individual contributions to cheese production. We used temporal temperature gradient gel electrophoresis (TTGE) to identify different bacterial species present in several dairy products, including members of the genera Lactobacillus, Lactococcus, Leuconostoc, Enterococcus, Pediococcus, Streptococcus, and Staphylococcus. The TTGE technique is based on electrophoretic separation of 16S ribosomal DNA (rDNA) fragments by using a temperature gradient. It was optimized to reveal differences in the 16S rDNA V3 regions of bacteria with low-G+C-content genomes. Using multiple control strains, we first set up a species database in which each species (or group of species) was characterized by a specific TTGE fingerprint. TTGE was then applied to controlled dairy ecosystems with defined compositions, including liquid (starter), semisolid (home-made fermented milk), and solid (miniature cheese models) matrices. Finally, the potential of TTGE to describe the bacterial microflora of unknown ecosystems was tested with various commercial dairy products. Subspecies, species, or groups of species of lactic acid bacteria were distinguished in dairy samples. In conclusion, TTGE was shown to distinguish bacterial species in vitro, as well as in both liquid and solid dairy products.
The transformation of milk to cheese involves a complex and dynamic microbial ecosystem in which numerous biochemical reactions occur. Two main groups of bacteria are involved in cheese manufacture and ripening. The first group consists of starter bacteria (mainly Lactococcus) that are added to milk during cheese manufacture. About 109 CFU of starter bacteria per g is present in the final product. The second group consists of adventitious microorganisms (secondary microflora) from the environment which contaminate the milk or cheese curd during manufacture and ripening (5, 33). This group includes numerous species of lactic acid bacteria (Lactobacillus, Pediococcus, Enterococcus, and Leuconostoc) and surface cheese bacteria (Micrococcus and Staphylococcus). During the production of pressed-curd cheeses in our experimental dairy, the size of the secondary microflora population is up to 107 CFU/g after 1 month of ripening, and these microorganisms may become the dominant viable microorganisms in cheese. The numerous hydrolytic enzymes expressed by this secondary microflora presumably affect proteolysis and lipolysis during cheese ripening and thus may contribute to cheese maturation (18, 25, 32, 42). However, the effects of adventitious microflora on cheese quality could be species or even strain dependant (54), and characterization of this microflora in cheese is thus an important industrial issue.
Routine methods to enumerate microorganisms in dairy products are currently based on conventional microbial techniques. These methods, which rely on bacterial growth in selective media, may fail to identify bacteria that cannot multiply outside the cheese environment. Indeed, cultivation-dependent approaches may bias our view of microbial diversity (1, 21). Furthermore, bacterial identification by this technique stops at the genus level. More precise bacterial identification techniques use taxonomic and discriminating methods, including biochemical tests, 16S ribosomal DNA (rDNA) sequencing, sodium dodecyl sulfate-polyacrylamide gel electrophoresis of proteins (43), randomly amplified polymorphic DNA fingerprinting (46), and Fourier transform infrared spectroscopy (2). However, these methods are labor-intensive and time-consuming.
Recently, more rapid molecular methods have been developed to analyze diversity within bacterial communities (41). These methods are based on direct analysis of DNA in the environment and do not require cell cultivation. They include single-stranded conformational polymorphism analysis (26), denaturing gradient gel electrophoresis (DGGE), and temporal temperature gradient gel electrophoresis (TTGE) (36). All of these approaches involve extraction of nucleic acids (DNA or RNA), amplification of genes encoding 16S rRNA, and analysis of PCR products by a genetic fingerprinting technique (39).
DGGE is based on electrophoretic separation of DNA molecules that are the same length but have different nucleotide sequences (27). It was first used to detect single-base DNA sequence variations (17). In this technique, PCR-amplified double-stranded DNA is subjected to electrophoresis under denaturing conditions (achieved by a solvent gradient); migration depends on the degree of DNA denaturation. TTGE is a related but simpler method, in which a temperature gradient rather than a solvent gradient is used to denature the DNA (4). Both DGGE and TTGE are now methods of choice for environmental microbiologists and have been used to determine the genetic diversities of natural microbial communities such as the communities in biofilms (36), soil (14, 19), ocean depths (48), hot springs (16, 45), lakes (55), a biodegraded wall painting (44), and fermented foods (3, 9, 8). Two complete reviews of DGGE and TTGE have been published recently (37, 38).
Here we describe the use of TTGE to detect and identify lactic acid bacteria and surface cheese bacteria in dairy products. We optimized TTGE conditions and created a bacterial reference set. Using the reference set as the standard, we confirmed the feasibility of using TTGE for bacterial identification in controlled dairy ecosystems (starter, fermented milk, and washed-curd miniature cheese models). TTGE was then used to identify dairy microflora in various commercial dairy ecosystems, including commercial starters, fermented milk samples, and different types of cheeses.
MATERIALS AND METHODS
Selection of bacterial strains for the reference set and genomic DNA extraction.
We selected 48 different bacterial species or subspecies (from bacteria with low-G+C-content genomes) belonging to the genera Lactobacillus, Lactococcus, Leuconostoc, Enterococcus, Pediococcus, Streptococcus, and Staphylococcus, as shown in Table 1. All these bacteria may be found in milk ecosystems. To ensure the reproducibility and significance of patterns obtained by TTGE, three strains were generally selected from each species group (in some cases, only one or two strains are available). Genomic DNA was prepared as described previously (11).
TABLE 1.
Bacterial strains used for the reference set and sequence analysis
| Species or subspecies | Strain(s) in reference seta | Sequence analysis
|
||
|---|---|---|---|---|
| Strain | GenBank accession no. | Predicted Tm (°C) | ||
| Lactobacillus gasseri | CNRZ222, CNRZ1946, CNRZ1503 | DSM20243 | M58820 | 71.55 |
| Lactobacillus johnsonii | CNRZ251, CNRZ1937, CNRZ462 | ATCC 33200T | AJ002515 | 71.55 |
| Lactobacillus plantarum | CNRZ211, CNRZ1246, CNRZ1565 | DSM20205 | M58827 | 71.7 |
| Lactobacillus pentosus | CNRZ1858, CNRZ1550, CNRZ1562 | JCM1588 | D79211 | 71.7 |
| Lactobacillus fermentum | CNRZ1609, CNRZ63, CNRZ1615 | ATCC 14931T | M58819 | 72.7 |
| Lactobacillus brevis | CNRZ1608, CNRZ735, CNRZ742 | ATCC 14869 | M58810 | 72.75 |
| Lactobacillus crispatus | CNRZ1927, CNRZ1925, CNRZ1924 | DSM20584T | Y17362 | 72.9 |
| Lactobacillus gallinarum | CNRZ1931, CNRZ1932, CNRZ1933 | ATCC 3319 | AJ242968 | 72.9 |
| Lactobacillus acidophilus | CNRZ1922, CNRZ204, CNRZ1927 | CNRZ204T | NDb | 72.9 |
| Lactobacillus amylovorus | CNRZ1928, CNRZ1929, CNRZ1930 | DSM20531 | M58805 | 72.9 |
| Lactobacillus helveticus | CNRZ323, CNRZ1148, CNRZ1110 | NCDO2712T | X61141 | 72.9 |
| Lactobacillus delbrueckii subsp. bulgaricus | CNRZ495, CNRZ752, CNRZ1159 | JCM1002T | AB007908 | 73.1 |
| Lactobacillus delbrueckii subsp. delbrueckii | CNRZ225, CNRZ231 | NCDO213T | X52654 | 73.2 |
| Lactobacillus delbrueckii subsp. lactis | CNRZ245, CNRZ1829, CNRZ332 | DSM20072 | M58823 | 73.2 |
| Lactobacillus rhamnosus | CNRZ212, CNRZ205, CNRZ1668 | CNRZ212T | ND | 76.05 |
| Lactobacillus paracasei | CNRZ62, CNRZ1853, CNRZ1219 | ATCC 334 | D86517 | 76.05 |
| Lactobacillus casei | CNRZ313, CNRZ1393 | ATCC 393 | M23928 | 76.15 |
| Lactobacillus reuteri | CNRZ230, CNRZ1827, CNRZ1657 | DSM20016T | X76328 | 76.2 |
| Enterococcus casseliflavus | LMG13518T, CNRZ 1935, CNRZ 1937 | NCIMB11449 | Y18161 | 72.3 |
| Enterococcus gallinarum | LMG13129T, CNRZ 1437, CNRZ 1203 | NCFB231 | Y18160 | 72.8 |
| Enterococcus faecium | LMG11423, EF18-4, CNRZ127c | NCFB942T | Y18294 | 72.85 |
| Enterococcus durans | E361, CNRZ132, EF12.1c | NCFB596T | Y18359 | 72.85 |
| Enterococcus hirae | LMG6399T, CE119, EF262c | DSM20160 | AJ276356 | 72.85 |
| Enterococcus faecalis | CNRZ1666, CE17, CNRZ415c | CIP103015T | ND | 73.8 |
| Leuconostoc fallax | 17Dc | DSM20189T | S63851 | 72.1 |
| Leuconostoc citreum | LMG9849T, 22R, 2Ec | ATCC 49370T | AF111948 | 72.2 |
| Leuconostoc lactis | LMG8894T, CNRZ1746, CNRZ1472 | DSM20202T | M23031 | 72.5 |
| Leuconostoc mesenteroides subsp. cremoris | CNRZ361T | DSM20346T | M23034 | 72.65 |
| Leuconostoc mesenteroides subsp. mesenteroides | CNRZ749T | DSM20343T | M23035 | 72.65 |
| Leuconostoc mesenteroides subsp. dextranicum | 10B, CRNZ77T, 10F, 50Mc | |||
| Leuconostoc pseudomesenteroides | LMG11482, LMG11483, LMG11499 | NCDO768T | X95979 | 72.7 |
| Leuconostoc carnosum | LMG11498T | LMG11498T | 11498-16 | 73.7 |
| Weissella paramesenteroides | LMG9852T | DSM20288T | M23033 | 73.2 |
| Pediococcus pentosaceus | E2079, E2071, CNRZ444c | E2079 | ND | 73.6 |
| Pediococcus acidilactici | E2075, E2070, CNRZ443c | E2075 | ND | 74.2 |
| Lactococcus garvieae | CNRZ1323, IBB66c | NCDO2156 | X54262 | 71.3 |
| Lactococcus raffinolactis | CNRZ1214, IBB131, CNRZ496c | NCDO617T | X54261 | 72.8 |
| Lactococcus plantarum | CNRZ1322, IBB76c | NCDO1869T | X542259 | 73.7 |
| Lactococcus lactis subsp. lactis | IL801, IL7, IL416, CNRZ144, CNRZ487 | ATCC 19435T | M58837 | 74.25 |
| Lactococcus lactis subsp. lactis biovar diacetylactis | CNRZ260, CNRZ257, CNRZ365 | IL1403 | X64887 | 74.25 |
| Lactococcus lactis subsp. cremoris | CNRZ105, CNRZ378, CNRZ113 | ATCC 19257 | M58836 | 74.25 |
| Streptococcus thermophilus | CNRZ1529, CNRZ1896, CNRZ1359 | DSM20617T | X68418 | 74.8 |
| Staphylococcus aureus | CNRZ3, 5.7.540, CNRZ875c | ATCC 12600T | D83357 | 72.22 |
| Staphylococcus epidermidis | 1.7.507, CNRZ873b, CNRZ748bc | ATCC 14990T | D83363 | 72.3 |
| Staphylococcus xylosus | CNRZ1665T, C39d5, C39d7c | ATCC 29971T | D83374 | 72.6 |
| Staphylococcus saprophyticus | S. sa, ED2, CNRZ291c | CNRZ911BT | ND | 73.1 |
| Staphylococcus caseolyticus | CNRZ249, CNRZ471, CNRZ470 | ATCC 13548T | D83359 | 73.45 |
| Staphylococcus hyicus | CNRZ874, CIP8158T | ATCC 11249T | D83368 | 73.60 |
CIP, Collection of the Institute Pasteur; LMG, Collection of the Laboratorium voor Microbiologie; CNRZ, Collection of the Centre National de la Recherche Zootechnique; IL, Collection of Génétique Microbienne (INRA, Jouy-en-Josas, France); ATCC, American Type Culture Collection; DSM, Deutshe Sammlung von Mikroorganismen und Zellkulturen; JCM, Japan Collection of Microorganisms; NCIMB, National Collection of Industrial and Marine Bacteria; NCFB, National Collection of Food Bacteria; NCDO, National Collection of Dairy Organisms.
ND, not deposited.
Strains EF18-4, E361, EF12.1, CE119, EF262, CE17, 17D, 22R, 2E, 10B, 10F, 50M, E2079, E2071, E2075, E2070, IBB66, IBB131, IBB76, 5.7.540, 1.7, 507, C39d5, C39d7, S. sa, and ED2 were solated from cheese (INRA collection).
Selection of the discriminating DNA region used for TTGE analyses, dendrogram design, and Tm determination.
To discriminate species by TTGE, we selected the V3 area of 16S rDNA (in Escherichia coli, this area corresponds to positions 339 through 539) (13, 50). To determine the discriminating potential of the V3 sequence, pairwise distances between the species used in this study were calculated, and dendrograms were constructed by using the neighbor-joining method included in the GeneBase software (Applied Maths, Sint-Martens-Latem, Belgium). We also used WinMelt software (Bio-Rad, Marne La Coquette, France), which calculates the melting temperature (Tm) of any sequence (28), to predict the migration of different V3 species on TTGE gels. The sequences used in the predictive analysis (Table 1) were obtained either from the CNRZ collection database (Elodie Lepage, Unité de Recherches Laitières et Génétique Appliquée) or from the GenBank database.
PCR amplification.
TTGE samples were prepared by performing two successive PCRs (nested PCR [20]) with the Gene Amp system (model 2400; Perkin-Elmer, Courtaboeuf, France). First, a 700-bp fragment of the 16S rDNA gene including the V3 region was amplified. The reaction mixture (100 μl) contained reaction buffer (10 mM Tris-HCl, 1.5 mM MgCl2, 50 mM KCl [final concentrations]), each deoxynucleoside triphosphate at a concentration of 200 μM, 60 pmol of primer W01 (5′-AGA GTT TGA TC[AC] TGG CTC-3′), 60 pmol of primerW012 (5′-TAC GCA TTT CAC C[GT]C TAC A-3′), ∼50 ng of bacterial DNA, and 2.5 U of Taq DNA polymerase (Q-BIOgene, Illkirch, France). The amplification program was 96°C for 4 min; 30 cycles of 96°C for 10 s, 50°C for 30 s, and 72°C for 1 min; and finally, 72°C for 2 min. Second, the 700-bp fragment was used to amplify the V3 region with the following primers (S. J. Turner, G. D. Lewis, D. J. Saul, C. S. Baker, and A. Rodrigo, N. Z. Microbiol. Soc. Annu. Meet., poster paper, 1998): HDA1-GC (5′-CGC CCG GGG CGC GCC CCG GGC GGG GCG GGG GCA CGG GGG GAC TCC TAC GGG AGG CAG CAG T-3′; the GC clamp sequence is in bold) and HDA2 (5′-GTA TTA CCG CGG CTG CTG GCA-3′). The reaction mixture (100 μl) consisted of a reaction buffer (10 mM Tris-HCl, 1.5 mM MgCl2, 50 mM KCl [final concentrations]), each deoxynucleoside triphosphate at a concentration of 200 μM, 60 pmol of each primer, 1 μl of the amplified 700-bp fragment, and 2.5 U of Taq DNA polymerase. The amplification program was 94°C for 4 min; 30 cycles of 94°C for 30 s, 58°C for 30 s, and 68°C for 1 min; and finally, 68°C for 7 min. The sizes and quantities of PCR products were determined by 2% agarose gel electrophoresis (Seakem CTG agarose; TEBU, Le Perray-en-Yvelines, France).
TTGE analysis.
PCR products obtained from V3 region amplification were subjected to TTGE analyses. TTGE was performed by using the Dcode universal mutation detection system (Bio-Rad) and gels that were 16 cm by 16 cm by 1 mm. Polyacrylamide (8%) gels were prepared and run with 1× TAE buffer diluted from 50× TAE buffer (2 M Tris base, 1 M glacial acetic acid, 50 mM EDTA). Gels were prepared with 8% (wt/vol) acrylamide stock solutions (acrylamide-bisacrylamide; 37.5:1) and a final urea concentration of 7 M. TTGE parameters and gradient temperatures were optimized to separate the bacterial species studied (species with low-G+C-content genomes). The final electrophoresis conditions were 41 V for 16 h with an initial temperature of 63°C and a final temperature of 70°C (the temperature was increased 0.4°C per h). Five-microliter samples of PCR products were deposited in wells. To avoid nonhomogenous temperature effects, samples were not deposited in the outermost wells. A magnetic stirrer was used to mix the buffers and improve the temperature gradient homogeneity. After runs, gels were stained for 15 min with ethidium bromide (0.5 μg/ml of 1× TAE buffer), rinsed for 20 min in 1× TAE buffer, and photographed on a UV transillumination table.
Gel analysis and reference database setup.
TTGE gels were standardized by including a V3 identification ladder made up of four reference species (Lactococcus garvieae, Lactococcus raffinolactis, Enterococcus faecalis, and Lactococcus lactis) (see Fig. 3, lanes marked “M”). An ordered data set was generated by using GelCompar software (Applied Maths), a data-processing tool. For this purpose, the photographed gels were converted into a file image, which was then analyzed by GelCompar. This software standardizes TTGE profiles to minimize migration differences between gels (46). Data for ∼135 strains corresponding to 48 species or subspecies were integrated into the TTGE database.
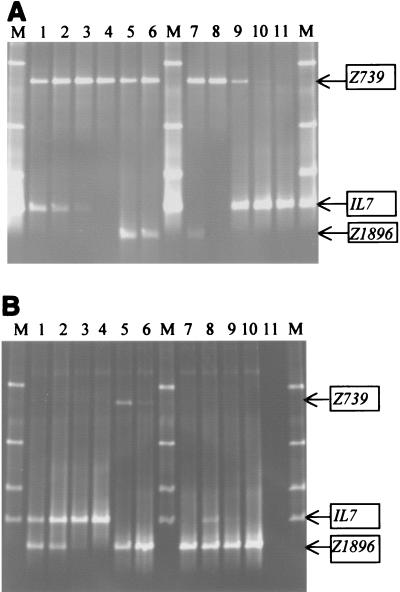
FIG. 3.
TTGE sensitivity in mixed cultures as a function of relative DNA concentrations. V3 16S rDNA fragments were PCR amplified from mixtures of two DNA samples present at different proportions and subjected to TTGE analyses. In the mixtures, the relative proportions of DNAs extracted from the two species varied from 1:1 to 1:0.001. The positions of the bands for the three reference strains used in this study are indicated on the right (Z739, CNRZ739; IL7, IL7; Z1896, CNRZ1896). (A) Lanes M, identification ladder; lane 1, CNRZ739 plus IL7; lane 2, CNRZ739 plus IL7 (1:10); lane 3, CNRZ739 plus IL7 (1:100); lane 4, CNRZ739 plus IL7 (1:1,000); lane 5, CNRZ739 plus CNRZ1896; lane 6, CNRZ739 plus CNRZ1896 (1:10); lane 7, CNRZ739 plus CNRZ1896 (1:100); lane 8, CNRZ739 plus CNRZ1896 (1:1,000); lane 9, IL7 plus CNRZ739 (1:10); lane 10, IL7 plus CNRZ739 (1:100); lane 11, IL7 plus CNRZ739 (1:1,000). (B) Lanes M, identification ladder; lane 1, IL7 plus CNRZ1896; lane 2, IL7 plus CNRZ1896 (1:10); lane 3, IL7 plus CNRZ1896 (1:100); lane 4, IL7 plus CNRZ1896 (1:1,000); lane 5, CNRZ1896 plus CNRZ739 (1:10); lane 6, CNRZ1896 plus CNRZ739 (1:100); lane 7, CNRZ1896 plus CNRZ739(1:1,000); lane 8, CNRZ1896 plus IL7 (1:10); lane 9, CNRZ1896 plus IL7 (1:100); lane 10, CNRZ1896 plus IL7 (1:1,000); lane 11, negative control.
Determination of TTGE sensitivity.
We examined TTGE sensitivity by using mixtures containing different proportions of two purified DNA samples. DNA was isolated from three common dairy starter species: Lactococcus lactis strain IL416, Streptococcus thermophilus strain CNRZ1896, and Lactobacillus plantarum strain CNRZ1572. The DNA concentration was measured by determining the optical density at 260 nm.
To determine the sensitivity of the technique and the capacity to detect the presence of a minor bacterial species, the following DNA mixtures were subjected to TTGE analyses: 725 ng of species A DNA plus 725 ng of species B DNA; 725 ng of species A DNA plus 72.5 ng of species B DNA; 725 ng of species A DNA plus 7.25 ng of species B DNA; and 725 ng of species A DNA plus 0.725 ng of species B DNA.
Bacterial starter culture preparation.
A starter culture was prepared under aseptic conditions by using sterile milk (Elle & Vire milk powder, reconstituted at a concentration of 10% [wt/vol] in sterile water and autoclaved at 110°C for 10 min) as the medium; this medium was inoculated with three different strains (Lactococcus lactis strain IL416, Leuconostoc mesenteroides strain 10F, and Lactobacillus plantarum strain CNRZ1572). The starter culture was examined to determine live bacterial counts, and DNA was extracted for TTGE analysis (see below).
Fermented milk production.
Fermented milk preparations were produced under aseptic conditions with sterile milk (prepared as described above). Fermentation was carried out at 22°C for 24 h either with a single strain (fermented milk 1 was prepared by using Lactococcus lactis strain IL416) or with several strains (fermented milk 2 was prepared by using Lactococcus lactis strain IL416, Leuconostoc mesenteroides strain 10F, Lactobacillus plantarum strain CNRZ1572, and Streptococcus thermophilus strain CNRZ1896). Bacterial counts were determined, and DNA was extracted for TTGE analysis.
Model miniature cheese production.
The model miniature washed-curd cheeses were prepared under controlled bacteriological conditions according to a protocol developed in our laboratory (22) by using sterile techniques and autoclaved equipment. Cheeses were made from whole microfiltered milk (Marguerite, Villefranche sur Saône, France), which is characterized by an excellent microbial quality (<20 CFU of mesophilic bacteria per ml). Since the risks of cheese contamination are limited, this model is a good tool for TTGE validation.
We manufactured eight cheeses. In series A, four cheeses were prepared by using Lactococcus lactis strain IL416 (1% [wt/vol] in milk) as the starter and different concentrations of Leuconostoc mesenteroides strain 10F as the adjunct culture (1% in cheese A1, 0.1% in cheese A2, 0.01% in cheese A3, and 0.001% in cheese A4). The cultures were prepared and inoculated into milk as described previously (23). Bacterial counting and extraction of DNA for TTGE were performed 1 day after production.
In series B, four other cheeses were prepared by using either Lactococcus lactis strain IL416 or Lactococcus lactis subsp. cremoris strain AM2 as the starter. Strain IL416 is characterized by its robust properties (i.e., it is poorly autolytic) (23), whereas strain AM2 is highly autolytic, presumably due to the presence of a prophage in its genome (6). Cheeses B1 and B3 were prepared by using strains AM2 and IL426, respectively, and cheeses B2 and B4 were prepared by using strains AM2 and IL416, respectively, plus Leuconostoc mesenteroides strain 10F. After 28 days of ripening, bacterial counts were determined, and DNA was extracted from the cheeses.
Determination of bacterial counts.
Dairy products (10 g) were emulsified in 100 ml of sterile 2% (wt/vol) trisodium citrate (Prolabo, Fontenay sous bois, France) and homogenized by using a mechanical blender (T-25 IKA Ultra-turrax; 19,000 rpm for 45 s; Labo Moderne, Paris, France) to disrupt lactococcal chains (30). Serial dilutions were prepared in sterile 1% (wt/vol) peptone (Prolabo) and plated on selective agar medium with a spiral plater (Spiral System, Cincinnati, Ohio). Starter lactococci were counted on M17 agar after 48 h of incubation at 30°C (47). Lactobacillus cells were counted on modified MRS agar (pH adjusted to 5.2) by incubation for 72 h in anaerobic conditions (12). The size of the Leuconostoc population was estimated on MSE agar after 48 h of incubation at 30°C (31).
Extraction of genomic DNA in dairy products.
Dairy products (5 g) were dissolved in 40 ml of sterile 2% (wt/vol) trisodium citrate and homogenized (19,000 rpm) by using the Ultra-turrax until the solutions were opaque. To each sample, 50 mg of pronase (Boerhinger, Mannheim, Germany) and 100 μl of β-mercaptoethanol were added, and this was followed by 3 h of incubation at 37°C. The bacteria were washed twice by centrifugation at 13,000 × g for 10 min. The pellets were resuspended first in sterile water and then in 10 ml of T1 buffer (1 M sorbitol, 0.1 M EDTA; pH 8). The cells were recentrifuged and finally resuspended in 500 μl of T1 buffer, transferred into Eppendorf tubes, and cooled for 10 min in ice. The cells were lysed by using glass beads (diameter, 150 to 200 μm; Sigma, Saint Quentin Fallavier, France) in the presence of T1 buffer (six cycles consisting of 30 s of vortexing at high speed and 1 min of storage in ice). After settling, the supernatant (∼400 μl) was removed and stored for 10 min in ice. DNA was then extracted by the phenol chloroform method as described previously (11). The DNA pellet was dissolved in 100 μl of Tris-EDTA, and concentrations were determined by 0.8% agarose gel electrophoresis.
Commercial dairy products.
The following liquid, semisolid, and solid milk products were used for studies: (i) a commercial mesophilic cheese starter (France); (ii) two commercial fermented milk preparations (France); and (iii) commercial cheeses, including raw milk and pasteurized Camembert cheeses, Brie (soft cheese) (France), Emmental, Comté, and Beaufort (Swiss type cheeses) (France), processed cheese (France), and a fresh artisan-made cheese (Ferme de Viltain, Jony-en-Josas, France). Bacterial counting and extraction of DNA for TTGE were performed as described above.
Cloning and sequencing of TTGE fragments.
Bands were excised from the TTGE gels obtained with commercial products and purified as described previously (44). The eluted DNA was reamplified with primers HDA1 (lacking the GC clamp) and HDA2. PCR products were then purified (Kit GIBCO, Invitrogen Life Technologies, Cergy Pontoise, France), cloned by using a Topo cloning kit (Zero Blunt; Invitrogen BV, Groningen, The Netherlands), and transformed into Escherichia coli, as described by the manufacturer. Cloning of the PCR products was necessary because TTGE analysis revealed weak bands in addition to the excised bands after reamplification. The resulting plasmids were used as a matrix to reamplify the insert with primers HDA1-GC and HDA2. The amplicons were subjected to TTGE analysis to confirm their relative positions. The cloned fragments that comigrated with the original bands were then sequenced by using an ABI PRISM dye terminator kit (7) and primer M13 (Invitrogen BV). Sequences were compared to the Ribosomal Database Project sequences (29) for species assignment.
RESULTS
TTGE database setup.
Forty-eight species were used to establish the TTGE database (Fig. 1). Most of these species generated a single specific band. Additional bands with lower intensities were sometimes present; these were probably artifacts of PCR, as their presence varied with the reaction conditions. In initial experiments, we found that direct PCR amplification of the V3 region of pure strains often gave rise to parasite bands in TTGE gels. To alleviate this problem, we used the nested PCR approach (see Materials and Methods), which produced clean V3 profiles. We checked that strains belonging to the same species (generally, three strains were tested for each species) had similar TTGE fingerprints (data not shown). In all cases, pure cultures gave rise to one major V3 band on TTGE gels.
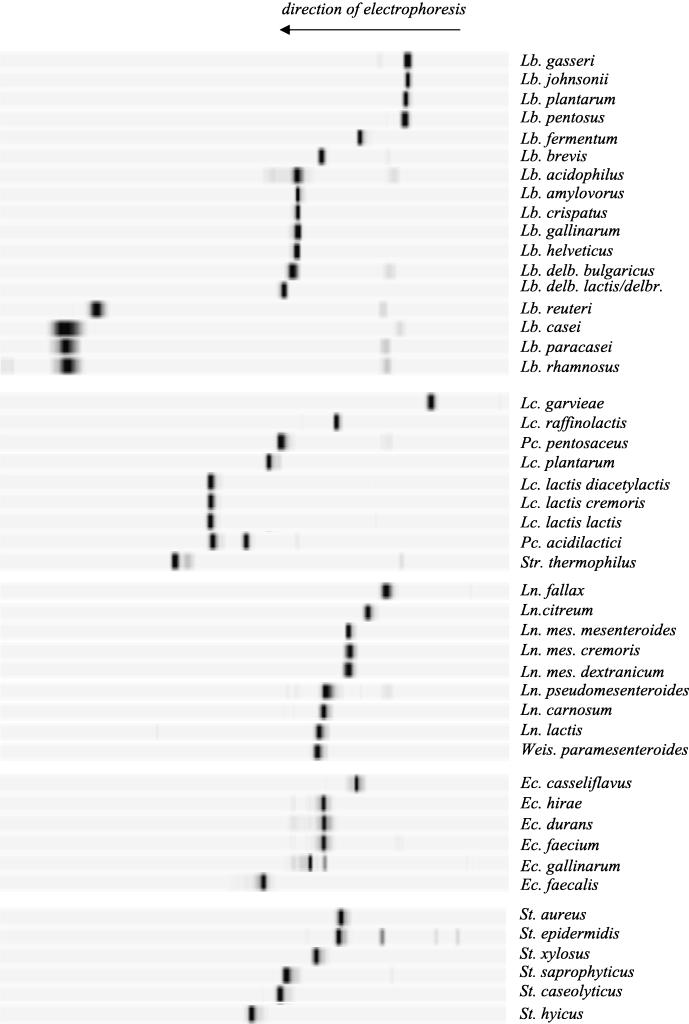
FIG. 1.
Species database compiled from TTGE profiles of V3 16S rDNA fragments of purified control strains. V3 fragments were separated by TTGE on a denaturing acrylamide (8%, wt/vol) gel. Gels were standardized by using GelCompar software (Applied Maths). The profiles are presented in groups by genus. Each species is characterized by a specific TTGE fingerprint. Lb., Lactobacillus; Lc., Lactococcus; Pc., Pediococcus; Str., Streptococcus; Ln., Leuconostoc; Weis., Weissella; Ec., Enterococcus; St., Staphylococcus; delb., delbrueckii; mes., mesenteroides.
The V3 sequence variability suggests that TTGE can differentiate between bacteria belonging to different genera. For example, Lactococcus lactis, Lactobacillus casei, and Leuconostoc mesenteroides were distinguished in tests in which pure cultures were used (Fig. 1). The ability to resolve more closely related species depends on V3 variability, which can be predicted either from V3 sequence analysis (Fig. 2) or from calculated V3 Tm values (Table 1). The level of discrimination for control bacteria is reported below for each genus.
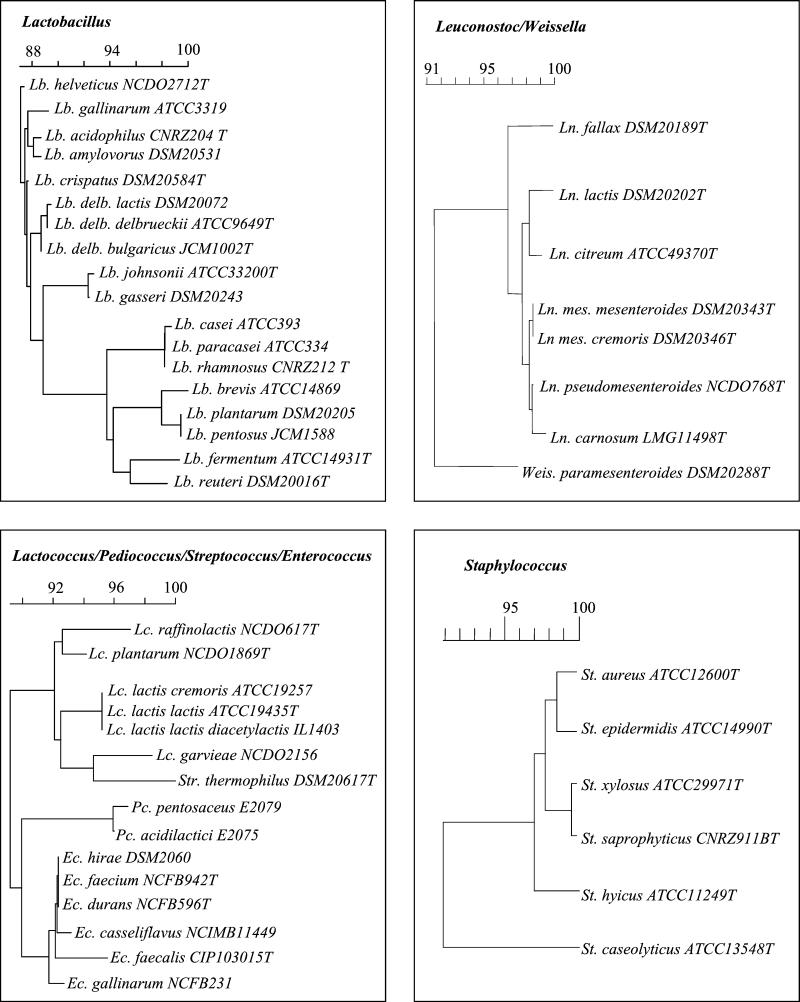
FIG. 2.
Schematic presentation of V3 region relatedness. Pairwise distances were calculated from the V3 region of 16S rDNA. The dendrogram was constructed by using the neighbor-joining method included in the GeneCompar software (Applied Maths). Sequences used for this analysis were obtained either from the CNRZ collection database (Elodie Lepage, Unité de Recherches Laitières et Génétique Appliquée) or from the GenBank database. delb., delbrueckii; mes., mesenteroides. For an explanation of other abbreviations see the legend to Fig. 1.
Lactobacillus.
Experimental results (Fig. 1) were generally consistent with V3 sequences predictions. TTGE did not distinguish between members of the Lactobacillus casei group (Lactobacillus casei, Lactobacillus paracasei, and Lactobacillus rhamnosus) or between members of the Lactobacillus acidophilus group (Lactobacillus gallinarum, Lactobacillus crispatus, Lactobacillus amylovorus, Lactobacillus acidophilus, and the closely related species Lactobacillus helveticus) (Fig. 2). Lactobacillus pentosus and Lactobacillus plantarum have similar V3 sequences (Fig. 2) which comigrate, as do the sequences of Lactobacillus johnsonii and Lactobacillus gasseri. Note that the Lactobacillus plantarum and Lactobacillus gasseri V3 sequences differ (Fig. 2), but the Tm values of these organisms are very similar (Table 1). This explains their quasi-similar TTGE profiles. The following species and subspecies were distinguishable by TTGE: Lactobacillus reuteri, Lactobacillus brevis, Lactobacillus fermentum, Lactobacillus delbrueckii subsp. bulgaricus, and Lactobacillus delbrueckii subsp. lactis.
Lactococcus.
The TTGE method makes it possible to differentiate four known species of lactococci (Lactococcus lactis, Lactococcus garvieae, Lactococcus plantarum, and Lactococcus raffinolactis). However, the V3 fragments of the closely related organisms Lactococcus lactis subsp. lactis, Lactococcus lactis subsp. lactis subsp. diacetylactis, and Lactococcus lactis subsp. cremoris comigrated (Fig. 1).
Leuconostoc.
Leuconostoc fallax and Leuconostoc citreum are distinguishable on TTGE gels. However, we could not distinguish among Leuconostoc mesenteroides subsp. mesenteroides, Leuconostoc mesenteroides subsp. cremoris, and Leuconostoc mesenteroides subsp. dextranicum, between Leuconostoc lactis and Weissella paramesenteroides, or between Leuconostoc carnosum and Leuconostoc pseudomesenteroides. In the latter case, the two species have different calculated Tm values (Table 1) and would be predicted to migrate at different positions.
Enterococcus.
Strains of Enterococcus faecalis, Enterococcus gallinarum, and Enterococcus casseliflavus were distinguishable by TTGE (Fig 1). In contrast, Enterococcus faecium, Enterococcus durans, and Enterococcus hirae all migrated at the same position, as expected based on sequence similarities (Fig. 2).
Staphylococcus.
The six species of the genus Staphylococcus gave rise to distinguishable signals (Fig. 1). However, the positions of Staphylococcus aureus and Staphylococcus epidermidis V3 fragments were very close on TTGE gels, as predicted by their Tm values (Table 1).
In a few cases, despite species differences and V3 sequence divergence, V3 fragments were found to comigrate. For example, the V3 fragments of Leuconostoc lactis, Staphylococcus xylosus, and Weissella paramesenteroides comigrated, as did those of Enterococcus faecium, Leuconostoc pseudomesenteroides, and Lactobacillus brevis and those of Pediococcus pentosaceus and Staphylococcus caseolyticus (Fig. 1). In some cases, comigration was predicted by the Tm values of the V3 fragments (this was predicted for Lactobacillus brevis and Leuconostoc pseudomesenteroides and for Pediococcus pentosaceus and Staphylococcus caseolyticus [Table 1]). However, the calculated Tm of Weissella paramesenteroides, which is very different from that of Leuconostoc lactis or Staphylococcus xylosus (Table 1), is inconsistent with experimental results. One possible explanation for this discrepancy is that the calculated Tm given by the algorithm may differ from the real Tm (http://biochem.roche.com/lightcycler/lc_support/pdfs/lc_6.pdf.).
Sensitivity of TTGE technique.
The sensitivity of TTGE for detection of minority bacterial populations was examined. DNAs extracted from pure bacterial cultures were combined and analyzed by TTGE; each sample contained DNAs derived from two strains in a different proportion (Fig. 3). The limit of detection of the minority species was determined. In all cases, a clear limit of detection was observed when the minority species accounted for 1:100 or less of the total DNA concentration. Detection of species by TTGE may be limited either by low DNA concentrations or by the presence of high concentrations of competing DNA. Tests performed with DNA from a single strain at dilutions that gave no signal in mixed samples did give rise to a band when the DNA was used as the single substrate (data not shown). We therefore consider it likely that competition for the PCR primers by the dominant DNA species is a limiting factor for TTGE sensitivity.
Application of TTGE to controlled dairy ecosystems.
To test the potential of the technique in situ, TTGE was performed with dairy samples whose bacterial compositions were known (Fig. 4). Extracts of one starter culture (Fig. 4, lane 1) and two fermented milk cultures (lanes 6 and 11) were analyzed. The TTGE bands were compared with the database species for identification. For the starter culture, TTGE clearly identified the presence of Lactococcus lactis, Lactobacillus plantarum, and Leuconostoc mesenteroides. Fermented milk 1 was found to contain Lactococcus lactis. Fermented milk 2 was prepared by using a mixture of four strains: Lactococcus lactis IL416, Leuconostoc mesenteroides 10F, Lactobacillus plantarum CNRZ1572, and Streptococcus thermophilus CNRZ1896. The first three strains were each present at concentrations between 2 × 108 and 2 × 109 CFU/g of dairy product (after 1 day) and were detectable by TTGE. However, Streptococcus thermophilus was present at a concentration of only to 2 × 106 CFU/g in this experiment and was not detectable by TTGE. These in situ results are in agreement with the detection limits determined above (Fig. 3).
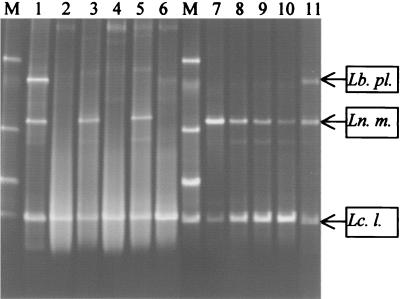
FIG. 4.
Identification of bacterial species present in controlled dairy products. TTGE was performed with V3 16S rDNA fragments that were PCR amplified from extracts of starter cultures, fermented milk, and cheese products. Each dairy product had a known bacterial composition. After standardization of the gel with GelCompar software, bands were identified by comparison with the reference database. The positions of the bands for the known species Lactobacillus plantarum (Lb. pl.), Leuconostoc mesenteroides (Ln. m.), and Lactococcus lactis (Lc. l.) are indicated on the right. Lane M, identification ladder; lane 1, starter culture (Lactococcus lactis, Leuconostoc mesenteroides, Lactobacillus plantarum); lane 2, model miniature cheese B1 (Lactococcus lactis strain AM2); lane 3, model miniature cheese B2 (Lactococcus lactis strain AM2 plus Leuconostoc mesenteroides strain 10F); lane 4, model miniature cheese B3 (Lactococcus lactis strain IL416); lane 5, model miniature cheese B4 (Lactococcus lactis strain IL416 plus Leuconostoc mesenteroides strain 10F); lane 6, fermented milk 1 (Lactococcus lactis strain IL416); lane 7, model miniature cheese A1 (3 × 109 Lactococcus CFU/g plus 4 × 109 Leuconostoc CFU/g); lane 8, model miniature cheese A2 (3 × 109 Lactococcus CFU/g plus 6 × 108 Leuconostoc CFU/g); lane 9, model miniature cheese A3 (2 × 109 Lactococcus CFU/g plus 7 × 107 Leuconostoc CFU/g); lane 10, model miniature cheese A4 (2 × 109 Lactococcus CFU/g plus 2 × 107 Leuconostoc CFU/g); lane 11, fermented milk 2 (2 × 109 Lactococcus CFU/g plus 8 × 108 Leuconostoc CFU/g plus 2 × 108 Lactobacillus CFU/g plus 2 × 106 Streptococcus CFU/g).
The capacity to distinguish species in a solid cheese matrix containing known bacteria was examined. In the four nonripened cheeses (designated cheeses A1, A2, A3, and A4), the proportion of lactococci to leuconostocs varied from 1:1 (each strain present at a concentration of 109 CFU/g) to 1:0.01. Both strains were readily detectable by TTGE (Fig. 4, lanes 7 to 10) in the 1-day-old cheeses. These results demonstrate that TTGE sensitivity is not reduced when the technique is applied to this complex system.
In the four ripened cheeses (designated cheeses B1, B2, B3, and B4), the bacterial counts after 28 days of ripening confirmed the autolytic properties of Lactococcus lactis strain AM2 (concentrations in cheeses B1 and B2, less than 106 CFU/g) and the robust properties of strain IL416 (concentration in cheeses B3 and B4, 109 CFU/g). Leuconostoc was present at concentrations of 107 and 108 CFU/g in cheeses containing AM2 and IL416, respectively. All bacteria in the four test cheeses were clearly identified by TTGE analysis (Fig. 4, lanes 2 to 5). Interestingly, despite its poor viability, Lactococcus lactis strain AM2 was readily detected by TTGE.
Application of TTGE to complex and unknown dairy ecosystems.
We used the TTGE database established with control strains to identify major bacterial populations present in several commercial dairy products, including a commercial starter culture, fermented milk cultures, and both industrial and artisan-made cheeses (Fig. 5). The commercial starter culture was found to contain a single band assigned to Lactococcus lactis. Classical plating methods confirmed the presence of Lactococcus lactis and the absence of other species. TTGE analysis of two commercial fermented milk preparations (designated A and B) revealed the presence of Lactobacillus delbrueckii subsp. bulgaricus and Streptococcus thermophilus; identification of these species was confirmed by plating on selective media. In addition, fermented milk B is described as a preparation that contains Lactobacillus casei; TTGE confirmed the presence of this species.
FIG. 5.
Identification of bacterial species present in commercial dairy products. TTGE was performed with V3 16S rDNA fragments that were PCR amplified from extracts of a commercial available starter, fermented milk preparations, and cheese products. After standardization of band migration with the GelCompar software, species were identified by comparison with known species in the reference database. Profile analyses are discussed in the text. Some bands (bands a through h) were excised, cloned, sequenced, and subjected to a Blast analysis for identification. These bands correspond to Lactobacillus casei (band a), Streptococcus thermophilus (band b), Lactobacillus casei (band c), Lactobacillus delbrueckii subsp. lactis (band d), Lactobacillus casei (band e), Buttiauxella agrestis (band f), Lactococcus lactis (band g), and Lactobacillus plantarum (band h). Unassigned bands i through k were compared to an extended reference set (data not shown) and identified as Hafnia alvei (bands i and j) and either Stenotrophomonas maltophilia or Escherichia coli (band k). del., delbrueckii. See the legend to Fig. 1 for an explanation of the other abbreviations.
TTGE analysis of eight cheeses revealed various profiles comprising between two and eight bands per product (Fig. 5). Most of the bands were identified by using our database reference. For example, an artisan-produced fresh cheese contained three identified bacterial species (Lactococcus lactis, Lactobacillus plantarum, and Leuconostoc mesenteroides) and one unassigned band (see below for identification). It is interesting that the total live bacterial count at the time of extraction of DNA for TTGE was less than 2 × 105 CFU/g (data not shown). Thus, the species detected by TTGE are likely to correspond to the starter composition of this fresh cheese. Cheeses having similar production procedures (e.g., Brie and Camembert cheeses, or Emmental, Comté, and Beaufort cheeses) produced common TTGE bands (corresponding to Lactococcus lactis, Leuconostoc mesenteroides, and Lactobacillus plantarum in soft cheeses and to Lactobacillus casei, Streptococcus thermophilus, and Lactobacillus delbrueckii in Swiss- type cheeses). Raw milk Camembert cheese showed the most complex profiles; eight bands were detected, compared to five for pasteurized Camembert cheese. The related Brie cheese, which had a short ripening time, had a simpler composition, as just two species, Lactococcus lactis and Streptococcus thermophilus, were detected by TTGE. We used two approaches to confirm the accuracy of TTGE assignments. In several cases, we performed plate counting to confirm the presence of the organisms identified by TTGE (data not shown). Bands obtained for Emmental, Beaufort, and raw milk Camembert cheeses were excised, cloned, and identified by DNA sequencing (Fig. 5, bands a to h). Where species assignments were made, bands were confirmed to be bands produced by the expected species. We also cloned and sequenced one unassigned band (Fig. 5, band f) from raw milk Camembert cheese, which corresponded to a gram-negative bacterium, Buttiauxella agrestis. An unassigned band found in the raw milk Camembert cheese and fresh artisan-made cheese profiles (Fig. 5, bands i and j) corresponds to Hafnia alvei, a gram-negative bacterium commonly encountered in cheeses. Moreover, the unassigned band (band k) of the pasteurized milk Camembert cheese corresponds either to Stenotrophomonas maltophilia or Escherichia coli, which are also gram-negative bacteria.
The results described above demonstrate the feasibility of using TTGE for detection of dominant species in various dairy ecosystems. TTGE is thus a potentially useful means of monitoring populations for both laboratory and commercial analyses.
DISCUSSION
In this work, we explored the uses and limits of TTGE in microbial ecology to describe biodiversity in dairy products. TTGE has proven to be a powerful and simple method for identifying species of bacteria in both liquid and solid dairy food environments. This method allowed differentiation generally at the species level (or at the level of groups of species) and sometimes at the subspecies level. For example, the capacity of TTGE to differentiate among Lactobacillus acidophilus, Lactobacillus delbrueckii subsp. bulgaricus, and Lactobacillus delbrueckii subsp. lactis species is of particular interest for the dairy industry. TTGE is both more sensitive and faster than conventional molecular and bacteriological methods of strain identification. Using TTGE with known bacterial samples, we set up a species database of bacteria with low G+C genome contents that could be used as a reference by microbiologists who study lactic acid bacteria. This is the first time that an exhaustive species database has been set up to allow rapid identification of bacteria in complex food ecosystems. We are currently widening the database to other species. By using different migration conditions, dairy microorganisms that are high-G+C-content species are also being analyzed.
The genomic DNA extraction protocol is efficient for PCR amplification when the starting material is dairy products. Moreover, potential contaminants in the complex milk substrate (such as exopolysaccharides or lipids) do not inhibit PCRs (53). The TTGE profiles for dairy ecosystems are relatively simple (less than 10 bands) compared to those for other ecosystems (e.g., the digestive tract [50] or soil [15]). By using the TTGE reference database, it was possible to directly identify a species as a bacterial component of various dairy products (milk, cheese, and fermented milk). Bands that are distinct from the reference database bands can be excised directly from the gel and sequenced (Fig. 5); this allows unknown species to be identified and also expands the TTGE species reference set.
In some cases, TTGE detected microflora in cheeses (e.g., autolytic or noncultivable strains) that could not be identified by traditional microbiological techniques. The capacity of TTGE to detect dead bacteria is particularly relevant to its application to cheeses, as autolytic strains are commonly used in cheese production (6). However, we note the following limitations of the TTGE system. (i) TTGE provides a description of the dominant bacterial species in a complex ecosystem. Minority bacterial species cannot be detected if they account for less than 1% of the most dominant species. Our results are in agreement with those obtained with other complex media (15, 35, 36). Thus, the use of TTGE to detect very minor species, such as pathogens, would require the use of highly specific primers (see reference 51 for a description of detection of minor species in human feces). (ii) In some cases, related species have identical V3 sequences and cannot be distinguished. In other cases, species have different V3 sequences but the same Tm (35) and thus migrate at the same position. Other, more discriminating areas are needed to differentiate between these strains, either in 16S rDNA regions (34, 49) or in other functional genes (52). (iii) We sometimes observed multiple bands for a single species, which may have represented PCR artifacts (50), 16S rDNA heterogeneity (10, 40), or heteroduplex formation (24). Formation of multiple bands may be particularly problematic in complex ecosystems, because such bands can result in an overestimate of the number of species present. PCR artifacts can be minimized by using high-quality primers and high-fidelity polymerases and by modifying PCR conditions to avoid the formation of artifacts (see Materials and Methods).
In conclusion, we believe that TTGE is an excellent tool for describing the dominant species in dairy ecosystems. This method could be used for typing cheeses according to their technologies, their origins, or their regional characteristics. The establishment of a molecular cheese fingerprint could be of considerable interest to industry, especially as the method is inexpensive and the setup is simple.
Acknowledgments
We are grateful to E. Lepage, P. Quénée, O. Firmesse, and S. Furlan (Collection CNRZ) for providing the strains. We especially thank E. Lepage for technical advice. We also thank C. Bach for fermented milk manufacture and D. Hemme for helpful discussions.
REFERENCES
- 1.Akkermans, A. D. L., M. S. Mirza, H. J. M. Harmsen, H. J. Blok, P. R. Heron, A. Sessitsch, and W. M. Akkermans. 1994. Molecular ecology of microbes: review of promises, pitfalls, and true progress. FEMS Microbiol. Rev. 15:185-194. [Google Scholar]
- 2.Amiel, C., L. Mariey, M. C. Curk-Daubié, P. Pichon, and J. Travert. 2000. Potential of Fourier infrared spectroscopy (FTIR) for discrimination and identification of dairy lactic acid bacteria. Lait 80:445-449. [Google Scholar]
- 3.Ampe, F., N. ben Omar, C. Moizan, C. Wacher, and J. P. Guyot. 1999. Polyphasic study of the spatial distribution of microorganisms in Mexican pozol, a fermented maize dough, demonstrates the need for cultivation-independent methods to investigate traditional fermentations. Appl. Environ. Microbiol. 65:5464-5473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Børresen-Dale, A. L., S. Lystad, and A. Langerød. 1997. Temporal temperature gradient gel electrophoresis (TTGE) compared with denaturing gradient gel electrophoresis (DGGE) and constant denaturing gel electrophoresis (CDGE) in mutation screening. Bioradiation 99:12-13. [Google Scholar]
- 5.Chapman, H. R., and M. S. Sharpe. 1990. Microbiology of cheese, p. 203-289. In R. K. Robinson (ed.), Dairy microbiology, the microbiology of milk products, 2nd ed., vol. 2. Elsevier, London, United Kingdom. [Google Scholar]
- 6.Chapot-Chartier, M.-P., C. Deniel, M. Rousseau, L. Vassal, and J.-C. Gripon. 1994. Autolysis of two strains of Lactococcus lactis during cheese ripening. Int. Dairy J. 4:251-269. [Google Scholar]
- 7.Cibik, R., E. Lepage, and P. Tailliez. 2000. Molecular diversity of Leuconostoc mesenteroides and Leuconostoc citreum isolated from traditional French cheeses as revealed by RAPD fingerprinting, 16S rDNA sequencing and 16S rDNA fragment amplification. Syst. Appl. Microbiol. 23:267-278. [DOI] [PubMed] [Google Scholar]
- 8.Cocolin, L., M. Manzano, C. Cantoni, and G. Comi. 2001. Denaturing gradient gel electrophoresis analysis of the 16S rRNA gene V1 region to monitor dynamic changes in the bacterial population during fermentation of Italian sausages. Appl. Environ. Microbiol. 67:5113-5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coppola, S., G. Blaiotta, D. Ercolini, and G. Moschetti. 2001. Molecular evaluation of microbial diversity occurring in different types of Mozzarella cheese. J. Appl. Microbiol. 90:414-420. [DOI] [PubMed] [Google Scholar]
- 10.Dählloff, I., H. Baillie, and S. Kjelleberg. 2000. rpoB-based microbial community analysis avoids limitations inherent in 16S rRNA gene intraspecies heterogeneity. Appl. Environ. Microbiol. 66:3376-3380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de los Reyes-Gavilan, C., G. K. Y. Limsowtin, P. Tailliez, L. Séchaud, and J. P. Accolas. 1992. A Lactobacillus helveticus-specific DNA probe detects restriction fragment length polymorphisms in this species. Appl. Environ. Microbiol. 58:3429-3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Man, J. D., M. Rogosa, and M. E. Sharpe. 1960. A medium for the cultivation of lactobacilli. J. Appl. Bacteriol. 23:130-135. [Google Scholar]
- 13.Ercolini, D., G. Moschetti, G. Blaiotta, and S. Coppola. 2001. Behavior of variable V3 region from 16S rDNA of lactic acid bacteria in denaturing gradient gel electrophoresis. Curr. Microbiol. 42:199-202. [DOI] [PubMed] [Google Scholar]
- 14.Felske, A., B. Engelen, U. Nübel, and H. Backhaus. 1996. Direct ribosome isolation from soil to extract bacterial rRNA for community analysis. Appl. Environ. Microbiol. 62:4162-4167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Felske, A., A. Wolterink, R. Van Lis, and A. D. Akkermans. 1998. Phylogeny of the main bacterial 16S rRNA sequences in Drentse A grassland soils (The Netherlands). Appl. Environ. Microbiol. 64:871-879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ferris, M. J., G. Muyzer, and D. M. Ward. 1996. Denaturing gradient gel electrophoresis profiles of 16S rRNA-defined populations inhabiting a hot spring microbial mat community. Appl. Environ. Microbiol. 62:340-346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fischer, S. G., and L. S. Lerman. 1983. DNA fragments differing by single base-pair substitutions are separated in denaturing gradient gels: correspondence with melting theory. Proc. Natl. Acad. Sci. USA 80:1579-1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fox, P. F., P. L. H. McSweeney, and C. M. Lynch. 1998. Significance of non-starter lactic acid bacteria in Cheddar cheese. Aust. J. Dairy Technol. 53:83-89. [Google Scholar]
- 19.Gelsomino, A., A. C. Keijzer-Wolters, G. Cacco, and J. D. van Elsas. 1999. Assessment of bacterial community structure in soil by polymerase chain reaction and denaturing gradient gel electrophoresis. J. Microbiol. Methods 38:1-15. [DOI] [PubMed] [Google Scholar]
- 20.Heuer, H., M. Krsek, P. Baker, K. Smalla, and E. M. Wellington. 1997. Analysis of actinomycete communities by specific amplification of genes encoding 16S rRNA and gel-electrophoretic separation in denaturing gradients. Appl. Environ. Microbiol. 63:3233-3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hugenholtz, P., B. M. Goebel, and N. R. Pace. 1998. Impact of culture-independent studies on the emerging phylogenetic view of bacterial diversity. J. Bacteriol. 180:4765-4774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hynes, E., J.-C. Ogier, and A. Delacroix-Buchet. 2000. Protocol for the manufacture of miniature washed-curd cheeses under controlled microbial conditions. Int. Dairy J. 10:733-737. [Google Scholar]
- 23.Hynes, E., J.-C. Ogier, and A. Delacroix-Buchet. 2001. Proteolysis during ripening of miniature washed-curd cheeses manufactured with different strains of starter bacteria and a Lactobacillus plantarum adjunct culture. Int. Dairy J. 11:587-597. [Google Scholar]
- 24.Jensen, M. A., and N. Straus. 1993. Effect of PCR conditions on the formation of heteroduplex and single-stranded DNA products in the amplification of bacterial ribosomal DNA spacer regions. PCR Methods Appl. 3:186-194. [DOI] [PubMed] [Google Scholar]
- 25.Lane, C. N., and P. F. Fox. 1996. Contribution of starter and adjunct lactobacilli to proteolysis in cheddar cheese during ripening. Int. Dairy J. 6:715-728. [Google Scholar]
- 26.Lee, D. H., Y. G. Zo, and S. J. Kim. 1996. Nonradioactive method to study genetic profiles of natural bacterial communities by PCR-single-strand-conformation polymorphism. Appl. Environ. Microbiol. 62:3112-3120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lerman, L. S., S. G. Fischer, I. Hurley, K. Silverstein, and N. Lumelsky. 1984. Sequence-determined DNA separations. Annu. Rev. Biophys. Bioeng. 13:399-423. [DOI] [PubMed] [Google Scholar]
- 28.Lerman, L. S., and K. Silverstein. 1987. Computational simulation of DNA melting and its application to denaturing gradient gel electrophoresis. Methods Enzymol. 155:482-501. [DOI] [PubMed] [Google Scholar]
- 29.Maidak, B. L., J. R. Cole, C. T. Parker, Jr., G. M. Garrity, N. Larsen, B. Li, T. G. Lilburn, M. J. McCaughey, G. J. Olsen, R. Overbeek, S. Pramanik, T. M. Schmidt, J. M. Tiedje, and C. R. Woese. 1999. A new version of the RDP (Ribosomal Database Project). Nucleic Acids Res. 27:171-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martley, F. G., and R. C. Lawrence. 1972. Cheddar cheese flavour. II. Characteristics of single strain starter associated with good or poor flavour development. N. Z. J. Dairy Sci. Technol. 7:38-44. [Google Scholar]
- 31.Mayeux, J. V., W. E. Sandine, and P. R. Elliker. 1962. A selective medium for detecting Leuconostoc in mixed-strain starter cultures. J. Dairy Sci. 45:655. [Google Scholar]
- 32.McSweeney, P. L. H., P. F. Fox, J. A. Lucey, K. N. Jordan, and T. M. Cogan. 1993. Contribution of the indigenous microflora to the maturation of Cheddar cheese. Int. Dairy J. 3:613-634. [Google Scholar]
- 33.McSweeney, P. L. H., E. M. Walsh, P. F. Fox, T. M. Cogan, F. D. Drinan, and M. Castelo-Gonzales. 1995. A procedure for the manufacture of Cheddar cheese under controlled bacteriological conditions and the effect of adjunct lactobacilli in cheese quality. Ir. J. Agric. Food Res. 33:183-192. [Google Scholar]
- 34.Mori, K., K. Yamazaki, T. Ishiyama, M. Katsumata, K. Kobayashi, Y. Kawai, N. Inoue, and H. Shinano. 1997. Comparative sequence analyses of the genes coding for 16S rRNA of Lactobacillus casei-related taxa. Int. J. Syst. Bacteriol. 47:54-57. [DOI] [PubMed] [Google Scholar]
- 35.Murray, A. E., J. T. Hollibaugh, and C. Orrego. 1996. Phylogenetic compositions of bacterioplankton from two California estuaries compared by denaturing gradient gel electrophoresis of 16S rDNA fragments. Appl. Environ. Microbiol. 62:2676-2680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Muyzer, G., E. C. de Waal, and A. G. Uitterlinden. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59:695-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Muyzer, G., T. Brinkhoff, U. Nübel, C. Santegoeds, H. Schäfer, and C. Wawer. 1997. Denaturing gradient gel electrophoresis (DGGE) in microbial ecology, p. 1-27. In A. D. L. Akkermans, J. D. van Elsas, and F. J. de Bruijn (ed.), Molecular microbial ecology manual. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 38.Muyzer, G., and K. Smalla. 1998. Application of denaturing gradient gel electrophoresis (DGGE) and temperature gradient gel electrophoresis (TTGE) in microbial ecology. Antonie Leeuwenhoek 73:127-141. [DOI] [PubMed] [Google Scholar]
- 39.Muyzer, G. 1999. DGGE/TTGE, a method for identifying genes from natural ecosystems. Curr. Opin. Microbiol. 2:317-322. [DOI] [PubMed] [Google Scholar]
- 40.Nübel, U., B. Engelen, A. Felske, J. Snaidr, A. Wieshuber, R. I. Amann, W. Ludwig, and H. Backhaus. 1996. Sequence heterogeneities of genes encoding 16S rRNAs in Paenibacillus polymyxa detected by temperature gradient gel electrophoresis. J. Bacteriol. 178:5636-5643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Øvereas, L., and V. Torsvik. 1998. Microbial diversity and community structure in two different agricultural soil communities. Microb. Ecol. 36:303-315. [DOI] [PubMed] [Google Scholar]
- 42.Peterson, S. D., and R. T. Marshall. 1990. Non-starter lactobacilli in Cheddar cheese. A review. J. Dairy Sci. 73:1395-1410. [Google Scholar]
- 43.Pot, B., C. Hertel, W. Ludwig, P. Deschee-Maeker, K. Kersters, and K. H. Schleifer. 1993. Identification and classification of Lactobacillus acidophilus, L. gasseri and L. johnsonii strains by SDS-PAGE and rRNA-targeted oligonucleotide probe hybridization. J. Gen. Microbiol. 139:513-517. [DOI] [PubMed] [Google Scholar]
- 44.Rölleke, S., G. Muyzer, C. Wawer, G. Wanner, and W. Lubitz. 1996. Identification of bacteria in a biodegraded wall painting by denaturing gradient gel electrophoresis of PCR-amplified gene fragments coding for 16S rRNA. Appl. Environ. Microbiol. 62:2059-2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Santegoeds, C. M., S. C. Nold, and D. M. Ward. 1996. Denaturing gradient gel electrophoresis used to monitor the enrichment culture of aerobic chemoorganotrophic bacteria from a hot spring cyanobacterial mat. Appl. Environ. Microbiol. 62:3922-3928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tailliez, P., P. Quénée, and A. Chopin. 1996. Estimation de la diversité parmi les souches de la collection CNRZ: application de la RAPD à un groupe de lactobacilles. Lait 76:147-158. [Google Scholar]
- 47.Terzaghi, B. E., and W. E. Sandine. 1975. Improved medium for lactic streptococci and their bacteriophages. Appl. Microbiol. 29:807-813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Teske, A., C. Wawer, G. Muyzer, and N. B. Ramsing. 1996. Distribution of sulfate-reducing bacteria in a stratified fjord (Mariager Fjord, Denmark) as evaluated by most-probable-number counts and denaturing gradient gel electrophoresis of PCR-amplified ribosomal DNA fragments. Appl. Environ. Microbiol. 62:1405-1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vasquez, A., S. Ahrné, B. Pettersson, and G. Molin. 2001. Temporal temperature gradient gel electrophoresis (TTGE) as a tool for identification of Lactobacillus casei, Lactobacillus paracasei, Lactobacillus zeae and Lactobacillus rhamnosus. Lett. Appl. Microbiol. 32:215-219. [DOI] [PubMed] [Google Scholar]
- 50.Walter, J., G. W. Tannock, A. Tilsala-Timisjarvi, S. Rodtong, D. M. Loach, K. Munro, and T. Alatossava. 2000. Detection and identification of gastrointestinal Lactobacillus species by using denaturing gradient gel electrophoresis and species-specific PCR primers. Appl. Environ. Microbiol. 66:297-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Walter, J., C. Hertel, G. W. Tannock, C. M. Lis, K. Munro, and W. P. Hammes. 2001. Detection of Lactobacillus, Pediococcus, Leuconostoc, and Weissella species in human feces by using group-specific PCR primers and denaturing gradient gel electrophoresis. Appl. Environ. Microbiol. 67:2578-2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wawer, C., and G. Muyzer. 1995. Genetic diversity of Desulfovibrio spp. in environmental samples analyzed by denaturing gradient gel electrophoresis of [NiFe] hydrogenase gene fragments. Appl. Environ. Microbiol. 61:2203-2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wheeler Alm, E., and D. A. Stahl. 1996. Extraction of microbial DNA from aquatic sediments, p. 1-29. In A. D. L. Akkermans, J. D. van Elsas, and F. J. de Bruijn (ed.), Molecular microbial ecology manual. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 54.Williams, A. G., S. E. Withers, and J. M. Banks. 2000. Energy sources of non-starter lactic acid bacteria isolated from Cheddar cheese. Int. Dairy J. 10:17-23. [Google Scholar]
- 55.Zwart, G., R. Huisman, M. P. van Agterveld, Y. van de Peer, P. de Rijk, H. Eenhoorn, G. Muyzer, E. J. van Hannen, and H. J. Laanbroek. 1998. Divergent members of the bacterial division Verrucomicrobiales in a template freshwater lake. FEMS Microbiol. Ecol. 25:159-169. [Google Scholar]