Abstract
The quantity of microorganisms that may be transferred to a food that comes into contact with a contaminated surface depends on the density of microorganisms on the surface and on the attachment strengths of the microorganisms on the materials. We made repeated contacts between pieces of meat and various surfaces (stainless steel and conveyor belt materials [polyvinyl chloride and polyurethane]), which were conditioned with meat exudate and then were contaminated with Listeria monocytogenes, Staphylococcus sciuri, Pseudomonas putida, or Comamonas sp. Attachment strengths were assessed by the slopes of the two-phase curves obtained by plotting the logarithm of the number of microorganisms transferred against the order number of the contact. These curves were also used to estimate the microbial population on the surface by using the equation of A. Veulemans, E. Jacqmain, and D. Jacqmain (Rev. Ferment. Ind. Aliment. 25:58-65, 1970). The biofilms were characterized according to their physicochemical surface properties and structures. Their exopolysaccharide-producing capacities were assessed from biofilms grown on polystyrene. The L. monocytogenes biofilms attached more strongly to polymers than did the other strains, and attachment strength proved to be weaker on stainless steel than on the two polymers. However, in most cases, it was the population of the biofilms that had the strongest influence on the total number of CFU detached. Although attachment strengths were weaker on stainless steel, this material, carrying a smaller population of bacteria, had a weaker contaminating capacity. In most cases the equation of Veulemans et al. revealed more bacteria than did swabbing the biofilms, and it provided a better assessment of the contaminating potential of the polymeric materials studied here.
When bacteria attach to surfaces and colonize them, their phenotypes change. They show, for instance, a decreased susceptibility to disinfectant (41, 42) and an increased exopolysaccharide production (46), and the community they form is called a biofilm. Biofilms are an integral part of our environment. In some cases they can be used in bioreactors to process waste; in other cases they cause malfunctions of one kind or another. The formation of biofilms creates major problems in such activities as water distribution (biofilms form in the distribution networks) (2, 10), health care (11, 13), and the food industry (7, 8, 14). A study by Haeghebaert et al. (24) has suggested that 40.5% of all food-borne infection outbreaks registered in France in 1996 were linked to contamination by equipment, highlighting the hazard that biofilms represent. In this study we examined biofilms liable to cause problems in the meat industry. Many studies have shown that when animals are slaughtered, microbial flora remains on the surface of the carcasses (5, 21), and despite the decontamination processes carried out (scorching the skin or using hot water, organic acid, or phosphate solutions), part of the microflora survives (3, 52). This resident flora comes into contact with the surfaces of equipment in the cutting halls (4, 22, 37). It can detach from the carcass and then contaminate sound products placed on the equipment. Conveyor belts are among the surfaces which, coming into contact with food, can remain heavily contaminated even after hygiene operations (4, 27, 31, 36). In this study, therefore, we set out to discover how much conveyor belts contribute to contaminating the food that comes into contact with them.
The quantity of microorganisms transferred from an inert surface to a food likely depends on the properties of the biofilm: the surface density of the microbial population, the structure of the biofilm, its capacity to produce exopolysaccharides, and the attachment strength of the microbial cells, which is probably linked to the previous properties. Here we have attempted to estimate these magnitudes. It is known that the conventional methods for counting microorganisms on surfaces, such as swabbing, do not give a good assessment of the microbial population's surface density (35, 39). Therefore, like Richard and Piton (39), we used the principle of the method proposed by Veulemans et al. (50) who assessed microbial populations on hands. Veulemans et al. (50) took successive prints from the same area of a hand by using adhesive tape and then made prints from the adhesive tape on agar. From the slopes of the straight lines plotting the logarithm of the number of CFU detached by each print against the order number of the print (first, second, and third, etc.) we can calculate the number of microorganisms on the skin. We also used the slopes of the straight lines so obtained to characterize the microorganisms' strength of attachment to the surfaces, as suggested by Eginton et al. (17, 18), who applied biofilms directly to agar. The steeper the slope, the weaker the microorganisms' attachment strength. The attachment strengths (which reflect the strength of the cells' attachment both to the inert substrate and to each other) were also assessed by comparing the population detached by swabbing with the figure obtained by the equation of Veulemans et al. (50). Lastly, given the role played in the attachment process by the physicochemical properties of the surfaces involved (6), we examined those of both biofilms and beef, to see whether there might be a link between these and the attachment of the biofilm microbial cells to the meat.
In this study, as well as conveyor belt materials (polyvinyl chloride [PVC] and polyurethane [PU]), we used stainless steel as a reference material because, in view of its high degree of disinfectibility and resistance to wear, chemical shock, and heat shock (27), stainless steel is widely recognized as an excellent material for the food industry. Before the materials were inoculated, they were conditioned with meat exudate so as to create conditions more like the actual conditions on open surfaces in the meat industry. As well as Listeria monocytogenes, we used bacterial genera such as Staphylococcus that are part of the microflora most commonly found in the meat industry (33) and a strain isolated in a food factory that was supposed to be a Comamonas testosteroni strain, a reclassification of Pseudomonas acidovorans (47). The food-borne pathogen L. monocytogenes was chosen because it may cause severe diseases with high mortality rates (20 to 30%) (40) and may have the ability to establish itself durably in food industry premises in spite of hygienic operations (9, 23, 28, 34, 48).
(This work forms part of G. Midelet's Ph.D. thesis research at the University of Bourgogne, Dijon, France.)
MATERIALS AND METHODS
Bacterial strains and culture conditions.
One of the strains, referred to as CCL 24 (a small gram-negative, oxidase-positive, catalase-positive rod), was difficult to identify: the closest taxon to it, according to the APILAB Plus v.3.3.3 database (bio Mérieux, Marcy l'Etoile, France), was C. testosteroni. Since the percent identification was only 34.7, we decided to name this strain Comamonas sp.
L. monocytogenes A, also referred to as CCL 128, and Comamonas sp. CCL 24 were isolated from a dairy environment, and Staphylococcus sciuri CCL 101 was isolated from the floor of the premises of a catering operation. Pseudomonas putida CCL 140 was kindly provided by M. W. Griffiths, University of Guelph, Guelph, Canada.
Cultures were maintained for no more than 1 month on tryptone soy agar (TSA) slopes (Difco, Le Pont de Claix, France) at 3°C. Slopes were inoculated from long-term cold storage.
Test surfaces.
Three materials were used (15 by 30 mm): slides of stainless steel (2 RB finish, AISI 304 British steel; Inox Industrie, Aulnay-sous-Bois, France), PVC (NONEX 2 M 1320; Ammeraal, Seclin, France), and PU (ROPANYL 2 M 1795; Ammeraal).
The slides were washed as described in an earlier report (29). The stainless steel was autoclaved, and the PU and PVC were sterilized by immersion in 300 ml of a 0.2% (vol/vol) peracetic acid solution (OXYGAL NEP; Penngar, Vaas, France) for 5 min at room temperature. The slides were rinsed in 300 ml of sterilized ultrapure water and dried in a laminar airflow hood.
Meat exudate.
Exudate from beef was obtained by thawing a frozen shoulder of beef. Ten kilograms of meat was cut into 5- by 5-cm pieces and placed in two stainless steel containers. The meat was then covered with aluminum foil, weighed down with six stainless steel plates of 1.4 kg each, and frozen at −20°C. It took 48 h at 10°C to thaw the meat, which produced 800 ml of exudate. Fourteen-milliliter portions of the exudate were kept at −20°C and used within the following 4 months. After the tubes were thawed and the meat exudate was centrifuged (40,000 × g, 15 min), the supernatant was filtered through a 0.22-μm-pore-size Stericup filter with a 1-μm-pore-size prefilter (Millipore, Saint Quentin en Yvelines, France).
The composition of the meat exudate was 0.5% nonprotein nitrogen, 10% protein, 0.2% lipid, 0.1% carbohydrate, 1.2% ashes, and 88% water. The analysis was performed by The Institut Scientifique d'Hygiène Alimentaire, Longjumeau, France, by using classical methods. Protein and nonprotein nitrogen were analyzed with the Kjedahl method, and lipid was analyzed with the ethero chloridric acid method. Ashes were weighed after incineration at 550°C. Carbohydrate content was calculated by the difference between the dry weight and the sum of the protein, lipid, and ash contents.
Biofilm development.
Refrigerated cultures were transferred to TSA slopes and incubated for 24 h at 25°C for S. sciuri and Comamonas sp., 30°C for P. putida, and 37°C for L. monocytogenes. Bacteria were washed twice by centrifugation (2,800 × g, 10 min) in 9 ml of physiological saline.
The concentrations of the suspensions were adjusted to 5 × 107 CFU ml−1 (optical density at 600 nm [OD600] = 0.15 in 1.5-cm-diameter tubes). Slides (stainless steel, PVC, or PU) were stuck in the bottoms of 50-cm-diameter petri dishes with double-sided adhesive tape (Tesa; Foto Film, Thiais, France). To prevent any dehydration of the biofilm during incubation, each 50-cm-diameter petri dish was then placed in a 120-cm-diameter petri dish containing 25 ml of water. Seven milliliters of meat exudate was deposited onto the slides, which were placed in a humidity cabinet (LHL 212 ME, Espec; Bioblock, Illkirch, France) at 25°C and 95% relative humidity (RH). The meat exudate was then eliminated, and 7 ml of the bacterial suspension was deposited onto the slides, which were kept at 25°C and 95% RH for 3 h to allow adhesion. The nonadhering bacteria were removed by pouring on 25 ml of peptone solution (pancreatic peptone) (1 g of Bacto peptone/liter; Difco) before incubation at 25°C for 20 h. Again, planktonic bacteria were removed by pouring on 50 ml of sterilized ultrapure water prior to contact angle measurements or 25 ml of peptone solution prior to the other determinations.
Swabbing and bacterial counts.
A dry swab was wiped over the biofilm once, then submerged in an Eppendorf tube containing 1 ml of sterilized peptone solution, then wiped a second time, broken, and placed in the Eppendorf tube. CFU were counted with a Spiral plater (Spiral System DS; Interscience, Saint Nom La Bretèche, France) on appropriate dilutions of the bacterial suspension. Counts were performed on TSA, which was then incubated at the optimum temperature for bacterial growth.
Contact angle measurements.
The contact angles (θ) were determined at room temperature by the sessile drop method with a goniometer (Krüss, Palaiseau, France), two polar liquids (water and formamide) (Sigma, Saint Quentin Fallanier, France), and one nonpolar liquid (di-iodomethane) (Sigma). Drops of 5 to 10 μl were deposited with a syringe. Contact angles were measured on opposite sides of each drop. Contact angles were measured on the biofilm, meat, cleaned materials, materials conditioned with meat exudate, and materials conditioned and rinsed, i.e., treated exactly as for biofilm development but with no bacterial cells. Around 40 measurements were taken for each liquid and each material or biofilm. We did not present here the calculations of free surface energy based on the contact angle values since these calculations are controversial (49).
Biofilm development in microtiter plates prior to exopolysaccharide quantification by enzyme-linked lectinsorbent assay (ELLA).
In 8 wells of a sterile 96-well microtiter plate (Luxlon; CML, Angers, France), we placed 100-μl volumes of meat exudate. The wells in the microtiter plate were incubated stationary at 25°C and 95% RH for 1 h and 15 min. The meat exudate was eliminated, and 100-μl volumes of bacterial suspension were deposited in the wells and then incubated at 25°C and 95% RH for 3 h to allow adhesion. The nonadhering bacteria were then removed by washing each well with 200 μl of peptone solution before further incubating the plate at 25°C and 95% RH for 20 h. The nonbiofilm bacteria were again removed by washing with 300 μl of sterilized ultrapure water. The remaining bacteria constituted the 1-day biofilm on which exopolysaccharide production was quantified.
Quantification by ELLA of exopolysaccharides produced by the biofilms.
The quantity of exopolysaccharides produced by the 1-day biofilms on polystyrene was assessed by using an ELLA method (30).
Two types of lectins were used: concanavalin A (ConA), which binds to d-glucose and d-mannose residues, and wheat germ agglutinin (WGA), which detects residues of N-acetyl-d-glucosamine and N-acetyl neuraminic acid (or sialic acid).
In short, peroxidase-labeled lectin solution was added to the wells colonized by the biofilm at various increasing concentrations. The microtiter plates were placed at room temperature for 1 h to allow the lectin to bind to the saccharide motifs of the biofilm exopolysaccharides. The peroxidase-labeled lectin solutions were removed. After three successive washes to eliminate unbound enzyme conjugate, the linked peroxidase conjugate was visualized after adding 2,2′-azino-bis(3-ethylbenzthiazoline-6-sulfonic acid) (ABTS) substrate solution (KPL, Gaithersburg, Md.). The reaction mixture was allowed to develop for 15 min in darkness, and absorbance was measured at 405 nm with a microplate reader (Fluostar; BMG LabTechnologies, Champigny sur Marne, France).
As with enzyme-linked immunosorbent assay techniques, the quantity of polysaccharide adsorbed to the well surfaces was estimated by plotting the OD405 against the logarithm of the concentration of the added peroxidase-labeled lectin: the more polysaccharide present, the less peroxidase-labeled lectin was needed to reach the plateau of the sigmoidal curve. A cutoff value of 0.8 was chosen to estimate the amount of exopolysaccharide produced by the 1-day biofilms.
Microscopic observations. (i) Biofilm structure.
Bacterial biofilms were stained by depositing a 5-μg/ml 4′,6′-diamidino-2-phenylindole (DAPI) (Sigma) solution on the biofilms and leaving them in darkness for 15 min. The biofilms were then rinsed with 25 ml of peptone solution and then immediately examined in a wet state under an epifluorescence microscope (magnification, ×40) (Axioskop; Zeiss, Le Pecq, France) connected to a charge-coupled device camera (Jaim 50; Adersa, Palaiseau, France) and a digital image acquisition system. With the image analysis software employed (Inducompts; Adersa), we were able to determine the number of objects (ranging from single cells to clusters). We divided the objects into five classes (surface areas of ≤44 μm2, 45 to 89 μm2, 90 to 134 μm2, 135 to 179 μm2, and ≥180 μm2) according to their surface areas. Ten fields of view were analyzed on each replicate. The field-of-view area was 7.65 × 104 μm2.
(ii) Localization of exopolysaccharides.
We used two lectins, ConA and WGA, conjugated with Alexa 488 fluorochrome. One hundred microliters of Alexa 488 ConA (Interchim; Montluçon) at a concentration of 200 μg/liter or Alexa 488 WGA (Interchim; Montluçon) at a concentration of 10 μg/ml was deposited on the 1-day biofilms, and the biofilms were left in darkness for 1 h. The biofilms were rinsed with 25 ml of sterilized ultrapure water before being examined under an epifluorescence microscope (excitation, 490 nm; emission, 520 nm) connected to a charge-coupled device camera.
Quantification of microorganisms transferred from biofilm to meat.
A piece of beef, 5 by 5 cm, topped with a 500-g weight, was placed on the biofilm and was left for 30 s. This step was repeated 12 times at the same location with a new piece of beef each time. Then each piece of beef was put in a bottle containing 10 ml of peptone solution. The bottles were vortexed for 20 s before a bacterial count was made on TSA plates. A subsequent sonication of the bottle for 4 min at 30°C in a 28-kHz, two-transducer 150-W sonication bath (Delta 220; Deltasonic, Meaux, France) did not improve the removal of bacteria from the meat (data not shown). The counts were then used, as described below, to quantify the attachment strengths of the microorganisms and the densities of the biofilm populations. The counts were also added together to determine the quantity of CFU detached by the 12 contacts.
Assessment of attachment strengths. (i) Difference between population assessments by swabbing and by the equation of Veulemans et al. (50).
Veulemans swabbing difference (VSD) was defined as the difference between the bacterial population of a biofilm as calculated by the equation established by Veulemans et al. (50) and the same population according to a count after swabbing.
(ii) Slopes of curve plotting the logarithm of detached CFU against the order number of the contact between biofilm and beef.
By plotting the logarithm of the number of CFU detached by the meat against the order number of the contacts, we obtained a two-phase curve. We used the Superfit program (12) written in Mathematica, version 4.0 (Wolfram Research, Champaign, Ill.), to estimate by nonlinear regression slopes k1 and k2 of the 2 straight lines without fixing the breakpoint. The slopes obtained were similar to those calculated with a breakpoint set at transfer number 3. We therefore used this latter method to trace two straight lines of the equation 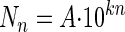 , where Nn is the number of CFU transferred, A is the original ordinate, n is the order number of the contact, and kn is either k1 (the slope of the first straight line) or k2 (the slope of the second straight line). k is a value that indicates attachment strength (17).
, where Nn is the number of CFU transferred, A is the original ordinate, n is the order number of the contact, and kn is either k1 (the slope of the first straight line) or k2 (the slope of the second straight line). k is a value that indicates attachment strength (17).
Assessment of the biofilm's bacterial population.
The number of bacteria present in the biofilm was calculated by using the formula given by Veulemans et al. (50): 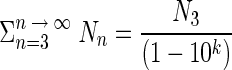 . The result of this formula was added to the number of bacteria detached by the first and second contact. This equation is explained as follows.
. The result of this formula was added to the number of bacteria detached by the first and second contact. This equation is explained as follows.
Veulemans et al. (50) and Egington et al. (17, 18) obtained a one-phase curve, and the regression analysis showed that the data fit with the general equation
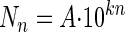 |
(1) |
which can also be written as follows:
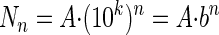 |
(2) |
where b = 10k. For the first contact
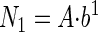 |
(3) |
so that 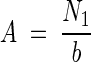 . For the nth contact, the equation becomes
. For the nth contact, the equation becomes
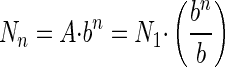 |
(4) |
which can also be written as follows:
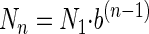 |
(5) |
so that
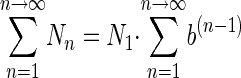 |
(6) |
but
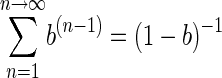 |
(7) |
according to the general binomial expression (1 + x + x2 + x3 + x4 +…) = (1 − x)−1, equation 7 becomes
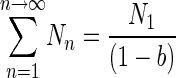 |
(8) |
In this study, this equation was only used to determine the population characterized by the second slope that was calculated with a breakpoint set at transfer number 3; therefore, the equation becomes
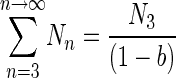 |
(9) |
and by replacing b with 10k in equation 9 we obtain
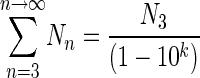 |
(10) |
Statistical analysis.
An analysis of variance was used to determine the significant differences between strains and between treatments. All calculations were performed with Statgraphics, version 3.3 (Sigma Plus, Paris, France), and statistical significance was evaluated at a P value of <0.05.
RESULTS
Contact angles.
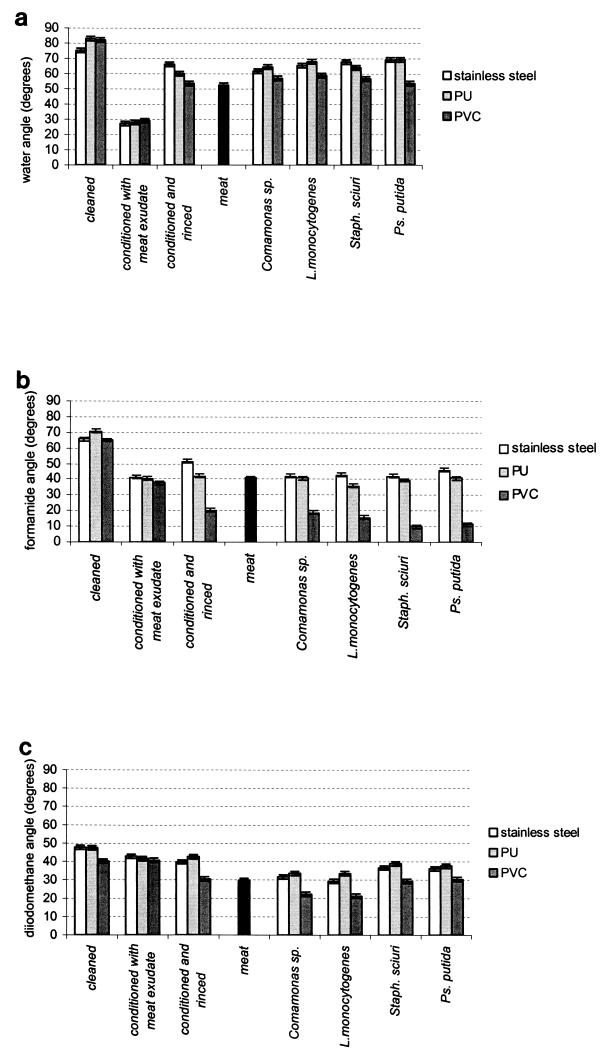
On all three materials after cleaning and sterilization, the angles of contact with water (Fig. 1a) were above 75°. These materials were therefore hydrophobic.
FIG. 1.
Contact angles measured on the surfaces of cleaned materials; conditioned materials; conditioned and rinsed materials; biofilms of Comamonas sp., L. monocytogenes, S. sciuri (Staph. sciuri), and P. putida (Ps. putida) on stainless steel, PVC, and PU; and on the surfaces of pieces of beef. Contact angles with water (a), formamide (b), and di-iodomethane (c) are shown. The bars represent the confidence intervals. The results are from two duplicate experiments and 10 measurements per experiment.
On the conditioned materials, the angles of contact with water and with formamide (Fig. 1a and b) were much smaller (<45°). The nature of the material had little effect on the contact angles (Fig. 1). The conditioning film altered the extreme surface of all three materials in almost exactly the same way: all became hydrophilic.
Of the three situations studied (cleaned materials, conditioned materials, and conditioned and rinsed materials), it was the conditioned and rinsed materials that generated surface properties closest to those of surfaces with bacterial biofilms. In all cases of conditioned and rinsed surfaces and biofilms, the angles with formamide on PVC were always smaller than those on PU and stainless steel. For conditioned and rinsed materials and biofilms of Comamonas sp. and L. monocytogenes also, the angles with di-iodomethane were significantly smaller on PVC than on PU or stainless steel.
There were observable similarities between biofilms of S. sciuri and P. putida, which both had contact angles on PVC close to 10° with formamide and close to 35° with di-iodomethane, and between biofilms of Comamonas sp. and L. monocytogenes, which both had contact angles of 20° with these two liquids. Furthermore, the Comamonas sp. and L. monocytogenes biofilms had contact angles with formamide that were more similar to those of conditioned and rinsed materials than were the biofilms of the other two strains.
Lastly, for the pieces of beef, the angles of contact with all three liquids were close to those of the conditioned and rinsed materials and biofilms.
Bacterial population and structure of biofilms.
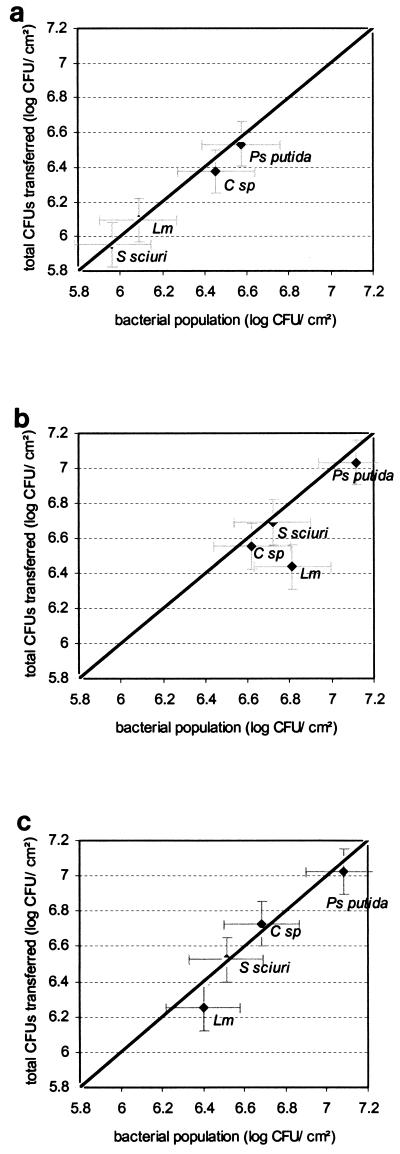
The P. putida biofilms had the largest bacterial populations (calculated with the equation of Veulemans et al. [50]) regardless of the substrate material (Fig. 2). On stainless steel only, biofilms of gram-negative bacteria had significantly larger populations than did the gram-positive biofilms. Biofilms on polymers had bacterial populations larger than those on stainless steel except for those of Comamonas sp., whose populations did not differ significantly between substrates.
FIG. 2.
Total CFU transferred to beef by 12 contacts with biofilms of Comamonas sp. (C sp), L. monocytogenes (Lm), S. sciuri, and P. putida (Ps putida) as a function of the bacterial populations of the same biofilms calculated by using the equation of Veulemans et al. (50) on stainless steel (a), on PU (b), and on PVC (c). The bars represent the confidence intervals of two duplicate experiments. The straight line is the bisector.
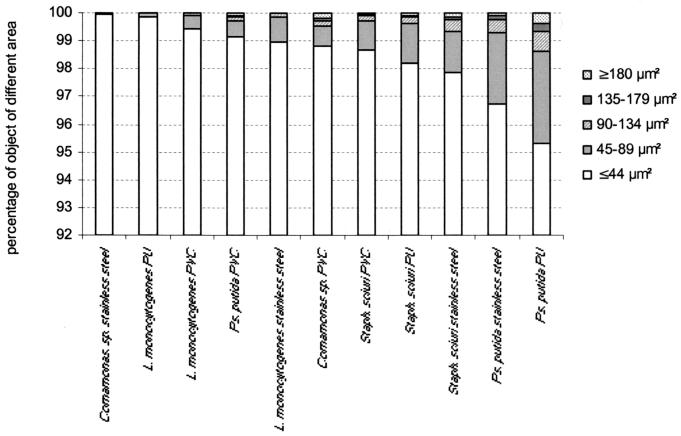
The percentage of objects >45 μm2 in size was higher in biofilms of S. sciuri and P. putida regardless of the material (with one exception, P. putida on PVC) (Fig. 3). In contrast, biofilms of Comamonas sp. and L. monocytogenes included very few objects larger than 45 μm2.
FIG. 3.
Percentage of objects in each of the five object area classes defined for analyzing the two-dimensional structure of biofilms of Comamonas sp., L. monocytogenes, S. sciuri (Staph. sciuri), and P. putida (Ps. putida) on stainless steel, PU, and PVC. The results are from two duplicate experiments.
Exopolysaccharide-producing capacity of biofilms.
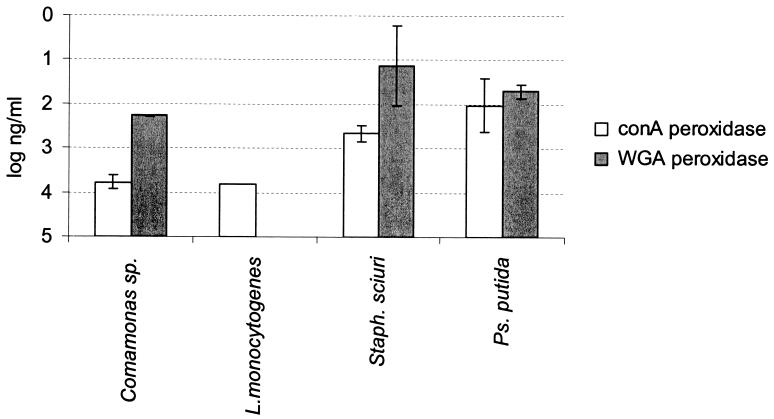
Biofilms of S. sciuri and P. putida grown on polystyrene produced more sugars detectable with peroxidase-labeled ConA (P = 0.04) and peroxidase-labeled WGA (P = 0.001) than did the biofilms of Comamonas sp. and L. monocytogenes (Fig. 4).
FIG. 4.
Comparison of the logarithm of the concentration of ConA and WGA required to achieve an OD405 of 0.8 for biofilms of Comamonas sp., L. monocytogenes, S. sciuri (Staph. sciuri), and P. putida (Ps. putida). The bars represent the individual standard deviations from one duplicate experiment. When exopolysaccharides were not detectable, the threshold value was used to perform the statistical analysis.
Characterization of attachment strengths of biofilm cells by using VSD and slopes k1 and k2.
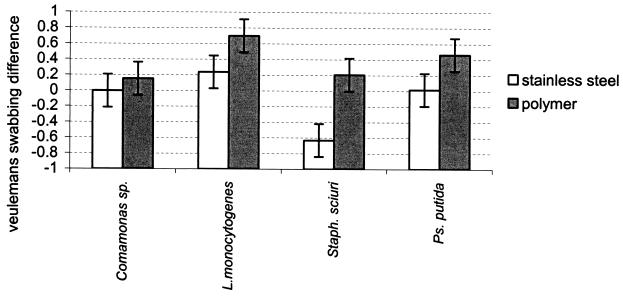
No significant differences in VSD were observed between the two polymers. However, taking the two polymers together, with three of the four strains (S. sciuri, L. monocytogenes, and P. putida), VSDs were significantly greater with polymers than with stainless steel (P = 0.002) (Fig. 5). Only biofilms of Comamonas sp. showed VSDs that did not significantly differ between the polymers and the stainless steel substrates.
FIG. 5.
Differences between the logarithm of the bacterial populations of biofilms of Comamonas sp., L. monocytogenes, S. sciuri (Staph. sciuri), and P. putida (Ps. putida) grown on stainless steel and polymers, calculated by using the equation of Veulemans et al. (50), and the logarithm of the bacterial populations of the same biofilms, calculated from a count after swabbing (VSD). The bars represent the confidence intervals from two duplicate experiments.
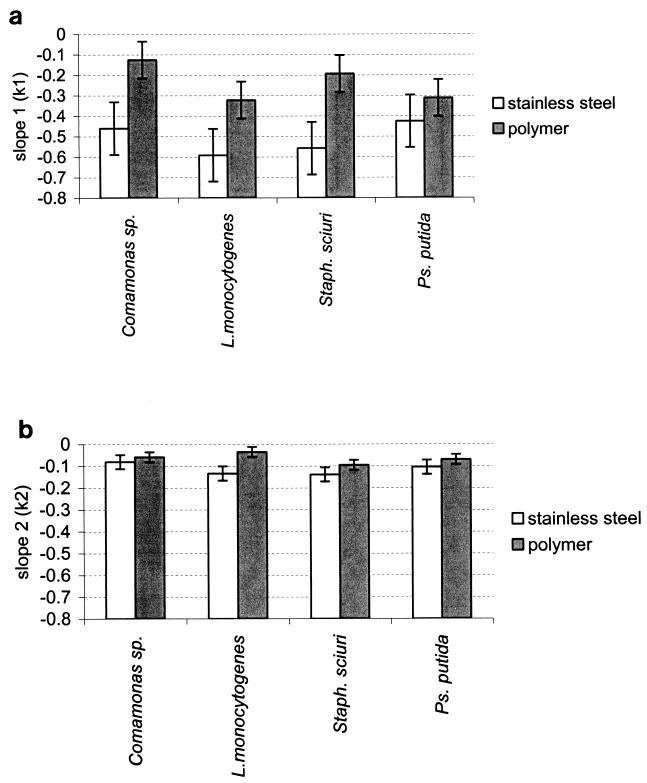
In all cases, the values of k1 and k2 were higher (low attachment strength) on stainless steel than on polymers (Fig. 6). Regarding k1, these differences were significant for all strains except P. putida, and regarding k2, these differences were significant only for L. monocytogenes.
FIG. 6.
Slopes of two-phase curves obtained by plotting the logarithm of the number of CFU transferred to beef as a function of the number of serial contacts with biofilms of Comamonas sp., L. monocytogenes, S. sciuri (Staph. sciuri), and P. putida (Ps. putida) grown on stainless steel and polymers. (a) Slope 1 (k1) characterizes the first 3 contacts, and (b) slope 2 (k2) characterizes the last contacts. The bars represent the confidence intervals from two duplicate experiments.
Regarding the nature of the strain, the mean of the VSDs obtained from L. monocytogenes biofilms on the two polymers was the greatest, though only the difference between Comamonas sp. and S. sciuri was significant. The mean of the k2 obtained from transfers from L. monocytogenes biofilms on the two polymers onto beef was the lowest (high attachment strength) of the values obtained; however, the differences with the other strains were only significant with S. sciuri.
Quantity of CFU transferred by the 12 contacts.
The total quantity of bacteria transferred onto beef by the 12 contacts was significantly higher for biofilms formed on the two polymers than for those formed on stainless steel (Fig. 2). The quantity of CFU transferred to the meat by the 12 contacts was in most cases very close to the number of bacteria in the population calculated with the Veulemans equation (the points are very close to the bisector), with the exception of the L. monocytogenes biofilms, where the difference between these two values was greatest on the polymers.
DISCUSSION
The physicochemical properties of the material surfaces were very different before and after conditioning by meat exudate. The cleaned materials studied here were hydrophobic, whereas materials that had been conditioned or conditioned and rinsed were hydrophilic. There was a marked difference in contact angles between conditioned materials and conditioned and rinsed materials. Rinsing, therefore, removed a high proportion of the meat exudate components, probably carbohydrates (20) and other hydrosoluble compounds. Among these situations, the conditioned and rinsed materials had surface properties close to those of meat and those of the biofilm surfaces. Or it is therefore fair to assume that the biofilms' surface properties, as assessed by the contact angles, mainly depended on the uncolonized part of the surface. In fact, the part of the surface not colonized by microbial cells amounts to about 80 to 95% of the surface area. In another study, using epifluorescence microscopy with lectins (ConA and WGA) marked with fluorochrome, we observed that the exopolysaccharides of P. putida and S. sciuri (the strains with the highest capacity to form exopolysaccharides) biofilms were visible only on the edges of the microbial cells (the N-acetylglucosamine of the wall of gram-positive cells, which has an affinity with WGA [44], was not detected in L. monocytogenes and therefore probably not detectable in S. sciuri, which is also a gram-positive bacterium). The contact angles on materials with biofilms were therefore more a reflection of the uncolonized surface, so they could not provide useful information about the affinity the microbial cells might have had for the meat.
We then characterized the bacterial biofilms in terms of bacterial population, capacity to produce exopolysaccharides, and structure. In biofilms on stainless steel, the bacterial population was higher with gram-negative strains than with gram-positive strains. This matches the results of Sommer et al. (45) and Wirtanen and Mattila-Sandholm (51), who used glass and stainless steel, respectively, as substrates for biofilms. On polymers, by contrast, only P. putida had a larger population than the other strains. In fact, the strong colonizing capacity of bacteria of the genus Pseudomonas is often mentioned in the literature (25, 33, 38).
Biofilms of S. sciuri and P. putida had higher exopolysaccharide-producing capacities than the other two strains. This was perhaps why the similarities between the physicochemical properties of surfaces with biofilms and those of conditioned and rinsed materials were less marked with biofilms of S. sciuri and P. putida than with biofilms of Comamonas sp. and L. monocytogenes (provided exopolysaccharide production on the different materials used was proportionate to that produced on polystyrene). Further, a higher proportion of clusters was seen among the objects making up the biofilms of S. sciuri and P. putida than in the biofilms of the other two strains. This might be due to the greater exopolysaccharide-producing capacity of these strains. A number of studies had shown that exopolysaccharides are necessary for microcolonies to form (1, 15, 32). Extracellular polymeric substances in general are known to play a part in the formation of bacterial aggregates (19), and calcium bridges can link the anionic polysaccharide chains, creating a network that traps the microbial cells (19). However, the clusters obtained in our study were not very large. This could be attributed to the very high inoculum level used to make the biofilms. Sommer et al. (45) showed with Pseudomonas sp. that a low level of adherent population on glass slides led to the formation of a small number of very large microcolonies while a high proportion of the adherent population produced a large number of small microcolonies.
However, in our study, the L. monocytogenes biofilms consisted mainly of single cells, as is usually observed with this bacterium in a pure culture (26, 43).
We then used two methods to assess the attachment strengths of the microorganisms in the biofilms. The first method compared the quantity of microorganisms detached by a nonaggressive treatment with the calculated population taken to be closest to the reality. Dewanti and Wong (16) calculated the proportion of cells detached by agitation compared to that assessed after surfaces were scraped and swabbed. Using this principle we calculated the VSD. It had been shown that swabbing alone, although relatively aggressive, did not provide a basis for determining the total population on the surface (35, 39).
The second method for assessing the attachment strength of a bacterial population was described by Eginton et al. (17). In our study it revealed two different bacterial populations. One, which was detached easily, was characterized by k1. The other, which was harder to detach, was characterized by k2. This latter population was the larger of the two (87 to 98.5% of the population calculated by the Veulemans equation). It was probably the same population studied by Eginton et al. (17), who removed the first population by very vigorous agitation just before making contacts between the biofilms and TSA. There was no correlation between the two slopes. Comamonas sp. had a k1 of −0.46 and a k2 of −0.08 on stainless steel, and the same strain had a k1 of −0.08 and a k2 of −0.05 on PU. The values of k1 also varied much more than the values of k2.
These two methods showed that attachment strengths were greater on polymers than on stainless steel and that this difference was more significant with L. monocytogenes biofilms. However, in most cases it was the density of the biofilm population that had the strongest influence on the number of cells transferred to the meat. A higher proportion of the biofilm bacteria were transferred to the meat with polymer substrates than with stainless steel (and as we have seen, biofilms on polymers had larger populations than those on stainless steel). Only the L. monocytogenes strain on polymers differed from the other strains in that the measured quantity of bacteria transferred by 12 contacts was markedly less than the biofilm population estimated by the equation of Veulemans et al. (50). This corroborates the highest attachment strength observed with this strain. Despite the similarities observed between L. monocytogenes and Comamonas sp. (physicochemical properties of surfaces colonized, low exopolysaccharide production, and low proportion of clusters), the biofilm cells had different attachment strengths. It would probably be useful to modify the structure of the biofilms by changing the culture conditions (inoculum level and proportion of glucose in the culture medium, etc.) to find out whether, for a given strain, there is a link between the biofilms' structure, their extracellular polymeric substance contents and viscoelasticity, and the attachment strengths of the cells. In this way one could make a hypothesis about cohesion, i.e., the cells' attachment to each other and their attachment to inert surfaces. Other factors that should be investigated in the future are the temperature of the surfaces and the fat content or residue of the surfaces.
Probably, given the weak attachment strengths observed on stainless steel, this material is also easier to clean than the polymers. This gives stainless steel a further advantage in addition to the qualities already mentioned. The polymers, on the other hand, are known to be materials that spoil quickly with mechanical and chemical wear and can thus shelter bacteria that resist cleaning and disinfection processes. Under the conditions we studied, it is likely that L. monocytogenes strains are harder to remove by hygienic operations than the other bacterial strains from the polymers we used (PVC and PU).
Lastly, making repeated contacts between a biofilm and meat (in our study) or agar (17, 18), despite the wide spread of the results, allows one to roughly assess the attachment strengths of cells in a biofilm and quantify the population, giving a more accurate value than can be achieved with swabbing. Thus, on the basis of the results obtained, use of the equation of Veulemans et al. gave a better assessment of the bacterial population of biofilms on polymers than did swabbing. Despite the nature of the material and the nature of the bacterial strain, both of which had an influence on attachment strength, it was the density of a biofilm's bacterial population that had the greatest influence on the quantity of CFU transferred to the meat by 12 contacts. Given that, the method used here to accurately count biofilm population may be useful for studying the contaminant capacity of the surfaces of equipment used in the food industry.
Acknowledgments
We are grateful to M. Cornu for great help in mathematics. Special thanks to D. Chassaing for technical help and to H. Coleman for the English translation.
REFERENCES
- 1.Allison, D. G., and I. W. Sutherland. 1987. The role of exopolysaccharides in adhesion of freshwater bacteria. J. Gen. Microbiol. 133:1319-1327. [Google Scholar]
- 2.Assanta, M. A., D. Roy, and D. Montpetit. 1998. Adhesion of Aeromonas hydrophila to water distribution system pipes after different contact times. J. Food Prot. 61:1321-1329. [DOI] [PubMed] [Google Scholar]
- 3.Bell, R. G. 1997. Distribution and sources of microbial contamination on beef carcasses. J. Appl. Microbiol. 82:292-300. [DOI] [PubMed] [Google Scholar]
- 4.Bizzaro, S., L. Deneuve, and J. L. Vendeuvre. 1990. Etude de la contamination microbienne des surfaces en entreprise. Viandes Prod. Carnes 11:220. [Google Scholar]
- 5.Borch, E., M. L. Kant-Muermans, and Y. Blixt. 1996. Bacterial spoilage of meat and cured meat products. Int. J. Food Microbiol. 33:103-120. [DOI] [PubMed] [Google Scholar]
- 6.Bos, R., H. C. van der Mei, and H. J. Busscher. 1999. Physico-chemistry of initial microbial adhesive interactions-its mechanisms and methods for study. FEMS Microbiol. Rev. 23:179-230. [DOI] [PubMed] [Google Scholar]
- 7.Carpentier, B., and O. Cerf. 1993. Biofilms and their consequences, with particular reference to hygiene in the food industry. J. Appl. Bacteriol. 75:499-511. [DOI] [PubMed] [Google Scholar]
- 8.Carpentier, B., A. C. L. Wong, and O. Cerf. 1998. Biofilms on dairy plant surfaces what's new? Bull. Int. Dairy Fed. 329:32-35.
- 9.Chasseignaux, E. 1999. Ecologie de Listeria monocytogenes dans les ateliers de transformation de viandes de volailles et de porcs. Ph.D. thesis. Université de Claude Bernard, Lyon, France.
- 10.Collivignarelli, C., G. Bertanza, and R. Pedrazanni. 2000. A comparison among different wastewater disinfection systems: experimental results. Environ. Technol. 21:1-6. [Google Scholar]
- 11.Cook, G. S., J. W. Costerton, and R. J. Lamont. 1998. Biofilm formation by Porphyromonas gingivalis and Streptococcus gordonii. J. Periodontal Res. 33:323-327. [DOI] [PubMed] [Google Scholar]
- 12.Cornu, M. 2000. Dynamique des populations bactériennes en cultures mixtes. Ph.D. thesis. Université de Claude Bernard, Lyon, France.
- 13.Costerton, W. J., K. J. Cheng, G. G. Gesey, T. I. Ladd, J. C. Nickel, M. Dasgupta, and T. J. Marrie. 1987. Bacterial biofilms in nature and disease. Annu. Rev. Microbiol. 41:435-464. [DOI] [PubMed] [Google Scholar]
- 14.Czechowski, M. H. 1991. Biofilms and surface sanitation in the food industry. Biodeterior. Biodegradation 8:453-454. [Google Scholar]
- 15.Danese, P. N., L. A. Pratt, and R. Kolter. 2000. Exopolysaccharide production is required for development of Escherichia coli K-12 biofilm architecture. J. Bacteriol. 182:3593-3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dewanti, R., and A. C. Wong. 1995. Influence of culture conditions on biofilm formation by Escherichia coli. Int. J. Food Microbiol. 26:147-164. [DOI] [PubMed] [Google Scholar]
- 17.Eginton, P. J., H. Gibson, J. Holah, P. S. Handley, and P. Gilbert. 1995. Quantification of the ease of removal of bacteria from surfaces. J. Ind. Microbiol. 15:305-310. [DOI] [PubMed] [Google Scholar]
- 18.Eginton, P. J., J. Holah, D. G. Allison, P. S. Handley, and P. Gilbert. 1998. Changes in the strength of attachment of microorganisms to surfaces following treatment with disinfectants and cleansing agents. Lett. Appl. Microbiol. 27:101-105. [DOI] [PubMed] [Google Scholar]
- 19.Flemming, H. C., J. Wingender, C. Mayer, V. Körstgens, and W. Borchard. 2000. Cohesiveness in biofilm matrix polymers, p. 88-105. In 59th Symposium of the Society for General Microbiology, Cambridge University Press, Cambridge, United Kingdom.
- 20.Garry, P. 1992. Propriétés physico-chimiques de surfaces en polyuréthane et conséquences sur l'encrassement et l'ahésion de Bacillus subtilis et Bacillus cereus. Ph.D. thesis. Université de Claude Bernard, Lyon, France.
- 21.Gill, C. O., and T. Jones. 2000. Microbiological sampling of carcasses by excision or swabbing. J. Food Prot. 63:167-173. [DOI] [PubMed] [Google Scholar]
- 22.Giovannacci, I., S. Queguiner, C. Ragimbeau, G. Salvat, J. L. Vendeuvre, V. Carlier, and G. Ermel. 2001. Tracing of Salmonella spp. in two pork slaughter and cutting plants using serotyping and macrorestriction genotyping. J. Appl. Microbiol. 90:131-147. [DOI] [PubMed] [Google Scholar]
- 23.Giovannacci, I., C. Ragimbeau, S. Queguiner, G. Salvat, J. L. Vendeuvre, V. Carlier, and G. Ermel. 1999. Listeria monocytogenes in pork slaughtering and cutting plants use of RAPD, PFGE and PCR-REA for tracing and molecular epidemiology. Int. J. Food Microbiol. 53:127-140. [DOI] [PubMed] [Google Scholar]
- 24.Haeghebaert, S., E. Delarocque Astagneau, V. Vaillant, A. Gallay, F. Le Querrec, and P. Bouvet. 1998. Les toxi-infections alimentaires collectives en France en 1996. Bull. Epidemiol. Hebd. 1998:36-39. [Google Scholar]
- 25.Hood, S. K., and E. A. Zottola. 1997. Adherence to stainless steel by foodborne microorganisms during growth in model food systems. Int. J. Food Microbiol. 37:145-153. [DOI] [PubMed] [Google Scholar]
- 26.Kalmokoff, M. L., J. W. Austin, X. D. Wan, G. Sanders, S. Banerjee, and J. M. Farber. 2001. Adsorption, attachment and biofilm formation among isolates of Listeria monocytogenes using model conditions. J. Appl. Microbiol. 91:725-734. [DOI] [PubMed] [Google Scholar]
- 27.Krysinski, E. P., L. J. Brown, and T. J. Marchisello. 1992. Effect of cleaners and sanitizers on Listeria monocytogenes attached to product contact surfaces. J. Food Prot. 55:246-251. [DOI] [PubMed] [Google Scholar]
- 28.Lawrence, L. M., and A. Gilmour. 1995. Characterization of Listeria monocytogenes isolated from poultry products and from the poultry-processing environment by random amplification of polymorphic DNA and multilocus enzyme electrophoresis. Appl. Environ. Microbiol. 61:2139-2144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leriche, V., and B. Carpentier. 1995. Viable but nonculturable Salmonella typhimurium within single and binary species biofilms in response to chlorine treatment. J. Food Prot. 58:1186-1191. [DOI] [PubMed] [Google Scholar]
- 30.Leriche, V., P. Sibille, and B. Carpentier. 2000. Use of an enzyme-linked lectinsorbent assay to monitor the shift in polysaccharide composition in bacterial biofilms. Appl. Environ. Microbiol. 66:1851-1856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lindsay, D., I. Geornaras, and A. Vonholy. 1996. Biofilms associated with poultry processing equipment. Microbios 86:105-116. [PubMed] [Google Scholar]
- 32.Mack, D., W. Fischer, A. Krokotsch, K. Leopold, R. Hartmann, H. Egge, and R. Laufs. 1996. The intercellular adhesin involved in biofilm accumulation of Staphylococcus epidermidis is a linear β-1,6-linked glucosaminoglycan: purification and structural analysis. J. Bacteriol. 178:175-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mettler, E., and B. Carpentier. 1998. Variations over time of microbial load and physicochemical properties of floor materials after cleaning in food industry premises. J. Food Prot. 61:57-65. [DOI] [PubMed] [Google Scholar]
- 34.Miettinen, M. K., K. J. Björkroth, and H. J. Korkeala. 1999. Characterization of Listeria monocytogenes from an ice cream plant by serotyping and pulsed field gel electrophoresis. Int. J. Food Microbiol. 46:187-192. [DOI] [PubMed] [Google Scholar]
- 35.Oulahal-lagsir, N. 2000. Procédé intégré de quantification de biofilms residuels des industries agro-alimentaires. Ph.D. thesis. University Claude Bernarol Lyon 1, Lyon, France.
- 36.Poulis, J. A., M. de Pijper, D. A. A. Mossel, and P. P. A. Dekkers. 1993. Assessment of cleaning and disinfection in the food industry with the rapid ATP-bioluminescence technique combined with the tissue fluid contamination test and a conventional microbiological method. Int. J. Food Microbiol. 20:109-116. [DOI] [PubMed] [Google Scholar]
- 37.Rahkio, M., and H. Korkeala. 1996. Microbiological contamination of carcasses related to hygiene practice and facilities on slaughtering lines. Acta Vet. Scand. 37:219-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ralyea, R. D., M. Wiedmann, and K. J. Boor. 1998. Bacterial tracking in a dairy production system using phenotypic and ribotyping methods. J. Food Prot. 61:1336-1340. [DOI] [PubMed] [Google Scholar]
- 39.Richard, J., and C. Piton. 1986. Semi-log model for interpreting the results of swabbing surfaces naturally contaminated. J. Appl. Bacteriol. 60:243-249. [DOI] [PubMed] [Google Scholar]
- 40.Rocourt, J. 1996. Risk factors for listeriosis. Food Control 7:195-202. [Google Scholar]
- 41.Ronner, A. B., and A. C. L. Wong. 1993. Biofilm development and sanitizer inactivation of Listeria monocytogenes and Salmonella typhimurium on stainless steel and Buna-n rubber. J. Food Prot. 56:750-758. [DOI] [PubMed] [Google Scholar]
- 42.Sagripanti, J. L., and A. Bonifacino. 2000. Resistance of Pseudomonas aeruginosa to liquid disinfectants on contaminated surfaces before formation of biofilms. J. AOAC Int. 83:1415-1422. [PubMed] [Google Scholar]
- 43.Sasahara, K. C., and E. A. Zottola. 1993. Biofilm formation by Listeria monocytogenes utilizes a primary colonizing microorganism in flowing systems. J. Food Prot. 56:1022-1028. [DOI] [PubMed] [Google Scholar]
- 44.Sizemore, R. K., J. J. Caldwell, and A. S. Kendrick. 1990. Alternate gram staining technique using a fluorescent lectin. Appl. Environ. Microbiol. 56:2245-2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sommer, P., C. Martin-Rouas, and E. Mettler. 1999. Influence of the adherent population level on biofilm population, structure and resistance to chlorination. Food Microbiol. 16:503-515. [Google Scholar]
- 46.Sutherland, I. W. 2001. Biofilm exopolysaccharides: a strong and sticky framework. Microbiology 147:3-9. [DOI] [PubMed] [Google Scholar]
- 47.Tamaoka, J., D.-M. Ha, and K. Komagata. 1987. Reclassification of Pseudomonas acidovorans den Dooren de Jong 1926 and Pseudomonas testosteroni Marcus and Talalay 1956 as Comamonas acidovorans comb. nov. and Comamonas testosteroni comb. nov., with an emended description of the genus Comamonas. Int. J. Syst. Bacteriol. 37:52-59. [Google Scholar]
- 48.Unnerstad, H., E. Barneman, J. Bille, M. L. Danielsson-Tham, E. Waak, and W. Tham. 1996. Prolonged contamination of a dairy with Listeria monocytogenes. Neth. Milk Dairy J. 50:493-499. [Google Scholar]
- 49.van der Mei, H. C., B. van de Belt-Gritter, G. Reid, H. Bialkowska-Hobrzanska, and H. J. Busscher. 1997. Adhesion of coagulase-negative staphylococci grouped according to physico-chemical surface properties. Microbiology 143:3861-3870. [DOI] [PubMed] [Google Scholar]
- 50.Veulemans, A., E. Jacqmain, and D. Jacqmain. 1970. Etude d'une méthode simple pour la détermination du degré de pollution des surfaces et la comparaison du pouvoir désinfectant de divers produits d'entretien. Rev. Ferment. Ind. Aliment. 25:58-65. [Google Scholar]
- 51.Wirtanen, G., and T. Mattila-Sandholm. 1993. Epifluorescence image analysis and cultivation of foodborne biofilm bacteria grown on stainless steel surfaces. J. Food Prot. 56:678-683. [DOI] [PubMed] [Google Scholar]
- 52.Yu, S. L., D. Bolton, C. Laubach, P. Kline, A. Oser, and S. A. Palumbo. 1999. Effect of dehairing operations on microbiological quality of swine carcasses. J. Food Prot. 62:1478-1481. [DOI] [PubMed] [Google Scholar]