Abstract
Many plant-associated bacteria synthesize the phytohormone indoleacetic acid (IAA). While IAA produced by phytopathogenic bacteria, mainly by the indoleacetamide pathway, has been implicated in the induction of plant tumors, it is not clear whether IAA synthesized by beneficial bacteria, usually via the indolepyruvic acid pathway, is involved in plant growth promotion. To determine whether bacterial IAA enhances root development in host plants, the ipdc gene that encodes indolepyruvate decarboxylase, a key enzyme in the indolepyruvic acid pathway, was isolated from the plant growth-promoting bacterium Pseudomonas putida GR12-2 and an IAA-deficient mutant constructed by insertional mutagenesis. The canola seedling primary roots from seeds treated with wild-type P. putida GR12-2 were on average 35 to 50% longer than the roots from seeds treated with the IAA-deficient mutant and the roots from uninoculated seeds. In addition, exposing mung bean cuttings to high levels of IAA by soaking them in a suspension of the wild-type strain stimulated the formation of many, very small, adventitious roots. Formation of fewer roots was stimulated by treatment with the IAA-deficient mutant. These results suggest that bacterial IAA plays a major role in the development of the host plant root system.
Bacteria that inhabit the rhizosphere may influence plant growth by contributing to a host plant's endogenous pool of phytohormones, such as auxins. Production of the auxin indoleacetic acid (IAA) is widespread among plant-associated bacteria (36). In phytopathogenic bacteria, such as Agrobacterium tumefaciens and pathovars of Pseudomonas syringae, IAA is produced from tryptophan via the intermediate indoleacetamide and has been implicated in the induction of plant tumors. Beneficial bacteria synthesize IAA predominantly by an alternate tryptophan-dependant pathway, through indolepyruvic acid; however, the role of bacterial IAA in plant growth promotion remains undetermined.
Promotion of root growth is one of the major markers by which the beneficial effect of plant growth-promoting bacteria is measured (15). Rapid establishment of roots, whether by elongation of primary roots or by proliferation of lateral and adventitious roots, is advantageous for young seedlings as it increases their ability to anchor themselves to the soil and to obtain water and nutrients from their environment, thus enhancing their chances for survival. Most root-promoting bacteria synthesize IAA, and their effect on plants mimics that of exogenous IAA.
Plants generally grow one or more primary roots from which lateral roots emerge by division of specific pericycle cells. Adventitious roots are a type of lateral root that arise from nonroot tissue, such as the tissue at the base of the stem or on cuttings. Whereas lateral and adventitious roots are induced by high concentrations of exogenous IAA, a feature exploited in horticulture by applying natural and synthetic auxins, primary root growth is stimulated by application of relatively low levels of IAA, typically between 10−9 and 10−12 M (2, 35, 39), and is inhibited by higher IAA concentrations, likely via auxin-induced ethylene (37).
Two different approaches have been taken to test for a similar trend in the effect of bacterial IAA on plant growth. In one method the effects of inoculating roots with bacterial mutants that produce different levels of IAA are compared. In the second approach the size of the inoculum of a single strain is varied; the rationale for this approach is that a higher inoculum density means that more IAA is available to the plant. While demonstrating that bacterial mutants that overproduce IAA have a root growth-inhibiting effect has been relatively straightforward (30, 42, 49), establishing a direct relationship between enhanced root growth and bacterial IAA has proven to be more elusive, given the difficulty of isolating bacterial mutants that are completely deficient in IAA synthesis (1, 8, 29, 31).
The rhizobacterium Pseudomonas putida GR12-2 is a strong candidate for development as a soil inoculant to enhance crop yields. Inoculation of canola, tomato, and other agriculturally important plants with this strain results in substantial promotion of seedling root growth (7, 18). Characteristics that may contribute to the ability of P. putida GR12-2 to enhance plant growth include the capacity to synthesize siderophores and thereby provide iron for the plant (7), the capacity to lower growth-inhibiting levels of ethylene in plant tissues by production of 1-aminocyclopropane-1-carboxylic acid (ACC) deaminase (16, 21), and the capacity to secrete IAA (49).
To determine if IAA is involved in the stimulation of root growth by P. putida GR12-2, the ipdc gene encoding indolepyruvate decarboxylase, which catalyzes a key step in the indolepyruvic acid pathway for IAA synthesis, was isolated, and an IAA-deficient mutant was constructed by insertional mutagenesis of ipdc. Changes in root development as a consequence of root interactions with this mutant were documented and compared to the effects of the wild-type bacterium.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
P. putida GR12-2 (28) was kindly provided by G. Brown of Agrium, Inc. (Saskatoon, Saskatchewan, Canada) and was routinely grown at 27°C in 3.6% tryptic soy broth (TSB) (Difco). The DF salts minimal medium of Dworkin and Foster (12) was also used for propagation of P. putida GR12-2 as indicated below. Escherichia coli DH5α (19) was grown at 37°C in 2% Luria-Bertani (LB) broth (Difco).
Isolation of the ipdc gene by PCR.
The following PCR primers were designed from the previously published Enterobacter cloacae FERM BP-1529 ipdc gene sequence (24) to span the entire open reading frame: forward primer 5′ GAAGGATCCCTGTTATGCGAACC 3′ and reverse primer 5′ CTGGGGATCCGACAAGTAATCAGGC 3′ (MOBIX, McMaster University, Hamilton, Ontario, Canada). A BamHI restriction site (underlined) was incorporated into the 5′ end of both the forward and reverse primers in order to facilitate subsequent subcloning of PCR products. These primers were used to amplify the ipdc gene from both lysed cells of P. putida GR12-2 and purified P. putida GR12-2 genomic DNA. PCR products were sequenced (MOBIX, McMaster University), and the sequence was analyzed by using the algorithms BLAST (National Center for Biotechnology Information [http://www.ncbi.nlm.nih.gov]) and ClustalW (EMBL [http://www.ebi.ac.uk]).
Marker exchange mutagenesis.
The ipdc sequence was subcloned into the suicide vector pJQ200 (40) and then disrupted by insertion of a gene for kanamycin resistance carried on a 2.3-kb EcoRI fragment from pHP45Ω-Km (14) into the unique PmlI site roughly in the middle of ipdc. The resultant plasmid, pJQIPDC-Kan, was first introduced into E. coli S17.1 (45) competent cells and selected for by resistance to gentamicin (30 μg/ml) and kanamycin (50 μg/ml), and then it was transferred to P. putida GR12-2 by filter mating. Transconjugants were identified by growth on Simmons citrate agar supplemented with kanamycin and then on DF salts minimal medium agar plus kanamycin for verification. To select for transconjugants in which the functional ipdc gene in the chromosome was replaced by the disrupted ipdc gene from the plasmid by double crossover between homologous ipdc sequences, colonies resistant to the lethal effects of the vector-encoded sacB gene product in the presence of sucrose were identified on tryptic soy agar containing kanamycin and 5% (wt/vol) sucrose. In addition, these colonies were plated on tryptic soy agar supplemented with gentamicin; lack of growth confirmed the absence of gentamicin acetyltransferase also encoded on the vector. Insertion of the kanamycin resistance gene into the chromosomal ipdc gene was verified by PCR and by Southern hybridization.
Quantification of IAA production.
Wild-type and IAA-deficient P. putida GR12-2 were propagated overnight in 5 ml of DF salts minimal media, and then 20-μl aliquots were transferred into 5 ml of DF salts minimal media supplemented with the following concentrations of l-tryptophan (from a filter-sterilized 2-mg/ml stock prepared in warm water; Sigma): 0, 50, 100, 200, and 500 μg/ml. After incubation for 42 h, the density of each culture was measured spectrophotometrically at 600 nm, and then the bacterial cells were removed from the culture medium by centrifugation (5,500 × g, 10 min). A 1-ml aliquot of the supernatant was mixed vigorously with 4 ml of Salkowski's reagent (150 ml of concentrated H2SO4, 250 ml of distilled H2O, 7.5 ml of 0.5 M FeCl3·6H2O [17]) and allowed to stand at room temperature for 20 min before the absorbance at 535 nm was measured. The concentration of IAA in each culture medium was determined by comparison with a standard curve.
To confirm that the mutant strain does not produce IAA when high concentrations of tryptophan are present, the IAA levels in the supernatants from wild-type and mutant cultures grown in the DF salts minimal medium containing 500 μg of l-tryptophan per ml were measured by high-performance liquid chromatography (HPLC). Filtered supernatant was analyzed by using a Hewlett-Packard model 1100 HPLC equipped with an Ultrasphere reverse-phase C18 column (5 μm; 4.6 by 150 mm); the mobile phase was acetonitrile-50 mM KH2PO4 (pH 3) (30/70) at a flow rate of 1 ml/min. Eluates were detected at 220 nm, and IAA was quantified by integrating the areas under the peaks; authentic IAA (Sigma) was used as a standard. The IAA produced by each strain was measured in triplicate.
Root elongation assay.
Cultures of wild-type and IAA-deficient P. putida GR12-2 were grown overnight from single colonies in 5 ml of DF salts minimal media without and with kanamycin, respectively. After approximately 24 h, 20 μl of each culture was transferred to 5 ml of DF salts minimal medium supplemented with tryptophan (200 μg/ml) to induce IAA production and incubated for an additional 42 h. The turbidities of the cultures were measured spectrophotometrically at 600 nm before the bacterial cells were separated from the culture medium by centrifugation. Each supernatant was immediately assayed to determine the IAA concentration by Salkowski's assay as described above. Cells were washed twice in 5 ml of sterile 0.03 M MgSO4, and the final suspension was adjusted to an absorbance at 600 nm of 0.5 with 0.03 M MgSO4.
Canola seeds (Hyola 401), kindly provided by J. Omielan (University of Guelph, Guelph, Ontario, Canada), were prepared and inoculated by the method outlined by Lifshitz et al. (28), with some modifications (7). Approximately 300 seeds, previously stored in a desiccator at 4°C, were surface sterilized by soaking them in 10 ml of 70% ethanol for 1 min and then in 10 ml of 1% sodium hypochlorite (bleach) for 10 min. To remove the residual bleach, the seeds were washed five times with sterile distilled water. For each treatment, approximately 100 seeds were transferred aseptically to sterile polystyrene petri dishes and incubated with 5 ml of either the wild-type or mutant bacterial suspension at room temperature for 1 h to allow the bacteria to bind to the seed coat and for seed imbibition. Seeds were also incubated in 5 ml of 0.03 M MgSO4 under the same conditions as a control.
Six seeds were aseptically placed in each growth pouch (Mega International, Minneapolis, Minn.), which had been previously filled with 10 ml of distilled water and autoclaved. For each treatment, 10 pouches were prepared. The pouches were placed upright in metal racks, with one treatment per rack to prevent cross-contamination, and there were two empty pouches at each end, of each rack. The racks were set in a plastic bin containing about 3 cm of deionized water and covered loosely with plastic wrap to prevent dehydration. The pouches were incubated in a growth chamber at 20°C with a daily cycle consisting of 12 h of darkness followed by 12 h of light (18 μmol of photons/m2/s). For the first 2 days, the seeds were kept in the dark by covering the pouches with aluminum foil. After 5 days, shoot and primary root lengths were measured and analyzed by two-way analysis of variance (F values are indicated below). Seeds that had failed to germinate 3 days after they were sown were marked, and shoots and roots that subsequently developed from these seeds were not measured.
Colonization assay.
To determine if the IAA-deficient mutant of P. putida GR12-2 is impaired in the ability to colonize roots, canola seeds were inoculated with either wild-type P. putida GR12-2 or the IAA-deficient mutant of P. putida GR12-2 or with a 1:1 mixture of the wild-type and mutant strains. Bacteria and seeds were prepared, inoculated, and incubated as described above for the root elongation assay. After 7 days, bacteria were extracted from six roots for each treatment (two roots from each of three growth pouches) by briefly rinsing the roots in sterile water and then suspending the roots in 1 ml of phosphate buffer (25 mM NaH2PO4, 25 mM Na2HPO4; pH 7.0) and shaking the preparations vigorously for 1 h at 4°C. Dilutions were prepared in phosphate buffer and spread onto LB agar to determine total bacterial counts and onto LB agar containing kanamycin (50 μg/ml) to determine mutant counts.
Rooting assay.
The effects of wild-type P. putida GR12-2 and the IAA-deficient mutant of P. putida GR12-2 on the development of adventitious roots on mung bean cuttings were assessed by using the method outlined by Mayak et al. (33). Mung bean (Vigna radiata) seeds were surface sterilized in a manner similar to that used for canola seeds as described above. Seeds were imbibed for 1 h in sterile distilled water, sown in sterile vermiculite, and then incubated in a growth chamber as described above. After 7 days, the portions of the seedlings above the surface of the vermiculite were excised with a razor blade and placed immediately in either water or a bacterial suspension prepared as follows.
Five milliliters of DF salts minimal medium was inoculated with the wild-type or the IAA-deficient mutant of P. putida GR12-2 and incubated overnight. Eighty microliters of each overnight culture was transferred to 20 ml of DF salts minimal medium containing tryptophan (200 μg/ml) and incubated for an additional 42 h. Cells were washed twice with sterile distilled water and resuspended in sterile water to an optical density at 600 nm (OD600) of 0.5. Three milliliters of each bacterial preparation was transferred to each of 10 borosilicate glass tubes (10 by 75 mm), and 10 additional tubes were filled with 3 ml of sterile distilled water. One mung bean cutting was placed in each tube (a total of 10 cuttings per treatment), and the tubes were placed in a rack, covered loosely with plastic wrap to prevent evaporation, and incubated in the growth chamber under the conditions described above. After 8 days, the cuttings were rinsed briefly with distilled water, and the numbers and lengths of adventitious roots were measured by using a magnifying glass; the data were analyzed by two-way analysis of variance (F values are indicated below).
Nucleotide sequence accession number.
The sequence of the P. putida GR12-2 ipdc gene reported in this paper has been deposited in the GenBank database under accession number AF285632.
RESULTS
Isolation of the ipdc gene.
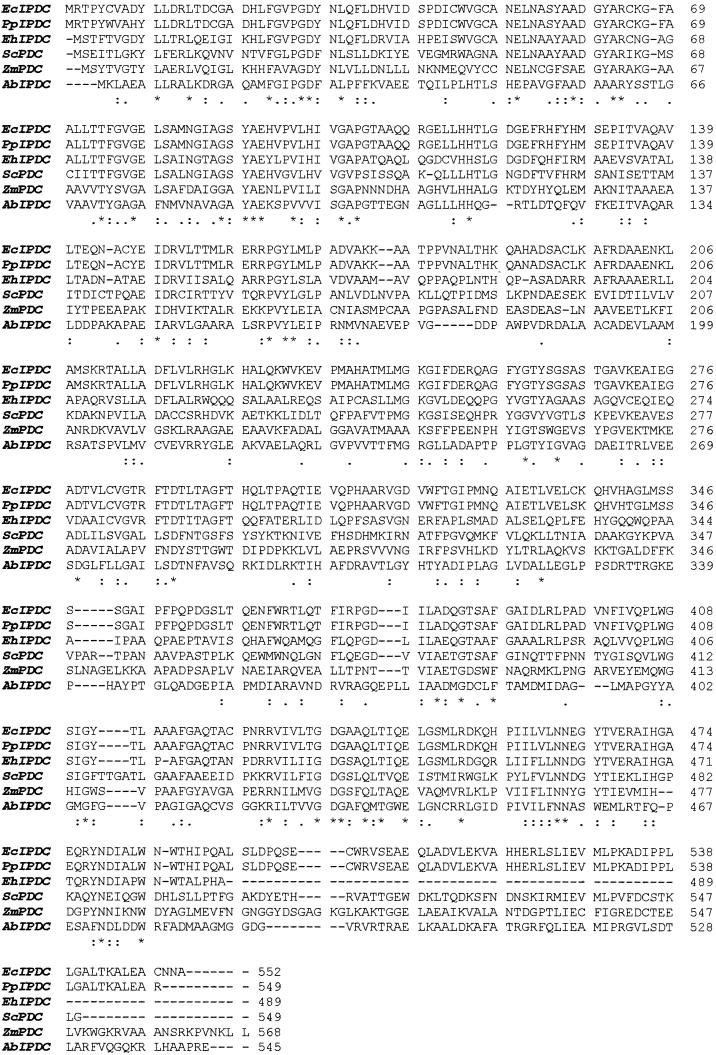
Hybridization of a P. putida GR12-2 genomic library (prepared by ligation of Sau3AI-digested fragments of total DNA with pUC19 and transformation of E. coli DH5α) with the indolepyruvate decarboxylase (ipdc) gene from E. cloacae FERM BP-1529 (24) revealed a single colony carrying 200 bp of the ipdc sequence. The sequence was 98% identical at the nucleotide level to the 3′ end of the E. cloacae FERM BP-1529 ipdc gene. The region downstream of the ipdc gene contains a putative transcription termination signal, identified by the presence of sequences capable of forming a stable stem-loop structure in the mRNA and a sequence 72% identical at the nucleotide level to an open reading frame in E. coli of unknown function (accession no. AE000327) on the opposite strand. Working on the assumption that the high degree of identity between the 3′ ends of the P. putida GR12-2 and E. cloacae FERM BP-1529 ipdc sequences could be extended to the entire gene, we designed PCR primers from the previously published E. cloacae FERM BP-1529 sequence to amplify the entire open reading frame. Electrophoresis of the PCR products obtained by using P. putida GR12-2 genomic DNA or cell lysates as templates revealed a single band of the expected size, about 1.7 kb. Sequence analysis of the PCR products and alignment of the nucleotide sequence with the ipdc gene from E. cloacae FERM BP-1529 confirmed that the P. putida GR12-2 ipdc gene had indeed been isolated. Furthermore, the translated amino acid sequence is similar to the amino acid sequences of other known bacterial indolepyruvate decarboxylases (Fig. 1).
FIG. 1.
Alignment of indolepyruvate decarboxylase (IPDC) amino acid sequences from P. putida GR12-2 (Pp), E. cloacae FERM BP-1529 (Ec), E. herbicola 299R (Eh), and A. brasilense Sp245 (Ab) and pyruvate decarboxylase (PDC) amino acid sequences from Z. mobilis (Zm) and S. cerevisiae (Sc). Asterisks indicate identical amino acids; colons and periods indicate strongly and weakly conserved amino acids, respectively.
The P. putida GR12-2 indolepyruvate decarboxylase is also similar to pyruvate decarboxylases from Zymomonas mobilis and Saccharomyces cerevisiae, exhibiting substantial levels of identity (33 and 36%, respectively) and similarity (51 and 53%, respectively) in the amino acid sequence (Fig. 1). Four of the six residues believed to be involved in substrate binding and catalysis of pyruvate decarboxylase are conserved in the indolepyruvate decarboxylase sequence; residues Asp29, His115, His116, and Glu468 of indolepyruvate decarboxylase from P. putida GR12-2 correspond to residues Asp27, His113, His114, and Glu473 in the active site of Z. mobilis pyruvate decarboxylase (11) (Fig. 1). In addition, most of the residues known to bind the cofactors Mg2+ and thiamine diphosphate in pyruvate decarboxylase are conserved in indolepyruvate decarboxylase; these residues include Glu52, Gly408, Asp435, Asn462, and Gly464.
Construction of an IAA-deficient mutant.
Vector pJQ200 (40) was used to deliver the ipdc sequence, disrupted by insertion of a gene for kanamycin resistance, into the genome of P. putida GR12-2. This plasmid has an origin of transfer (oriT) and mob genes from plasmid RP4, enabling transfer of the vector from E. coli S17.1 (45) into P. putida GR12-2 via conjugation. However, once in P. putida GR12-2, the vector cannot replicate because it has an origin of replication derived from pACYC184 that is functional only in enterobacteria (40). Thus, following transfer of the vector to P. putida GR12-2, kanamycin-resistant cells can arise only if the kanamycin resistance gene is inserted into the ipdc gene in the genome by homologous recombination between the ipdc sequences on the plasmid and in the chromosome. In addition, because gentamicin acetyltransferase and SacB are encoded on the vector, selection for the absence of these traits (that is, selection for sensitivity to gentamicin and resistance to the lethal effects of SacB in the presence of sucrose) selects against the incorporation of the entire plasmid into the genome that would result from a single crossover event.
Following the transfer of pJQIPDC-Kan from E. coli S17.1 to P. putida GR12-2 by conjugation, transconjugants were initially selected on Simmons citrate agar (on which E. coli donor cells cannot grow) containing kanamycin (on which nontransformed P. putida GR12-2 cells cannot grow). Several colonies were picked and subcultured on DF salts minimal agar (on which E. coli cannot grow) containing kanamycin to confirm that they were indeed derived from P. putida GR12-2. Growth on TSB agar containing kanamycin and 5% sucrose and the lack of growth on TSB agar containing gentamicin indicated that the kanamycin resistance gene, but not the remainder of the plasmid, had been inserted into the chromosome in all selected transconjugants. Replacement of the functional ipdc gene in the chromosome of P. putida GR12-2 with the ipdc gene disrupted by the kanamycin resistance gene from pJQIPDC-Kan was confirmed by PCR by using primers designed to amplify the ipdc gene and whole-cell lysates of transconjugants and wild-type P. putida GR12-2 as templates. The PCR products from transconjugants were 2.3 kb larger, corresponding to the size of the kanamycin resistance gene fragment, than the PCR products from the wild-type strain (data not shown); the PCR products were confirmed to contain the ipdc sequence by Southern hybridization (data not shown). In addition, Southern hybridization confirmed the presence of a larger EcoRI fragment carrying the ipdc gene in the chromosome of the mutant strain than in the chromosome of the wild-type bacterium (data not shown).
Quantification of IAA production.
In the absence of tryptophan supplements, both the IAA-deficient mutant and wild-type P. putida GR12-2 produced very low levels of IAA (Table 1). However, when both strains were grown in the presence of 50 μg of tryptophan per ml for approximately 42 h, wild-type P. putida GR12-2 responded by producing higher levels of IAA, while the mutant was not capable of producing significant amounts of IAA (Table 1). As the concentration of tryptophan in the growth medium was increased, IAA production by the wild-type strain increased. In contrast, IAA production by the mutant strain remained very low. Measurement of IAA by HPLC confirmed that in the presence of a high concentration of tryptophan (500 μg/ml), the wild-type bacterium produced high levels of IAA (68.3 ± 2.2 μg/ml/OD600 unit), while IAA was not detected in the medium from mutant cells (<1 μg/ml/OD600 unit). The apparent slight increase in IAA concentration in the medium in mutant cultures containing 500 μg of tryptophan per ml, as measured colorimetrically by reaction with Salkowski's reagent (Table 1), may have been due to accumulation of indolepyruvic acid, which can also react with Salkowski's reagent, albeit to a much lesser extent than IAA (data not shown). Indolepyruvic acid is the product of catalysis of tryptophan by tryptophan transaminase, the first step in the IAA biosynthetic pathway, and is the substrate for indolepyruvate decarboxylase, which is not functional in the mutant. Growth of the mutant and wild-type P. putida GR12-2 was not affected by the addition of high levels of tryptophan to the medium (data not shown).
TABLE 1.
Production of IAA by wild-type and IAA-deficient mutant of P. putida GR12-2 in the presence of various concentrations of tryptophan
| Tryptophan concn (μg/ml) | IAA production (μg/ml/OD600 unit)a
|
|
|---|---|---|
| Wild type | Mutant | |
| 0 | 0.5 ± 0.1 | 0.5 ± 0.0 |
| 50 | 14.5 ± 0.5 | 0.5 ± 0.2 |
| 100 | 22.5 ± 2.0 | 0.8 ± 0.3 |
| 200 | 26.2 ± 0.3 | 1.0 ± 0.4 |
| 500 | 32.7 ± 2.9 | 2.0 ± 0.3 |
Average ± standard error from three separate experiments.
Analysis of root elongation.
IAA produced by P. putida GR12-2 has a significant impact on the ability of this bacterium to stimulate the growth of the primary roots of canola seedlings. Whereas the roots from seeds treated with wild-type P. putida GR12-2 were on average 35% longer than the roots from uninoculated control seeds after 5 days (P < 0.01; F1/8 = 30.1), the lengths of roots from seeds treated with the IAA-deficient mutant were not different from the lengths of roots from uninoculated control seeds (P < 0.01; F1/8 = 4.3) (Table 2). IAA produced by the wild-type strain had no effect on shoot length, as the lengths of shoots from seeds inoculated with wild-type P. putida GR12-2 were not different from the lengths of shoots from uninoculated seeds (F1/4 = 5.1) (Table 2). Thus, as expected, eliminating IAA production did not affect shoot length (F1/4 = 3.9).
TABLE 2.
Lengths of roots and shoots from canola seeds treated with wild-type or IAA-deficient mutant of P. putida GR12-2
| Bacterial treatment | Root length (mm)a | Shoot length (mm)b |
|---|---|---|
| None | 54.5 ± 2.8 | 35.2 ± 1.0 |
| Wild type | 74.2 ± 4.4 | 32.4 ± 2.1 |
| IAA mutant | 61.9 ± 1.9 | 32.8 ± 0.9 |
Average ± standard error from five separate experiments.
Average ± standard error from three separate experiments.
The loss of the capacity to synthesize IAA did not reduce the ability of IAA-deficient P. putida GR12-2 to colonize canola seedling roots. The population size attained by the mutant bacterium on the surface of roots 7 days after seed inoculation was similar to that attained by the wild-type strain when it was inoculated alone (Table 3). In addition, the relative population sizes were maintained on the roots when the seeds were coinoculated with equal amounts of the two strains (Table 3).
TABLE 3.
Colonization of wild-type and IAA-deficient P. putida GR12-2 on canola roots 7 days after seed inoculation
| Inoculum density (108 CFU/ml) | Inoculum ratio | Recovered population (106 CFU/root)a
|
Recovered ratio | ||
|---|---|---|---|---|---|
| Wild type | Mutant | Wild type | Mutant | ||
| 14.1 | 1:0 | 3.0 ± 0.5 | 1:0 | ||
| 12.8 | 0:1 | 3.2 ± 0.6 | 0:1 | ||
| 4.3 | 5.4 | 1:1.3 | 2.0 ± 0.2 | 2.3 ± 0.4 | 1:1.1 |
Mean ± standard error for bacteria recovered from six roots.
Rooting assay.
Mung bean cuttings that were excised above the roots after 7 days of growth in vermiculite and placed in either water or a bacterial suspension had visible roots at the base of the stem after 5 days. After 8 days, the cuttings in water had a few long roots (on average, about seven 3.4-mm-long roots growing just above the base) (Table 4). More than three times as many adventitious roots developed on cuttings placed in a suspension of wild-type P. putida GR12-2. Most of these roots were very small (less than 1 mm long) and were distributed over several centimeters up from the base of the stem; sometimes there were a few longer roots right at the base. Roots that developed in the suspension of the IAA-deficient mutant of P. putida GR12-2 were both abundant and long; this is likely the best situation for propagation of cuttings in the long term. Twice as many roots were present on these cuttings as on the cuttings growing in water, and the roots were generally longer than those that developed in the wild-type bacterial suspension (Table 4).
TABLE 4.
Effects of wild-type and IAA-deficient mutant of P. putida GR12-2 on the number and length of adventitious roots on mung bean cuttingsa
| Treatment | No. of roots/cutting | Root length (mm) |
|---|---|---|
| Water | 6.8 ± 1.0 | 3.4 ± 0.2 |
| IAA mutant | 13.8 ± 1.9 | 2.6 ± 0.1 |
| Wild type | 20.3 ± 2.2 | 1.6 ± 0.1 |
Similar results were obtained in replicate experiments. The values are means ± standard errors for 10 cuttings.
DISCUSSION
The amino acid sequence determined from the ipdc gene isolated from P. putida GR12-2 reveals a 552-amino-acid protein with a predicted molecular weight of approximately 60,000. This protein is not transcribed from an operon containing the other genes involved in the biosynthesis of IAA by the indolepyruvic acid pathway as it is transcribed from its own promoter (unpublished data) and has a transcription termination sequence just downstream of the translation stop codon. It is reasonable to conclude that the enzymes involved in the indolepyruvic acid pathway are not expressed from an operon because multiple copies of the gene encoding the first enzyme in the pathway, an aromatic aminotransferase, are often present in a single bacterium, and the enzyme prefers amino acid substrates other than tryptophan (22, 23, 46). Thus, this aromatic aminotransferase is not solely an IAA biosynthesis enzyme.
The sequence of indolepyruvate decarboxylase from P. putida GR12-2 is similar to the sequence of indolepyruvate decarboxylase from E. cloacae FERM BP-1529 (99% identity), which was isolated from the rhizosphere of cucumber (24), and to the sequence of the same protein from Erwinia herbicola 299R (57% identity and 71% similarity), an epiphytic bacterium isolated from pear (5). The sequences of the indolepyruvate decarboxylases from two Azospirillum brasilense strains, Sp245 and Sp7 (10, 50), are somewhat different from the sequences of these enzymes from the bacteria mentioned above, including P. putida GR12-2 (29% identity and 44% similarity), although identified conserved regions are present.
To understand how microbial IAA influences plant growth, mutants with significantly reduced levels of IAA have been generated for the phytopathogens P. syringae (9, 34), A. tumefaciens (29), and E. herbicola pv. gypsophilae (8) and for the plant growth-promoting bacterium Azosprillum lipoferum (1). Insertional mutagenesis of the ipdc gene was used here to successfully generate a mutant of P. putida GR12-2 that is deficient in IAA production even in the presence of tryptophan, conditions under which the wild-type strain produces copious amounts of IAA.
The loss of the ability to produce IAA following disruption of the ipdc gene confirms that P. putida GR12-2 produces IAA via the indolepyruvic acid pathway. This provides more support for the hypothesis (36) that plant growth-promoting bacteria, such as Azospirillum spp. (10) and E. cloacae (24), produce IAA via the indolepyruvic acid pathway, in contrast to plant pathogens, which seem to preferentially synthesize IAA via the indoleacetamide pathway (25, 48). Indeed, rendering the ipdc gene inactive by insertional mutagenesis and thereby eliminating IAA production by this pathway significantly reduces the ability of P. putida GR12-2 to promote primary root growth in canola seedlings. It is known from application of exogenous IAA (13, 47) or application of diluted culture extracts or low-density inocula of bacteria that produce high levels of IAA (20, 43) that low concentrations of IAA can stimulate primary root elongation; here we demonstrate directly that bacterial IAA plays a major role in promotion of root elongation when a bacterium is associated with its host plant.
IAA secreted by a bacterium may promote root growth directly by stimulating plant cell elongation or cell division or indirectly by influencing bacterial ACC deaminase activity. ACC deaminase, produced by many plant growth-promoting bacteria (16, 44), including P. putida GR12-2 (21), is involved in the stimulation of root elongation in seedlings (27). ACC deaminase hydrolyzes plant ACC, the immediate precursor of the phytohormone ethylene, and thereby prevents the production of plant growth-inhibiting levels of ethylene (38). Mutants of plant growth-promoting bacteria that do not produce ACC deaminase have lost the ability to stimulate root elongation (27). It is possible that IAA and ACC deaminase work in concert to stimulate root elongation. Exogenous IAA is known to increase transcription and activity of ACC synthase (37), which catalyzes the production of ACC in plants. ACC stimulates ACC deaminase activity in bacteria (21, 26).
The reduced ability of IAA-deficient P. putida GR12-2 to promote canola root growth cannot be attributed to defective colonization. Although Brandl and Lindow (6) showed that an IAA mutant of E. herbicola 299R was less competitive than the wild-type strain for colonization of bean leaves and pear flowers, the P. putida GR12-2 IAA mutant was able to colonize canola roots to the same extent as the wild-type strain following seed inoculation.
While low levels of IAA stimulate root elongation, high levels of bacterial IAA, whether from IAA-overproducing mutants or strains that naturally secrete high levels or from high-density inocula, stimulate the formation of lateral and adventitious roots (3, 4, 30, 33, 42, 49). P. putida GR12-2 cells that produce wild-type levels of IAA stimulated the formation of many short adventitious roots on mung bean cuttings, and an IAA-overproducing mutant stimulated the formation of even more adventitious roots than the wild-type strain (33). In contrast, the IAA-deficient mutant of P. putida GR12-2 stimulated the formation of fewer roots than the wild-type bacterium, and the roots were generally longer than those induced by the wild-type strain.
Initiation of adventitious and lateral roots may be mediated by IAA-induced ethylene. An ACC deaminase-negative mutant of P. putida GR12-2 which cannot reduce ethylene levels in plants stimulated the formation of more small adventitious roots than the wild-type strain (33). The increase in the number of roots on the cuttings correlated with an increase in ethylene production. The IAA-deficient mutant of P. putida GR12-2 may not stimulate the formation of ACC synthase and therefore ethylene synthesis in plants; thus, fewer adventitious roots were initiated on the cuttings.
High levels of exogenous or bacterial IAA, and therefore high levels of ethylene, have also been shown to inhibit elongation growth in roots (4, 30, 41, 42, 49). Thus, while adventitious roots that formed on mung bean cuttings inoculated with wild-type P. putida GR12-2 were very short (most were less than 1 mm long), the roots whose formation was stimulated by the IAA-deficient mutant strain were longer. The cuttings were exposed to a high inoculum density continuously for an extended period of time; therefore, cuttings treated with the wild-type bacterium were exposed to a high level of IAA.
From a practical point of view, treatment of cuttings with an IAA-deficient mutant may be a beneficial method for propagation of plants. Certainly, while many adventitious roots are desirable, longer roots with more surface area through which the plants can absorb nutrients and water from the soil would be advantageous. Treatment with the IAA-deficient mutant of P. putida GR12-2 provides just such a compromise between the many short roots whose formation is stimulated by the wild-type strain and the few long roots produced by treatment with water.
Production of IAA, a plant hormone that does not apparently function as a hormone in bacterial cells, may have evolved in bacteria because it is important in the bacterium-plant relationship. In this research we showed that bacterial IAA stimulates the development of the host plant root system. The advantage for root-associated bacteria is a rich supply of nutrients, as much of the metabolic products of the carbon fixed by plants is lost from roots and moves into the rhizosphere as exudates, lysates, and mucilage (32).
Acknowledgments
We thank Alberto Mendoza Herrera, Centro de Biotecnología Genómica, Reynosa, Tampico, Mexico, for HPLC measurements of IAA.
This work was supported by grants to B.R.G. and by a scholarship to C.L.P. from the Natural Sciences and Engineering Research Council of Canada and by an Ontario Graduate Scholarship in Science and Technology to C.L.P.
REFERENCES
- 1.Abdel-Salam, M. S., and W. Klingmüller. 1987. Transposon Tn5 mutagenesis in Azosprillum lipoferum: isolation of indoleacetic acid mutants. Mol. Gen. Genet. 210:165-170. [Google Scholar]
- 2.Alvarez, R., S. J. Nissen, and E. G. Sutter. 1989. Relationship between indole-3-acetic acid levels in apple (Malus pumila Mill.) rootstocks cultured in vitro and adventitious root formation in the presence of indole-3-butyric acid. Plant Physiol. 89:439-443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barbieri, P., and E. Galli. 1993. Effect on wheat root development of inoculation with an Azospirillum brasilense mutant with altered indole-3-acetic acid production. Res. Microbiol. 144:69-75. [DOI] [PubMed] [Google Scholar]
- 4.Beyerler, M., P. Michaux, C. Keel, and D. Haas. 1997. Effect of enhanced production of indole-3-acetic acid by the biological control agent Pseudomonas fluorescens CHA0 on plant growth, p. 310-312. In A. Ogoshi, K. Kobayashi, Y. Homma, F. Kodama, N. Kondo, and S. Akino (ed.), Plant growth-promoting rhizobacteria: present status and future prospects. OECD, Paris, France.
- 5.Brandl, M. T., and S. E. Lindow. 1996. Cloning and characterization of a locus encoding an indolepyruvate decarboxylase involved in indole-3-acetic acid synthesis in Erwinia herbicola. Appl. Environ. Microbiol. 62:4121-4128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brandl, M. T., and S. E. Lindow. 1998. Contribution of indole-3-acetic acid production to the epiphytic fitness of Erwinia herbicola. Appl. Environ. Microbiol. 64:3256-3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Caron, M., C. L. Patten, S. Ghosh, and B. R. Glick. 1995. Effects of the plant growth promoting rhizobacterium Pseudomonas putida GR12-2 on the physiology of canola roots. Plant Growth Regul. Soc. Am. Q. 23:297-302. [Google Scholar]
- 8.Clark, E., S. Manulis, Y. Ophir, I. Barash, and Y. Gafni. 1993. Cloning and characterization of iaaM and iaaH from Erwinia herbicola pathovar gypsophilae. Phytopathology 83:234-240. [Google Scholar]
- 9.Comai, L., and T. Kosuge. 1980. Involvement of plasmid deoxyribonucleic acid in indoleacetic acid synthesis in Pseudomonas savastanoi. J. Bacteriol. 143:950-957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Costacurta, A., V. Keijers, and J. Vanderleyden. 1994. Molecular cloning and sequence analysis of an Azospirillum brasilense indole-3-pyruvate decarboxylase gene. Mol. Gen. Genet. 243:463-472. [DOI] [PubMed] [Google Scholar]
- 11.Dobritzsch, D., S. König, G. Schneider, and G. Lu. 1998. High resolution crystal structure of pyruvate decarboxylase from Zymomonas mobilis. J. Biol. Chem. 273:20196-21204. [DOI] [PubMed] [Google Scholar]
- 12.Dworkin, M., and J. W. Foster. 1958. Experiments with some microorganisms which utilize ethane and hydrogen. J. Bacteriol. 75:592-601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Evans, M. L., H. Ishikawa, and M. A. Estelle. 1994. Responses of Arabidopsis roots to auxin studied with high temporal resolution: comparison of wild type and auxin-response mutants. Planta 194:215-222. [Google Scholar]
- 14.Fellay, R., J. Frey, and H. Krisch. 1987. Interposon mutagenesis of soil and water bacteria: a family of DNA fragments designed for in vitro insertional mutagenesis of Gram-negative bacteria. Gene 52:147-154. [DOI] [PubMed] [Google Scholar]
- 15.Glick, B. R., D. M. Karaturovic, and P. C. Newell. 1995. A novel procedure for rapid isolation of plant growth promoting pseudomonads. Can. J. Microbiol. 41:533-536. [Google Scholar]
- 16.Glick, B. R., D. M. Penrose, and J. Li. 1998. A model for the lowering of plant ethylene concentrations by plant growth-promoting bacteria. J. Theor. Biol. 190:63-68. [DOI] [PubMed] [Google Scholar]
- 17.Gordon, S. A., and R. P. Weber. 1951. Colorimetric estimation of indoleacetic acid. Plant Physiol. 26:192-195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hall, J. A., D. Peirson, S. Ghosh, and B. R. Glick. 1996. Root elongation in various agronomic crops by the plant growth promoting rhizobacterium Pseudomonas putida GR12-2. Isr. J. Plant Sci. 44:37-42. [Google Scholar]
- 19.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557-575. [DOI] [PubMed] [Google Scholar]
- 20.Harari, A., J. Kige, and Y. Okon. 1988. Involvement of IAA in the interaction between Azospirillum brasilense and Panicum miliaceum roots. Plant Soil 110:275-282. [Google Scholar]
- 21.Jacobson, C. B., J. J. Pasternak, and B. R. Glick. 1994. Partial purification and characterization of ACC deaminase from the plant growth-promoting rhizobacterium Pseudomonas putida GR12-2. Can. J. Microbiol. 40:1019-1025. [Google Scholar]
- 22.Kittel, B. L., D. R. Helinski, and G. S. Ditta. 1989. Aromatic aminotransferase activity and indoleacetic acid production in Rhizobium meliloti. J. Bacteriol. 171:5458-5466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koga, J., K. Syono, T. Ichikawa, and T. Adachi. 1994. Involvement of l-tryptophan aminotransferase in indole-3-acetic acid biosynthesis in Enterobacter cloacae. Biochim. Biophys. Acta 1209:241-247. [DOI] [PubMed] [Google Scholar]
- 24.Koga, J., T. Adachi, and H. Hidaka. 1991. Molecular cloning of the gene for indolepyruvate decarboxylase from Enterobacter cloacae. Mol. Gen. Genet. 226:10-16. [DOI] [PubMed] [Google Scholar]
- 25.Kuo, T., and T. Kosuge. 1970. Role of aminotransferase and indole-3-pyruvic acid in the synthesis of indole-3-acetic acid in Pseudomonas savastanoi. J. Gen. Appl. Microbiol. 16:191-204. [Google Scholar]
- 26.Li, J., and B. R. Glick. 2001. Transcriptional regulation of the Enterobacter cloacae UW 1-aminocyclopropane-1-carboxylate (ACC) deaminase gene (acdS). Can. J. Microbiol. 47:359-367. [DOI] [PubMed] [Google Scholar]
- 27.Li, J., D. H. Ovakim, T. C. Charles, and B. R. Glick. 2000. An ACC deaminase minus mutant of Enterobacter cloacae UW4 no longer promotes root elongation. Curr. Microbiol. 41:101-105. [DOI] [PubMed] [Google Scholar]
- 28.Lifshitz, R., J. W. Kloepper, M. Kozlowski, C. Simonson, J. Carlson, E. M. Tipping, and I. Zaleska. 1987. Growth promotion of canola (rapeseed) seedlings by a strain of Pseudomonas putida under gnotobiotic conditions. Can. J. Microbiol. 33:390-395. [Google Scholar]
- 29.Liu, S.-T., K. L. Perry, C. L. Schardl, and C. I. Kado. 1982. Agrobacterium Ti plasmid indoleacetic acid gene is required for crown gall oncogenesis. Proc. Natl. Acad. Sci. USA 79:2812-2816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Loper, J. E., and M. N. Schroth. 1986. Influence of bacterial sources of indole-3-acetic acid on root elongation of sugar beet. Phytopathology 76:386-389. [Google Scholar]
- 31.Manulis, S., L. Valinski, Y. Gafni, and J. Hershenhorn. 1991. Indole-3-acetic acid biosynthetic pathways in Erwinia herbicola in relation to pathogenicity on Gypsophila paniculata. Physiol. Mol. Plant Pathol. 39:161-171. [Google Scholar]
- 32.Martens, D. A., and W. T. Frankenberger. 1994. Assimilation of exogenous 2′-14C-indole-3-acetic acid and 3′-14C-tryptophan exposed to the roots of three wheat varieties. Plant Soil 166:281-290. [Google Scholar]
- 33.Mayak, S., T. Tirosh, and B. R. Glick. 1997. The influence of plant growth promoting rhizobacterium Pseudomonas putida GR12-2 on the rooting of mung bean cuttings, p. 313-315. In A. Ogoshi, K. Kobayashi, Y. Homma, F. Kodama, N. Kondo, and S. Akino (ed.), Plant growth-promoting rhizobacteria: present status and future prospects. OECD, Paris, France.
- 34.Mazzola, M., and F. F. White. 1994. A mutation in the indole-3-acetic acid biosynthesis pathway of Pseudomonas syringae pv. syringae affects growth in Phaseolus vulgaris and syringomycin production. J. Bacteriol. 176:1374-1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meuwley, P., and P.-E. Pilet. 1991. Local treatment with indole-3-acetic acid induces differential growth responses in Zea mays L. roots. Planta 185:58-64. [DOI] [PubMed] [Google Scholar]
- 36.Patten, C. L., and B. R. Glick. 1996. Bacterial biosynthesis of indole-3-acetic acid. Can. J. Microbiol. 42:207-220. [DOI] [PubMed] [Google Scholar]
- 37.Peck, S. C., and H. Kende. 1995. Sequential induction of the ethylene biosynthetic enzymes by indole-3-acetic acid in etiolated peas. Plant Mol. Biol. 28:293-301. [DOI] [PubMed] [Google Scholar]
- 38.Penrose, D. M., B. A. Moffat, and B. R. Glick. 2001. Determination of 1-aminocyclopropane-1-carboxylic acid (ACC) to assess the effects of ACC deaminase-containing bacteria on roots of canola seedlings. Can. J. Microbiol. 47:77-80. [DOI] [PubMed] [Google Scholar]
- 39.Pilet, P.-E., and M. Saugy. 1987. Effect on root growth of endogenous and applied IAA and ABA. Plant Physiol. 83:33-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Quandt, J., and M. F. Hynes. 1993. Versatile suicide vectors which allow direct selection for gene replacement in Gram-negative bacteria. Gene 127:15-21. [DOI] [PubMed] [Google Scholar]
- 41.Rahman, A., T. Amakawa, N. Goto, and S. Tsurumi. 2001. Auxin is a positive regulator for ethylene-mediated response in the growth of Arabidopsis roots. Plant Cell Physiol. 42:301-307. [DOI] [PubMed] [Google Scholar]
- 42.Sawar, M., and R. J. Kremmer. 1995. Enhanced suppression of plant growth through production of l-tryptophan compounds by deleterious rhizobacteria. Plant Soil 172:261-269. [Google Scholar]
- 43.Selvadurai, E. L., A. E. Brown, and J. T. G. Hamilton. 1991. Production of indole-3-acetic acid analogues by strains of Bacillus cereus in relation to their influence on seedling development. Soil Biol. Biochem. 23:401-403. [Google Scholar]
- 44.Shah, S., J. Li, B. A. Moffatt, and B. R. Glick. 1998. Isolation and characterization of ACC deaminase genes from two different plant growth-promoting rhizobacteria. Can. J. Microbiol. 44:833-843. [PubMed] [Google Scholar]
- 45.Simon, R., U. Priefer, and A. Pühler. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram-negative bacteria. Bio/Technology 1:784-791. [Google Scholar]
- 46.Soto-Urzua, L., Y. G. Xochinua-Corona, M. Flores-Encarnaciaon, and B. E. Baca. 1996. Purification and properties of aromatic amino acid aminotransferases from Azospirillum brasilense UAP 14 strain. Can. J. Microbiol. 42:294-298. [DOI] [PubMed] [Google Scholar]
- 47.Thimann, K. V., and R. H. Lane. 1938. After-effects of treatment of seed with auxin. Am. J. Bot. 25:535-543. [Google Scholar]
- 48.White, F. F., and S. F. Ziegler. 1991. Cloning of the genes for indoleacetic acid synthesis from Pseudomonas syringae pv. syringae. Mol. Plant-Microbe Interact. 4:207-210. [Google Scholar]
- 49.Xie, H., J. J. Pasternak, and B. R. Glick. 1996. Isolation and characterization of mutants of the plant growth-promoting rhizobacterium Pseudomonas putida GR12-2 that overproduce indoleacetic acid. Curr. Microbiol. 32:67-71. [Google Scholar]
- 50.Zimmer, W., M. Wesche, and L. Timmermans. 1998. Identification and isolation of the indole-3-pyruvate decarboxylase gene from Azospirillum brasilense Sp7: sequencing and functional analysis of the gene locus. Curr. Microbiol. 36:327-331. [DOI] [PubMed] [Google Scholar]