Abstract
Burkholderia cepacia complex (Bcc) bacteria reside in soil, plant rhizospheres, and water, but their prevalence and distribution in outdoor environments is not clear. We sampled a variety of soil and rhizosphere environments with which people may have contact: playgrounds, athletic fields, parks, hiking trails, residential yards, and gardens. A total of 91 sites was sampled in three large U.S. cities. In the first phase of the study, putative Bcc isolates were recovered on Burkholderia cepacia selective agar and trypan blue tetracycline medium and subsequently examined for biochemical reactivity and growth at 32 and 22°C. Isolates were further examined by PCR assays targeting Bcc-specific ribosomal DNA and recA gene sequences. Among the 1,013 bacterial isolates examined, 68 were identified as Bcc; 14 (15%) of 91 sampled sites yielded Bcc isolates. In the second phase, DNA was extracted directly from soil samples and examined with PCR assays targeting Bcc 16S rRNA gene sequences. Either 82 or 93% of the soil samples were positive for at least one Bcc genomovar, depending on the PCR assay system used. Cloning and sequencing were performed to check the specificity of the PCR assays. Sequence analysis of the 463-bp 16S rRNA inserts from eight clones indicated that all were from members of the Bcc. The four soil samples from which these clones were generated did not yield isolates identified as Bcc. Based on PCR detection, Bcc appears to be prevalent in soil from urban and suburban environments. Culture-based recovery of Bcc may underestimate environmental populations.
The Burkholderia cepacia complex (Bcc) has emerged in recent years as an important human opportunistic pathogen, particularly for people with cystic fibrosis (14, 15, 26). It also holds promise as an agent of biocontrol of many plant pathogens (7, 8, 20) and as a bioremediation organism for the degradation of a wide range of recalcitrant compounds (21, 23). Although several attempts have been made to distinguish between benign and human pathogenic strains of Bcc, clear demarcations between environmentally useful and clinically significant strains have not been found (11, 28, 34, 47, 49, 50). Moreover, a small but steady number of Bcc infections in cystic fibrosis patients each year occur from strains that have not been previously encountered in the clinical setting (26). These strains are presumed to come from the natural environment, further blurring the lines between natural, beneficial, and potentially hazardous Bcc strains.
The taxonomy of Bcc has undergone considerable change in the past several years. It is now known that bacteria previously considered to be B. cepacia are in fact nine distinct genomospecies (or genomovars). These include B. cepacia genomovar I, B. multivorans, B. cepacia genomovar III, B. stabilis, B. vietnamiensis, B. cepacia genomovar VI, B. ambifaria, B. anthina, and B. pyrrocinia (9, 42, 43).
To date, the Bcc has been investigated by medical and environmental microbiologists by different methods, including the development of different selective media (16, 19). Perhaps not surprisingly, estimations of Bcc populations from these studies have varied greatly: agricultural researchers have found substantial populations of Bcc in soil and rhizosphere environments (16, 22, 33), but medical researchers have isolated Bcc from the natural environment only rarely (6, 32). The recent taxonomic changes to the Bcc, coupled with the inherent difficulty in identifying this organism, have compounded the difficulties in communicating across disciplines about the presence of Bcc in natural environments.
The conflicting reports of the prevalence of Bcc in soil have become a huge source of controversy as researchers struggle to regulate the use of Bcc as an agent of biocontrol or bioremediation. If, as some have proposed, Bcc is rarely encountered in soils, then deliberately adding any strain of Bcc to soil may well constitute an unacceptable risk to vulnerable people. If, however, Bcc is commonly found in soils, the risk posed by augmenting existing populations through agricultural and engineering applications may be negligible.
Essential to the process of determining the occurrence of Bcc in soils was the use of the protocols mutually acceptable to the medical and environmental microbiology communities, including rigorous identification procedures that reflect the most current taxonomy. The objective of the present study was to determine the environmental prevalence of the Bcc. We identified the presence of Bcc in soil samples by a variety of both culture-based and growth-independent methods to determine whether Bcc is present in soil environments with which people may have frequent contact.
MATERIALS AND METHODS
Sampling.
Sample locations (n = 91) were chosen to represent a wide range of soil microbiological habitats within the urban and suburban areas of Philadelphia, Pa.; Cleveland, Ohio; and Portland, Oreg. Soil and rhizosphere samples were taken from places where people commonly contact soil, such as playgrounds, gardens, and golf courses (Table 1). A clean hand trowel was surface sterilized with 10% bleach solution (0.5% sodium hypochlorite), and soil was placed in a sterile plastic bag and kept cold (48).
TABLE 1.
Sample types and results of culture-based and growth-independent efforts to identify Bcc
| Sample type | No. of samples | No. PCR positive in one of the Bcc-specific assays/total no. (%) | No. of samples from which Bcc was isolated on selective media/total no. (%) | No. of Bcc isolates from TBT plates | No. of Bcc isolates from BCSA plates | Total no. of Bcc isolates |
|---|---|---|---|---|---|---|
| Grass or turf | 25 | 25/25 (100) | 7/25 (28) | 24 | 13 | 37 |
| Amended soil (compost, potting soil, or heavily enriched gardens) | 22 | 21/22 (95) | 1/22 (4.5) | 0 | 1 | 1 |
| Nonamended soil (forest floor, bare ground, paths, animal burrows) | 24 | 23/23a (100) | 3/24 (12.5) | 17 | 7 | 24 |
| Pond or creekside mud | 6 | 5/5a (100) | 2/6 (33) | 3 | 2 | 5 |
| Soil clinging to vegetables | 2 | 0/0a | 1/2 (50) | 1 | 0 | 1 |
| Nonsoil environments (sand, bark or sawdust mulch, or horse arena dust) | 12 | 7/12 (58) | 0/12 (0) | 0 | 0 | 0 |
| Total | 91 | 81/87 (93) | 14/91 (15.4) | 45 | 23 | 68 |
Four samples from the 91 sites contained inadequate soil for the DNA extraction procedure; PCRs were consequently not possible. A total of 87 samples were evaluated by PCR.
Soil samples were examined within 72 h of collection. Precautions were taken to ensure that soil samples were not contaminated during handling in the lab. The soil was mixed, and several grams were immediately frozen at −20°C for later use in the direct extraction of DNA and examination of the extracts with PCR. Soil water content was determined gravimetrically on 20 g of soil dried at 105°C for 48 h. For culturing, ca. 1 g of soil was placed in a preweighed tube containing 10 ml of sterile 0.1 M MgSO4 buffer. Soil suspensions were sonicated for 2 min with an ultrasonic cleaner (Model ME 4.6; Mettler Electronics Corp., Anaheim, Calif.) to dislodge bacteria from soil particles. Serial dilutions were plated onto two media selective for Bcc: trypan blue tetracycline agar (TBT) (16) and B. cepacia selective agar (BCSA) (19), both amended with 50 μg of nystatin/ml to inhibit fungal growth. Plates were incubated at room temperature (20 to 22°C) for 4 days (BCSA) or 5 days (TBT).
Soil samples containing plant roots were often divided into “bulk soil” and “rhizosphere” components. In the case of rhizosphere samples, plant roots were removed from the soil and shaken to dislodge any loosely adhering soil. The root was then cut into 1- to 4-cm lengths, placed in a preweighed dilution blank and sonicated and plated as described above. The root segments were then blotted dry and weighed.
Soil samples were also obtained from two vegetables purchased at a farm stand. The vegetables (beets and lima bean pods) and adhering soil were placed in sterile plastic bags. After transport to the lab, 15 ml of sterile buffer was added to each of the bags to dislodge the soil, and serial dilutions were made directly out of the bag.
Isolation of bacteria.
The number of bacteria in each major morphology type (colony color, size, and habit) was noted. Representatives of each recorded morphology type were isolated on the same media from whence they came. Isolated colonies were then grown in nonselective broth culture (Luria-Bertani [LB] medium, King’s medium B broth [KB], and tryptic soy broth) and stored at −80°C in 7% dimethyl sulfoxide until further analysis. For transport between laboratories, isolates were grown from frozen stock in 5 ml of nonselective media. Sterile transport swabs (BBL CultureSwab Plus; Becton Dickinson, Sparks, Md.) were swirled in the broth to inoculate and mailed by overnight courier. Bacteria were recovered from transport swabs by culturing onto nonselective (Mueller-Hinton [MH]) agar.
Identification of isolates.
Growth and morphology on MH agar were recorded after 24 to 48 h of incubation at both room temperature (RT; 20 to 22°C) and at 32°C. Bacteria were subcultured from MH agar onto BCSA and checked again for growth after 24 to 48 h of incubation at RT and at 32°C. Bacteria from MH agar were stored in LB broth with 15% glycerol at −80°C.
All isolates were tested for oxidase reactivity by using 1% tetramethyl p-phenylenediamine dihydrochloride. All isolates testing positive with a screening PCR assay (below) were further assessed for reactivity with lysine decarboxylase, o-nitrophenyl-β-d-galactoside (ONPG), and for oxidation-fermentation of sucrose and lactose (Remel, Lenexa, Kans.) as described previously (31). A subset of isolates was further tested by using the RapID NF Plus kit (Remel) according to the manufacturer's instructions.
All isolates were examined with a screening boil-lysis PCR assay that amplifies DNA from all species within the genus Burkholderia and the closely related genera Ralstonia and Pandoraea (27). In brief, a loopful of bacteria was recovered from MH agar and suspended in 500 to 1,000 μl of UV-irradiated sterile water in a 1.5-ml centrifuge tube and pelleted by centrifugation at 5,000 rpm for 5 min. The supernatant was decanted, and the pellet was resuspended in water, heated at 100°C for 20 min, and allowed to cool to room temperature. After centrifugation, 5 μl of supernatant was used as a template in a PCR with 16S ribosomal DNA (rDNA)-directed primers RHG-F (5′-GGGATTCATTTCCTTAGTAAC-3′) and RHG-R (5′-GCGATTACTAGCGATTCCAGC-3′) described previously (27). PCR assays included a water blank as a negative control; universal bacterial primers were used as a test for the amplificability of the DNA in each assay. In extensive previous testing by using known Bcc-positive and -negative control samples, this assay yielded occasional false positives when boil-lysis bacterial preparations were used as a template; however, no false negatives were detected (data not shown). Thus, DNA was purified from all boil-lysis PCR-positive isolates by using the Easy-DNA kit (Invitrogen, Carlsbad, Calif.) with modifications described previously (27), and the assay was repeated. Isolates remaining PCR-positive were assessed for biochemical reactivity (as described above) and underwent further PCR testing employing 16S rDNA- and recA-directed Bcc-specific primers as described previously (27, 29).
Extraction of DNA from soil samples.
Frozen aliquots of all soil samples were used to provide DNA for a series of PCR assays. DNA was extracted by using the Bio 101 FastDNA SPIN Kit for Soil (Q-Biogene, Carlsbad, Calif.). Soil samples were extracted according to the manufacturer's instructions with the recommended modifications for greater yield. Samples were extracted in duplicate. Every other extraction included a blank, in which 300 μl of DNA-free water was substituted for soil, and a spiked sample. Two types of spiked samples were prepared; some were made by adding Bcc cells to autoclaved soil, and some were made by adding Bcc cells to field soil. DNA was quantified by using a DNA fluorometer (Model TKO 100; Hoefer Scientific Instruments, San Francisco, Calif.) and Hoechst 3357 bisbenzamide dye. A standard of calf thymus DNA (Sigma Chemical Co., St. Louis, Mo.) at 25 ng/μl was used to calibrate the fluorometer each time it was used. Extracted, purified DNA was standardized to a concentration of 25 ng/μl and stored in water at −20°C until use.
Soil PCR assays.
A variety of previously described (2, 27) 16S rDNA-directed PCR assays were used to ascertain whether Bcc DNA was present in soil samples. Soil-extracted DNA samples were first tested with an assay designed to amplify bacterial DNA (27). Samples testing negative were retested by using more or less DNA; all negative samples were assayed at least twice. If positive, samples were tested with PCR with primers RHG-F and RHG-R (described above), which amplify DNA from all species within the genera Burkholderia, Ralstonia, and Pandoraea. Samples testing positive were further assessed with PCR assays that amplified subgroups of genomovars within the Bcc (2, 27).
The sensitivity and specificity of all primer pairs were ascertained by using DNA extracted from a set of 35 known strains, representing Bcc genomovars I to VII, as well as several other bacterial species (Table 2). Strains were grown from frozen stock overnight in 5 ml of LB broth (strain AMMD was grown in 5 ml of KB). Cells were centrifuged for 15 min at 14,000 × g and then resuspended in 100 μl of water. DNA was extracted from the pellet by using the Bio 101 FastDNA kit according to the manufacturer's instructions by using the Cell Lysis Solution TC. DNA was quantified and stored as described above.
TABLE 2.
Strains used to test sensitivity and specificity of PCR primer pairs
| Strain | Other names | LMG accession no.a | Origin (source, country)b | Genomovar |
|---|---|---|---|---|
| Burkholderia spp. | ||||
| cep 31 | ATCC 25416T | LMG 1222T | Onion, United States | I |
| cep 80 | ATCC 17759 | LMG 2161 | Soil, Trinidad | I |
| FC 461 | LMG 17997 | UTI, Belgium | I | |
| cep 509 | LMG 18821 | CF, Australia | I | |
| cep 144 | ATCC 17616 | LMG 17588 | Soil, United States | II |
| FC 445 | LMG 13010T | CF, Belgium | II | |
| FC 769 | CP-A1-1 | LMG 18825 | CF-e, United Kingdom | II |
| cep781 | C1576 | LMG 16660 | CF-e, United Kingdom | II |
| c5393 | LMG 18822 | CF, Canada | II | |
| cep 24 | PC 184 | LMG 18829 | CF-e, United States | III |
| FC 475 | BC7 | LMG 18826 | CF-e, Canada | III |
| FC 505 | K56-2 | LMG 18863 | CF-e, Canada | III |
| FC 511 | LMG 18830 | CF-e, Australia | III | |
| cep 565 | J2315 | LMG 16656 | CF-e, United Kingdom | III |
| c5424 | LMG 18827 | CF-e, Canada | III | |
| c6433 | LMG 18828 | CF-e, Canada | III | |
| FC 367 | LMG 14294 | CF, Belgium | IV | |
| FC 472 | LMG 14086 | Respirator, United Kingdom | IV | |
| FC 779 | LMG 18888 | Clinical, Belgium | IV | |
| c7322 | LMG 18870 | CF, Canada | IV | |
| cep 40 | PC 259 | LMG 18835 | CF, United States | V |
| FC 369 | LMG 10929T | Rice, Vietnam | V | |
| FC 441 | LMG 18836 | CGD, Canada | V | |
| Bcc 232 | CF | VI | ||
| Bcc 305 | CF | VI | ||
| AMMD | LMG 19182T | Pea rhizosphere, United States | VII | |
| Bcc 118 | CF, United States | VII | ||
| Bcc 267 | CF, Australia | VII | ||
| Burkholderia gladioli | LMG 2216 | |||
| Burkholderia caribensis | LMG 18531T | |||
| Ralstonia pickettii | LMG 5942T | |||
| Pseudomonas aeruginosa | ||||
| Pseudomonas stutzeri | ||||
| Serratia marcescens | ||||
| Achromobacter cycloclastes |
LMG, Laboratorium Microbiologie Ghent Culture Collection, Universiteit Ghent, Ghent, Belgium.
Abbreviations: CF, cystic fibrosis infection; CF-e, strain that has spread epidemically among patients with CF; CGD, infection of chronic granulomatous disease patient; UTI, urinary tract infection.
PCR assays were performed with 50-μl reaction mixtures containing 1× PCR buffer (Promega Corp., Madison, Wis.), 0.06% bovine serum albumin, 2 mM MgCl2 (Promega), each deoxynucleoside triphosphate at a concentration of 0.20 mM (Promega), the forward and reverse primers (each at a concentration of 0.20 mM), and 2 U of Taq DNA polymerase (Promega). The only exceptions were assays with primer pair PC-SSF-PC-SSR (described below; see Table 3), which used a cocktail containing 1.5 mM MgCl2. The amount of DNA per assay was 50 ng per reaction vessel in reactions with pure cultures and varied from 50 to 250 ng per reaction vessel in reactions with DNA extracted from soil. Typically, the reaction was first run with 150 ng of DNA; other amounts were tried if the initial run was negative.
TABLE 3.
Primer pairs used in 16S rDNA soil PCR assays
| Reference | Primer pair | Targeta | Annealing temp (°C) |
|---|---|---|---|
| LiPuma et al. (27) | UFPL, URPL | Kingdom Bacteria | 55 |
| RHG-F, RHG-R | Members of the Burkholderia, Pandoraea, and Ralstonia genera | 55 | |
| BC-GII, BC-R | Genomovar II | 54 | |
| BC-GV, BC-R | Genomovar V and some genomovar II | 55 | |
| PC-SSF, PC-SSR | Genomovars I, III, IV, and VII | 53 | |
| Bauernfeind et al. (2) | Eub16-1, CeMuVi-16-2 | Genomovars I to VII | 53 |
| Eub 16-1, Ce-16-2 | Genomovars I, III, IV, and VII | 56 | |
| Eub 16-1, MuVi-16-2 | Genomovars II, V, and VII | 53 |
In the interest of clarity, genomovar designations have been used instead of species names. Genomovar II, B. multivorans; genomovar IV, B. stabilis; genomovar V, B. vietnamiensis; genomovar VII, B. ambifaria.
For each primer pair, the Mg2+ concentration and the optimal annealing temperature were determined by using DNA extracted from pure cultures. Thermal cycler parameters used were slightly different than those published (2, 27) and are listed in Table 3. Assays employing primer pairs described by LiPuma et al. (27) used the following thermal cycler parameters: denatured for 3 min at 95°C, followed by 30 cycles of 1 min at 94°C, 1 min at the annealing temperature, and 1 min at 72°C. The final extension step was 4 min. Samples were held at 25°C until removed from the thermal cycler and placed at 4°C. Reactions with primer pairs described by Bauernfeind et al. (2) had the following parameters: 5 min of denaturing at 95°C, followed by 30 cycles of 30 s at 95°C, 30 s at the annealing temperature, and 45 s at 72°C. The final extension step was 7 min; samples were then held at 25°C. All PCR assays included a positive DNA control (50 μl of DNA from a pure culture of Bcc, which should amplify with the primer pair used), negative DNA control (50 μl of DNA from a pure culture of Bcc or a close relative which should not amplify with the primer pair used), and a water blank (includes all ingredients except DNA) (37).
All PCR products were separated from genomic DNA by gel electrophoresis on 1% agarose gels and visualized with ethidium bromide. A band on the gel was considered a positive reaction, even if faint. No bands were seen which were not at the same position on the gel as the positive control. One quarter of all reactions were repeated to assess reproducibility.
Limit of detection of PCR assays.
The limit of detection of the PCR assays was ascertained by adding Bcc to a clay loam (Jory series; 39.5% clay, 39.8% silt, and 20.7% sand). Dry, twice-autoclaved soil (1 g) was placed in a sterile tube, and 300 μl of a broth culture of Pseudomonas aeruginosa containing ca. 4 × 108 CFU was added to represent “background” bacterial populations. Serial dilutions of broth cultures of two Bcc strains (B. vietnamiensis FC 441 [LMG 18836]; B. ambifaria AMMD [LMG 19182]) were added in amounts ranging from 10 to 108 CFU per g of soil. The soil-broth mixture was gently mixed and allowed to stand 20 min. All dilutions of each broth culture were also plated on tryptic soy agar to determine the actual CFU/ml. Wet soil (300 mg) from each dilution was placed into an extraction tube and extracted, the DNA was quantified, and PCR assays were run as described earlier. Controls included a unit with soil, P. aeruginosa, and 100 μl of sterile water; a unit with soil and 400 μl of sterile water; and two units with sterile water only.
RFLP screening and DNA sequencing.
To verify that Bcc DNA was amplified in the foregoing PCR assays, selected amplicons were cloned and screened by restriction fragment length polymorphism (RFLP) analysis. Amplicons were generated from four soil samples with the primer pair Eub-16-1-CeMuVi-16-2, which amplifies Bcc genomovars I to VII. Amplicons were purified by ethanol precipitation and transformed into Escherichia coli JM109 by using the pGEM-T Easy Vector System (Promega Corp., Madison, Wis.) according to the manufacturer's instructions. Transformants were screened for inserts by using α-complementation with X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) and IPTG (isopropyl-β-d-thiogalactopyranoside); the insert size was determined by PCR with primer pair Eub-16-1-CeMuVi-16-2.
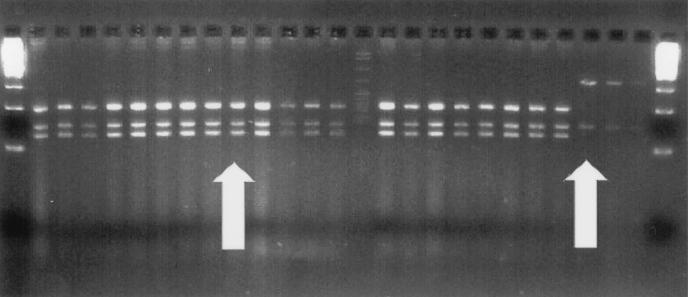
A total of 120 clones with the correctly sized insert were assessed by RFLP analysis with Sau96I (Promega). The amplicons from the PCR assay described above were used as a template. Each reaction vessel included 7.5 μl of water, 2 μl of 10× buffer (Promega), 10 μl (ca. 120 ng) of DNA, and 2.5 U of Sau96I. The reagents were mixed by pipetting and centrifuged briefly to collect the contents at the bottom of the tube and then incubated in a water bath at 37°C for 16 h. The reaction was stopped by adding 4 μl of 6× gel loading dye (0.0625 g of bromophenol blue, 0.0625 g of xylene cyanol, and 3.75 g of Ficoll in 25 ml of water) to each tube and centrifuging the mixture again briefly. DNA fragments were separated by gel electrophoresis in a 3% agarose gel (Metaphor; BioWhittaker Molecular Applications, Rockland, Maine) at 4°C and visualized with ethidium bromide (Fig. 1). The results were compared to a computer digest of published Bcc sequences (Wisconsin Package, version 10.1; Genetics Computer Group, Madison, Wis.) and to positive controls digested with the clones.
FIG. 1.
RFLP assay with Sau96I digest of 463-bp fragment of rrn gene. The left arrow points to Burkholderia pattern; the right arrow points to one of the non-Burkholderia patterns. The far left and far right lanes are molecular ladders, with 100-, 200-, and 300-bp bands.
Two clones that had the “Burkholderia” pattern were sequenced from each of the four soil samples (eight total). Three clones representing non-Bcc patterns were also sequenced. The clones were grown overnight in 3 ml of LB medium, and vector DNA was prepared by using the Eppendorf Perfectprep Plasmid Mini Kit (Hamburg, Germany) according to the manufacturer's instructions. Nucleotide sequence data were obtained by using T7 and SP6 primers. The sequencing was performed by using Taq dye terminator chemistry and an ABI cycle sequencer (Central Services Laboratory, Center for Gene Research and Biotechnology, Oregon State University, Corvallis, Oreg.). The resulting sequences were used to search for similarities among known sequences by using the basic local alignment tool (BLAST) at the National Center for Biotechnology Information (Bethesda, Md.).
RESULTS
Isolation of bacteria.
Putative Bcc colonies were recovered from both BCSA and TBT. The mean recovery on BCSA was 6.1 log10 CFU per g (dry weight) of soil; on TBT it was 6.2 log10 CFU per g (dry weight) of soil. The mean population recovered was not significantly different between Portland and Cleveland and between the two selective media (analysis of variance; P = 0.72). Colony counts from Philadelphia samples on TBT were lower than the TBT counts for other locations due to rampant fungal growth on the plates. Thus, in the Portland and Cleveland samples (obtained subsequently), TBT was amended with nystatin (50 μg/ml) to control fungal growth, and data from the Philadelphia samples were omitted from the comparison performed above. Some soil samples yielded more colonies when plated on TBT; others yielded more on BCSA. The color and morphology of the isolates were noted at each step; up to 10 different colony types were observed on each medium.
Rhizosphere samples had higher numbers of CFU per g (dry weight) of soil than did bulk soil samples. The mean differences in CFU between rhizosphere and bulk soil samples were 0.80 log CFU per g of soil on BCSA plates and 1.06 log CFU per g of soil on TBT plates. These differences were statistically significant (one-tailed t test assuming unequal variance: P = 0.0004 for BCSA and P = 0.018 for TBT). Overall, the rhizosphere samples did not yield more different types of colonies than did the bulk soil samples.
Identification of isolates.
A total of 1,260 bacterial colonies was chosen from TBT plates and BCSA plates and streaked for purity on the same media from which they came. Since the colonies were chosen as representatives of observed morphologies, selection was nonrandom. Of the 1,260 isolates originally selected, 114 did not survive isolation, 88 were not culturable in any of the nonselective broth media, and 23 died during storage at −80°C. Thus, 1,035 (i.e., 82% of the original total number of colonies selected) were available for further testing.
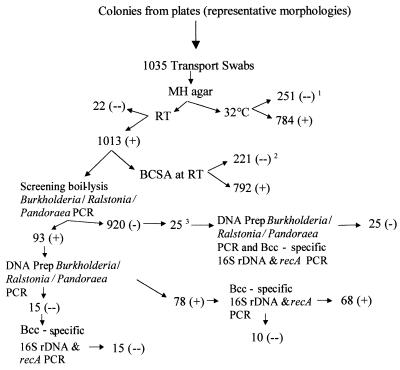
Of these 1,035 isolates, 22 could not be recovered from transport swabs when cultured onto MH agar at either RT or 32°C (Fig. 2). Another 229 that grew at RT did not grow when incubated at 32°C; ultimately, none of these demonstrating temperature sensitivity proved to be Bcc. Among the 1,013 isolates that grew on MH agar at RT, 221 could not be subcultured on BCSA at RT; again, none of these ultimately proved to be Bcc.
FIG. 2.
Identification of isolates. See the text for descriptions and explanation. Superscripts: 1, includes 22 colonies that also did not grow at RT (none of the remaining 229 were positive in screening PCR); 2, no colonies were positive in the screening PCR; 3, all 25 colonies grew on BCSA, had a slow and/or weak oxidase reaction, and had a colony morphology consistent with Bcc.
Screening PCR (by the boil-lysis method) specific for the genera Burkholderia, Ralstonia, and Pandoraea was positive for 93 of the 1,013 isolates. As a test of the sensitivity of the boil-lysis method, the screening PCR was repeated with genomic DNA prepared from 25 of the 920 PCR-negative isolates. Although all 25 demonstrated positive growth on BCSA, slow and/or weak oxidase reaction, and colony morphology consistent with Bcc, none was positive in the repeat screening PCR with prepared DNA. All 25 were also subsequently negative in 16S rDNA and recA-targeted PCR assays specific for all Bcc species.
All 93 isolates testing positive in the screening PCR assay by the boil-lysis method were retested by using prepared genomic DNA as a template. Fifteen of these were negative with repeat testing and remained negative with Bcc-specific PCR assays. Among the 78 testing positive with repeat screening PCR, 10 were negative with Bcc-specific PCR assays. The remaining 68 isolates were confirmed as members of the Bcc by using Bcc-specific 16S rDNA and recA targeted PCR assays (27, 29). Thus, 6.5% of the isolates screened and 5.4% of those initially selected from the two selective media ultimately proved to be Bcc species.
Recovery of Bcc.
Bcc was isolated from 14 of 91 (15%) of sample sites (Table 1). Bcc was recovered on both BCSA and TBT. More than twice as many Bcc isolates were recovered from TBT (n = 45; 66% of total) as from BCSA (n = 23; 34% of total), although most isolate-positive samples (78%) yielded results on both BCSA and TBT. The soil samples out of which Bcc was cultured were not significantly different from the others with regard to water content.
There was no clear rhizosphere enrichment effect. Bcc was isolated from 3 of 20 (15%) of rhizosphere samples. Similarly, bulk soil yielded Bcc in 14.7% of the samples (13 of 88). In only one sample was Bcc cultured from a rhizosphere sample when it was not cultured from the parallel soil sample; in another sample, Bcc was isolated from the bulk soil fraction but not the rhizosphere. Bcc was isolated from the rhizospheres of clover, grass, and an impatiens. It was not isolated from four other grass rhizosphere samples, six turf samples, or the rhizospheres of tomato (in soil and in potting mix), lettuce, dandelion, or wild geranium or from the rhizosphere of maize after harvest.
Extraction of DNA from soil samples.
DNA was extracted from 87 soil samples. The mean amount of DNA extracted was 3.6 μg of DNA per g (dry weight) of soil; the median was 17 ng per g (dry weight) of soil. The amount of DNA spanned 5 orders of magnitude (minimum, 0.12 ng of DNA per g [dry weight] of soil; maximum, 40.3 μg of DNA per g [dry weight] of soil), but all soil samples yielded at least a small amount of DNA. Some low values are not surprising given the inhospitality of some sampled environments (e.g., playground sand). Assuming a typical bacterial population of 109 bacteria per g of soil and 5 to 8 fg of DNA per cell, the extraction efficiency of the procedure, with the mean amount of DNA extracted, can be estimated to be between 28 and 45%.
No DNA was detected in extraction blanks when they were evaluated with the fluorometer. Spiked samples made by adding bacteria to nonautoclaved soil contained ca. 10 times more DNA than spiked samples constructed by using twice-autoclaved soil. DNA was successfully extracted from all spiked samples.
16S rDNA soil PCR assays.
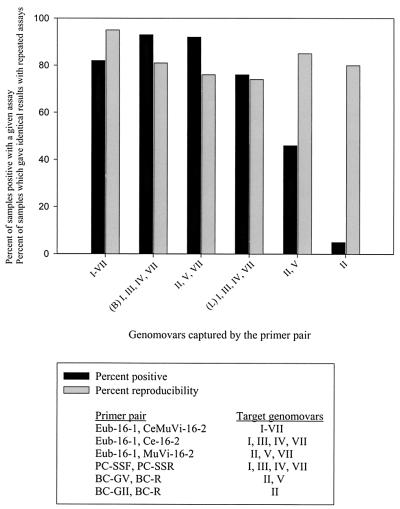
All 87 soil samples from which DNA had been extracted were evaluated by PCR assay, and all but one was positive for bacterial DNA. This sample was from the dry sand of a baseball diamond. Three more samples were found to be negative at the “genus” level by the Burkholderia-Ralstonia-Pandoraea PCR assay; these samples were from wet creekside sand, playground sand, and bark mulch. The remaining 83 soil samples were all evaluated with the six Bcc-specific PCR assays (Fig. 3). Overall, 93% of the 87 soil samples from which DNA had been extracted were positive in at least one of the Bcc-specific PCR assays described by Bauernfeind et al. (2), whereas 82% were positive in at least one of the assays described by LiPuma et al. (27). Many of the assays were initially negative and were repeated with more and/or less template DNA. The optimum amount of template solution did vary between samples, as has been observed previously (24).
FIG. 3.
Results of 16S rDNA PCR assays on DNA extracted from soil samples. “(B)” and “(L)” refer to primers designed by Bauernfeind et al. (2) and LiPuma et al. (27), respectively. Bars: ▪, percent samples determined to be positive by a given assay; ░⃞, percent samples that gave identical results in repeated assays.
One-quarter of all of the PCR assays were repeated to assess reproducibility. Reproducibility here means providing the same result in repeated assays (positive-positive and negative-negative). The overall reproducibility was 81%. Fully 72% of all of the changes were associated with faint bands, suggesting that in some cases the amount of template DNA, or the amount of a potentially inhibiting coextracted substance, was near a threshold concentration for detection.
Limit of detection.
The DNA extracted from the limit of detection experiment was evaluated by using the same PCR assay scheme. Low amounts (0.17 to 1.17 ng of DNA per g [dry weight] of soil) of DNA were extracted from the prepared soils. In all assays, the limit of detection was 105 CFU per g (dry weight) of soil. The amount of “background” bacteria present at this level was 4 × 108 CFU per g of soil, or approximately a 1/104 ratio. This is slightly less than the 109 CFU figure suggested by Cullen and Hirsch (10) as an appropriate estimate of the bacterial population in a “typical” gram of soil. Background populations are important, since the effect of diluting the target DNA into a larger pool of sample DNA is to lower the achievable detection sensitivity (24).
There were some slight differences based on the primer pair used and the strain used. The limit of detection could be improved to 50 CFU per g (dry weight) of soil by running two sequential cycles of PCR and by using the product of the first assay as a template for the second. Similar results were reported by Bell et al. (3) with sequential PCR. No amplification on any of the blanks was observed in this procedure. However, it was considered too vulnerable to PCR error to use with the soil samples (41).
Cloning and RFLP assays.
Clones (n = 120) were generated from four soil samples; each clone contained a vector with a 463-bp insert, which was the amplicon from PCR assays designed to capture Bcc genomovars I to VII. A PCR screen of these clones revealed that most (97.6%) had the correctly sized insert. A digest with Sau96I showed that 82.7% of the correctly sized inserts had the Burkholderia pattern (Table 4). In sum, in three of the four soil samples, >90% of the evaluated clones had the Burkholderia pattern. In one soil sample, only 9.5% of the clones had the Burkholderia pattern. None of these soil samples yielded isolates identified as Bcc.
TABLE 4.
Results from RFLP analyses and sequencing
| Soil sample location | Bcc cultured on selective media from sample | % Clones with Burkholderia RFLP pattern | Bcc DNA identified in sample |
|---|---|---|---|
| Vegetable garden | No | 96.4 | Yes |
| Edge of jogging-cycling path | No | 100 | Yes |
| Golf course | No | 90.0 | Yes |
| Flowerbed in botanical garden | No | 9.5 | Yes |
Of the 96 clones with the Burkholderia RFLP pattern, 8 were selected for sequencing. Two were selected from each soil sample. All of the sequences were identified as Bcc by using the BLAST program; species designations are difficult given the rapidly changing taxonomy and a 463-bp segment. Three clones displaying non-Burkholderia patterns were also sequenced. These were identified as a chimeric sequence, an unidentified soil clone, and a member of the genus Zoogloea.
DISCUSSION
Advancements in the taxonomy of the B. cepacia complex, coupled with the inherent difficulty in identifying these species, have made it difficult to interpret the literature on Bcc's prevalence in the soil environment. This study examined the prevalence of the B. cepacia complex in urban and suburban soil environments where people may contact soil, by using two different selective media and polyphasic (phenotypic and genotypic) identification protocols. DNA was also extracted directly from soil samples and examined by using various PCR assays specific for Bcc.
Isolation of members of the Bcc was attempted on two different selective media, BCSA and TBT. Both are reported to have good selectivity for Bcc; in previous studies, 93.6% of clinical isolates cultured on BCSA were Bcc (19), whereas 72% of colonies from environmental samples recovered on TBT were identified as P. cepacia (16). Our results showed substantially lower selectivity. Only 8.8% of isolates (nonrandomly selected) from TBT were identified as Bcc, as were only 2.9% of the isolates from BCSA. Overall, 5.4% of 1,260 isolates originally selected from both media ultimately were identified as Bcc.
The discrepancy between our results and previous studies with these media could stem from several issues. First, BCSA was developed for use in the clinical setting. The diversity of bacteria present in cystic fibrosis sputum is much more narrow than in soil; it is not surprising that many soil-living bacterial species besides Bcc can metabolize BCSA. Second, BCSA is a rich medium, amended with polymyxin B, vancomycin, and gentamicin. It is possible that the richness of BCSA placed too much metabolic stress on nutrient-deprived soil populations of Bcc. It could also be that the antibiotics used are either too selective or not selective enough. Environmental strains of Bcc may not have, or do not yet express, genes encoding antibiotic resistance that characterize clinical strains and consequently weren't able to grow on BCSA. Butler et al. (6) reported substantially lower antibiotic resistance by environmental strains. However, others report the isolation from soil of Bcc with considerable antibiotic resistance (39). Alternatively, it is possible that a variety of soil-living non-Bcc bacteria are able to overcome the antibiotic selectivity of BCSA. These other bacteria may have overwhelmed any Bcc that were present on the plates.
TBT was developed for use with environmental strains of B. cepacia (16) and, in fact, we found that more than twice as many Bcc isolates were recovered from TBT than from BCSA. This could be because of the smaller number of antibiotics used (tetracycline only) or perhaps because of the relative meagerness of the medium. However, 8.8%, even nonrandomly selected, is still a far smaller percentage of colonies that are Bcc than the published values of 72% (16).
A significant problem in evaluating previous reports of media designed to select Bcc from the environment is that the taxonomy of Bcc and, indeed, of the entire genus Burkholderia has changed rapidly. “B. cepacia” has gone from being considered a single Pseudomonas species to being a complex of no fewer than nine species (genomovars) within the genus Burkholderia in a few years' time. It is possible that some researchers may not have kept current with this increasing taxonomic complexity. Add to this the notorious difficulty in identifying Bcc with widely available biochemical test schemes (38, 40, 44), and it becomes difficult to know how much confidence to place in an identification of “B. cepacia” (46; E. Mahenthiralingam, N. Burton, S. Laevers, and P. Vandamme, Abstr. Int. Burkholderia cepacia Working Group, Bethesda, Md. [http://go.to/cepacia], 2000).
Some earlier studies have also reported isolating Bcc infrequently (6, 32). Together with our results, this suggests that Bcc from environmental sources is in fact not easy to recover on these selective media. It may be that other selective media such as PCAT (1) or non-culture-based methods are necessary for an accurate assessment of the prevalence of Bcc in the environment.
Interestingly, although more Bcc was isolated from TBT than from BCSA, all isolates that were ultimately identified as Bcc were able to grow on BCSA, suggesting that soil strains of Bcc were more capable of growing on BCSA after first growing on TBT. Reference strains of genomovars I to VII were successfully grown on TBT and BCSA, ruling out a categorical inability of any genomovar to be cultured on these media. It may be that some genomovars, or some strains, make the soil-media transition better than others, due to the assumption of the viable-but-not-culturable (VBNC) state, loss of antibiotic resistance, or other factors. Temperature may also be a useful screening tool, as all of the Bcc isolates were capable of growth at 32°C.
We specifically sampled the rhizosphere in 20 (18.7%) of 107 samples. Bcc is a known rhizosphere colonizer; populations of up to 105 CFU per g of root have been identified on the roots of peas (22). Other plant hosts known to support Bcc in the rhizosphere include maize (18, 33), tomatoes (39), wheat and lupine (1), and perennial ryegrass (35). It thus was theorized that populations of Bcc would be enriched by the presence of a plant root, and that otherwise low—and possibly undetectable—populations of Bcc would be detected in rhizosphere samples, but this was not demonstrated by our results. The rhizobacteria we used that could grow on the selective media were more abundant than bacteria in the bulk soil (by ca. 1 log unit per g of soil). However, Bcc was not isolated from the rhizosphere any more frequently than from the bulk soil samples. Other studies blended or ground the roots of plants and plated the root slurry (1, 5), instead of plating only the adhering soil, as we did. Blending and plating root slurry would have recovered endophytic populations in addition to populations external to the root, and endophytic populations of Bcc can be substantial (17).
The second half of our study sought to use growth-independent methods to detect the presence of Bcc in soil samples. An estimated 90 to 99% of bacteria in soil are not culturable by conventional methods (10), and it seemed possible that some Bcc strains might be included in the nonculturable majority. We thus directly extracted DNA from soils and evaluated the extracts for the presence of Bcc DNA by two independently developed sets of 16S rDNA-directed PCR assays. PCR assays targeting 16S rDNA genes have been used in other studies of soil bacterial populations (3, 12, 37). Advantages to using these sequences as a PCR target include the high copy number of rrn genes (six copies distributed on the three or four replicons typical of most strains of Bcc) (25). There are many published sequences of this gene, facilitating comparison with sequences generated in the course of this study. Finally, the primers give one product of one size with pure cultures, unlike some other primer pairs which target the recA gene (29). Among the disadvantages of the 16S rDNA gene is, principally, its highly conserved nature. There may well be other members of the β-proteobacteria that are similar to Bcc and that were not tested in the development of the primer pairs we used in this study. A highly conserved gene is also an unlikely source of easily gained differentiablity between genomovars within the B. cepacia complex, which by definition are 98 to 99% homologous in their 16S rDNA sequences.
The performance of the 16S rDNA PCR assays was very consistent. Reproducibility of results ranged from good (74%) to excellent (95%), depending on the primer pair. The results from the two PCR schemes generally supported one another, although there were differences between them. For instance, primer pairs developed by both Bauernfeind et al. (2) and LiPuma et al. (27) were designed to amplify as a group genomovars I and III, B. stabilis (IV), and B. ambifaria (VII). In assays with soil extracts, however, these primer pairs did not perform identically; 76% of samples were positive with the latter primers, whereas 93% were positive with the former. These differences could result from various sensitivities or specificities of the primer pairs to the target DNA or to various inhibiting contaminants that may have been coextracted with the DNA.
Most soil samples were positive for one of the Bcc-specific PCR assays; 82% were determined to be positive by one of the assays described by LiPuma et al., (27), and 93% were determined to be positive in at least one of the assays described by Bauernfeind et al. (2). This is much higher than the results of isolation from selected media; only 15% of samples yielded isolates that were identified as Bcc. There are several possibilities for the discrepancy. One possibility is the limit of detection of plating on selective media, as opposed to the limit of detection of the PCR assays. It is difficult to know what the limit of detection of our culturing effort actually was, given the nonrandom nature of colony selection; the PCR assays had a limit of detection of 105 CFU per g of soil. Different detection limits have been seen in clinical studies, where a patient may be “culture negative” but “PCR positive” for Bcc (45). If the numbers of the desired bacterium are low, they may be impossible to detect via plate culture, since they will be outpaced by more numerous or faster-growing organisms. It could also be that the bacteria are culturable but not able to grow with the selective agents in these two media, as previously discussed.
Alternatively, it could be that the bacteria are viable but not culturable. Bacteria in the VBNC state are thought to be common in a substrate-limited habitat such as soil (30). Since it has been shown that bacteria in the VBNC state do not lose pathogenicity or virulence (30), the ability to ascertain the presence of Bcc in this state could be very helpful in delineating the risk posed by environmental populations to susceptible people. It is also possible that the bacteria were in fact not present in the soil and that the PCR assays were amplifying Bcc DNA remaining from previous populations that had survived degradation by being adsorbed to soil colloids (4, 13, 36). The likelihood of large amounts of DNA remaining uncorrupted in soil is, however, not high.
A final explanation for the discrepancy between culture-based and growth-independent estimations of Bcc prevalence is that the PCR assays used are not sufficiently specific and are amplifying non-Bcc DNA. This is the case at least part of the time, as is shown in our cloning and sequencing data; in one soil sample, 90% of the tested clones had an RFLP pattern that corresponded to a member of the genus Zoogloea. This finding was subsequently confirmed by sequencing. It is important to note, however, that all four soil samples contained sequences which were definitely identified as part of the B. cepacia complex. The presence of some non-Bcc patterns in the RFLP digest was fully expected, since the PCR and cloning techniques we used have been shown to introduce errors (41). Nor is it particularly surprising, given the highly conserved nature of the 16S gene, that other bacterial DNA is able to give a positive signal in these PCR assays. It is thus possible that the non-Bcc DNA increased the likelihood of a false positive but that Bcc DNA was also present in many of the samples.
Perhaps the most informative aspect of this study is the difference between the culture-based and non-culture-based methods. Bcc was isolated from only 15% of soil samples on the two selective media. In contrast, 76 and 93% of the soil samples were determined to be positive for Bcc DNA by the two PCR assay systems. The screening and sequencing portion of the study demonstrated that Bcc DNA was being amplified in all four of the tested soil samples and accounted for the majority of the DNA amplified in three of the four samples. Also, no Bcc was isolated from any of these four samples in which Bcc DNA was conclusively present. The specificity of the soil PCR assays is not perfect, and it is possible that some “Bcc-positive” soil samples do not in fact contain Bcc DNA. It seems clear, however, that although the 16S PCR assays may overestimate the prevalence of Bcc in the environment, culturing on the currently available selective media underestimates Bcc populations.
Of the many possible explanations for the difference between culture-based and non-culture-based results, one of the simplest is that many Bcc isolates are not culturable on the media we used. Neither TBT, developed for environmental research, nor BCSA, developed for use in the medical field, was effective. It follows that the use of selective media may not be the best way to estimate the environmental prevalence of Bcc in soils and, further, that populations of Bcc in soils may be much higher than was previously estimated. In summary, our results demonstrate a relatively low recovery of Bcc from selective media but also indicate that Bcc DNA appears to be frequently encountered in urban soil environments. Assessment of the risk posed by indigenous and introduced Bcc may proceed much more rapidly with the continued refinement of growth-independent methods.
Acknowledgments
This work was supported by a grant from the Cystic Fibrosis Foundation. S.C.M.M. was also supported by fellowships from the Storkan-Hanes Foundation and the Howard Hughes Medical Institute.
REFERENCES
- 1.Balandreau, J., V. Viallard, B. Cournoyer, T. Coenye, S. Laevens, and P. Vandamme. 2001. Burkholderia cepacia genomovar III is a common plant-associated bacterium. Appl. Environ. Microbiol. 67:982-985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bauernfeind, A., I. Schneider, R. Jungwirth, and C. Roller. 1999. Discrimination of Burkholderia multivorans and Burkholderia vietnamiensis from Burkholderia cepacia genomovars I, III, and IV by PCR. J. Clin. Microbiol. 37:1335-1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bell, K. S., M. S. Kuyukina, S. Heidbrink, J. C. Philip, D. W. J. Aw, I. B. Ivshina, and N. Christofi. 1999. Identification and environmental detection of Rhodococcus species by 16S rDNA-targeted PCR. J. Appl. Microbiol. 87:472-480. [DOI] [PubMed] [Google Scholar]
- 4.Bertolla, F., and P. Simonet. 1999. Horizontal gene transfers in the environment: natural transformation as a putative process for gene transfers between transgenic plants and microorganisms. Res. Microbiol. 150:375-384. [DOI] [PubMed] [Google Scholar]
- 5.Bevivino, A., S. Sarrocco, D. Calmastri, S. Tabacchioni, C. Cantale, and L. Chiarini. 1998. Characterization of a free-living maize rhizosphere population of Burkholderia cepacia: effect of seed treatment on disease suppression and growth promotion of maize. FEMS Microbiol. Ecol. 27:225-237. [Google Scholar]
- 6.Butler, S. L., C. J. Doherty, J. E. Hughes, J. W. Nelson, and J. R. W. Govan. 1995. Burkholderia cepacia and cystic fibrosis: do natural environments present a potential hazard? J. Clin. Microbiol. 33:1001-1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cain, C. C., A. T. Henry, R. H. Waldo III, L. J. Casida, Jr., and J. O. Falkinham III. 2000. Identification and characteristics of a novel Burkholderia strain with broad-spectrum antimicrobial activity. Appl. Environ. Microbiol. 66:4139-4141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cartwright, D. K., W. S. Chilton, and D. M. Benson. 1995. Pyrrolnitrin and phenazine production by Pseudomonas cepacia, strain 5.5B, a biocontrol agent of Rhizoctonia solani. Appl. Microbiol. Biotechnol. 43:211-216. [Google Scholar]
- 9.Coenye, T., P. Vandamme, J. R. W. Govan, and J. J. LiPuma. 2001. Taxonomy and identification of the Burkholderia cepacia complex. J. Clin. Microbiol. 39:3427-3436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cullen, D. W., and P. R. Hirsch. 1998. Simple and rapid method for direct extraction of microbial DNA from soil for PCR. Soil Biol. Biochem. 30:983-993. [Google Scholar]
- 11.Darling, P., M. Chan, A. D. Cox, and P. A. Sokol. 1998. Siderophore production by cystic fibrosis isolates of Burkholderia cepacia. Infect. Immun. 66:874-977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dojka, M. A., P. Hugenholtz, S. K. Haack, and N. R. Pace. 1998. Microbial diversity in a hydrocarbon- and chlorinated-solvent-contaminated aquifer undergoing intrinsic bioremediation. Appl. Environ. Microbiol. 64:3869-3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Frostegard, A., S. Courtois, V. Ramisse, S. Clerc, D. Bernillon, F. le Gall, P. Jeannin, X. Nesme, and P. Simonet. 1999. Quantification of bias related to the extraction of DNA directly from soils. Appl. Environ. Microbiol. 65:5409-5420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Govan, J. R. W., and V. Deretic. 1996. Microbial pathogenesis in cystic fibrosis: mucoid Pseudomonas aeruginosa and Burkholderia cepacia. Microbiol. Rev. 60:539-574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Govan, J. R. W., J. E. Jughes, and P. Vandamme. 1996. Burkholderia cepacia: medical, taxonomic and ecological issues. J. Med. Microbiol. 45:395-407. [DOI] [PubMed] [Google Scholar]
- 16.Hagedorn, C., W. D. Gould, T. R. Bardinelli, and D. R. Gustavson. 1987. A selective medium for enumeration and recovery of Pseudomonas cepacia biotypes from soil. Appl. Environ. Microbiol. 53:2265-2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hallman, J., R. Rodriguez-Kabana, and J. W. Kloepper. 1999. Chitin-mediated changes in bacterial communities of the soil, rhizosphere, and within roots of cotton in relation to nematode control. Soil Biol. Biochem. 31:551-560. [Google Scholar]
- 18.Hebbar, K. P., D. Atkinson, W. Tucker, and P. J. Dart. 1992. Suppression of Fusarium monilforme by maize root-associated Pseudomonas cepacia. Soil Biol. Biochem. 24:1009-1020. [Google Scholar]
- 19.Henry, D. A., M. E. Campbell, J. J. LiPuma, and D. P. Speert. 1997. Identification of Burkholderia cepacia isolates from patients with cystic fibrosis and use of a simple new selective medium. J. Clin. Microbiol. 35:614-619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang, Y., and P. T. W. Wong. 1998. Effect of Burkholderia (Pseudomonas) cepacia and soil type on the control of crown rot in wheat. Plant Soil 203:103-108. [Google Scholar]
- 21.Juhasz, A. L., M. L. Britz, and G. A. Stanley. 1996. Degradation of high molecular weight polycyclic aromatic hydrocarbons by Pseudomonas cepacia. Biotechnol. Lett. 18:577-582. [Google Scholar]
- 22.King, E. B., and J. L. Parke. 1993. Biocontrol of aphanomyces root rot and pythium damping-off by Pseudomonas cepacia AMMD on four pea cultivars. Plant Dis. 77:1185-1188. [Google Scholar]
- 23.Krumme, M. L., K. N. Timmis, and D. F. Dwyer. 1993. Degradation of trichloroethylene by Pseudomonas cepacia G4 and the constitutive mutant strain G4 5223 PR1 in aquifer microcosm. Appl. Environ. Microbiol. 59:2746-2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kuske, C. R., K. L. Banton, D. L. Adorada, P. C. Stark, K. K. Hill, and P. J. Jackson. 1998. Small-scale DNA sample preparation method for field PCR detection of microbial cells and spores in soil. Appl. Environ. Microbiol. 64:2463-2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lessie, T. G., W. Hendrickson, B. D. Manning, and R. Devereux. 1996. Genomic complexity and plasticity of Burkholderia cepacia. FEMS Microbiol. Lett. 144:117-128. [DOI] [PubMed] [Google Scholar]
- 26.LiPuma, J. J. 1998. Burkholderia cepacia: management issues and new insights. Clin. Chest Med. 19:473-486. [DOI] [PubMed] [Google Scholar]
- 27.LiPuma, J. J., B. J. Dulaney, J. D. McMenamin, P. W. Whitby, T. L. Stull, T. Coenye, and P. Vandamme. 1999. Development of rRNA-based PCR assays for identification of Burkholderia cepacia complex isolates recovered from cystic fibrosis patients. J. Clin. Microbiol. 37:3167-3170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Livesley, M. A., I. A. Baxter, P. A. Lambert, J. R. W. Govan, P. W. Weller, D. E. Lacey, D. G. Allison, B. Giwercman, and N. Hoiby. 1998. Subspecific differentiation of Burkholderia cepacia isolates in cystic fibrosis. J. Med. Microbiol. 47:999-1006. [DOI] [PubMed] [Google Scholar]
- 29.Mahenthiralingam, E., J. Bischof, S. K. Byrne, C. Radomski, J. Davies, Y. Av-Gay, and P. Vandamme. 2000. DNA-based diagnostic approaches for identification of Burkholderia cepacia complex, Burkholderia vietnamiensis, Burkholderia multivorans, Burkholderia stabilis, and Burkholderia cepacia genomovars I and III. J. Clin. Microbiol. 38:3165-3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McDougald, D., S. A. Rice, D. Weichart, and S. Kjelleberg. 1998. Nonculturability: adaptation or debilitation? FEMS Microbiol. Ecol. 25:1-9. [Google Scholar]
- 31.McMenamin, J. D., T. M. Zaccone, T. Coenye, P. Vandamme, and J. J. LiPuma. 2000. Misidentification of Burkholderia cepacia in U.S. cystic fibrosis treatment centers: an analysis of 1,051 recent sputum samples. Chest 117:1661-1665. [DOI] [PubMed] [Google Scholar]
- 32.Mortensen, J., M. Fisher, and J. J. LiPuma. 1995. Recovery of Pseudomonas cepacia and other Pseudomonas spp from the environment. Infect. Control Hosp. Epidemiol. 16:30-32. [DOI] [PubMed] [Google Scholar]
- 33.Nacamulli, C., A. Bevivino, C. Dalmastri, S. Tabacchioni, and L. Chiarini. 1997. Perturbation of maize rhizosphere microflora following seed bacterization with Burkholderia cepacia MCI7. FEMS Microbiol. Ecol. 23:183-193. [Google Scholar]
- 34.Nelson, J. W., S. L. Butler, D. Krieg, and J. R. W. Govan. 1984. Virulence factors of Burkholderia cepacia. FEMS Immunol. Med. Microbiol. 8:89-98. [DOI] [PubMed] [Google Scholar]
- 35.Nijhuis, E. S., M. J. Jaat, I. W. E. Zeegers, C. Waalwijk, and J. A. van Veen. 1993. Selection of bacteria suitable for introduction into the rhizosphere of grass. Soil Biol. Biochem. 25:885-895. [Google Scholar]
- 36.Paget, E., L. J. Simonet, and P. Monrozier. 1992. Adsorption of DNA on clay minerals: protection against DNase I and influence on gene transfer. FEMS Microbiol. Lett. 97:31-40. [Google Scholar]
- 37.Pepper, I. L., and S. D. Pillai. 1994. Detection of specific DNA sequences in environmental samples via polymerase chain reaction, p. 707-725. In R. W. Weaver, S. Angle, P. Bottomley, D. Bezdick, S. Smith, A. Tabatabai, and A. Wollum (ed.), Methods of soil analysis, part 2. Microbiological and biochemical properties. Book series no. 5. Soil Science Society of America, Madison, Wis.
- 38.Segonds, C., T. Heulin, N. Marty, and G. Chabanon. 1999. Differentiation of Burkholderia species by PCR-restriction fragment length polymorphism analysis of the 16S rRNA gene and application to cystic fibrosis isolates. J. Clin. Microbiol. 37:2201-2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sfalanga, A., F. Di Cello, L. Mugnai, S. Tegli, R. Fani, and G. Surico. 1999. Isolation and characterization of a new antagonistic Burkholderia strain from the rhizosphere of healthy tomato plants. Res. Microbiol. 150:45-59. [DOI] [PubMed] [Google Scholar]
- 40.Shelley, D. B., T. Spilker, E. J. Graceley, T. Coenye, P. Vandamme, and J. J. LiPuma. 2000. Utility of commercial systems for identification of Burkholderia cepacia complex from cystic fibrosis sputum culture. J. Clin. Microbiol. 38:3112-3115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Speksnijder, A. G. C. L., G. A. Kowalchuk, S. de Jong, E. Kline, J. R. Stephen, and H. J. Laanbroek. 2001. Microvariation artifacts introduced by PCR and cloning of closely related 16S rRNA gene sequences. Appl. Environ. Microbiol. 67:469-472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vandamme, P., B. Holmes, M. Vancanneyet, T. Coenye, B. Hoste, R. Coopman, H. Reverts, S. Lauwers, M. Gillis, K. Kersters, and J. R. W. Govan. 1997. Occurrence of multiple genomovars of Burkholderia cepacia in cystic fibrosis patients and proposal of Burkholderia multivorans sp. nov. Int. J. Syst. Bacteriol. 47:1188-1200. [DOI] [PubMed] [Google Scholar]
- 43.Vandamme, P., E Mahenthiralingam, B. Holmes, T. Coenye, B. Hoste, P. De Vos, D. Henry, and D. P. Speert. 2000. Identification and population structure of Burkholderia stabilis sp. nov. (formerly Burkholderia cepacia genomovar IV). J. Clin. Microbiol. 38:1042-1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.van Pelt, C., C. M. Verduin, W. H. F. Goessens, M. C. Vos, B. Rummler, C. Segonds, F. Reubsaet, H. Verbrugh, and A. van Belkum. 1999. Identification of Burkholderia spp. in the clinical microbiology laboratory: comparison of conventional and molecular methods. J. Clin. Microbiol. 37:2158-2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Whitby, P. W., H. L. N. Dick, P. W. Campbell III, D. E. Tullis, A. Matlow, and T. L. Stull. 1998. Comparison of culture and PCR for dectection of Burkholderia cepacia in sputum samples of patients with cystic fibrosis. J. Clin. Microbiol. 36:1642-1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wigley, P., and N. F. Burton. 1999. Genotypic and phenotypic relationships in Burkholderia cepacia isolated from cystic fibrosis patients and the environment. J. Appl. Microbiol. 86:460-468. [DOI] [PubMed] [Google Scholar]
- 47.Winstanley, C., B. A. Hales, J. A. W. Morgan, M. J. Gallagher, S. D. Puthucheary, M. F. Cisse, and C. A. Hart. 1999. Analysis of fliC variation among clinical isolates of Burkholderia cepacia. J. Med. Microbiol. 48:657-662. [DOI] [PubMed] [Google Scholar]
- 48.Wollum, A.G., II. 1994. Soil sampling for microbiological analysis, p. 1-14. In R. W. Weaver, S. Angle, P. Bottomley, D. Bezdick, S. Smith, A. Tabatabai, and A. Wollum (ed.), Methods of soil analysis, part 2. Microbiological and biochemical properties. Book series no. 5. Soil Science Society of America, Madison, Wis.
- 49.Yohalem, D. S., and J. W. Lorbeer. 1994. Intraspecific metabolic diversity among strains of Burkholderia cepacia isolated from decayed onions, soil, and the clinical environment. Antonie Leeuwenhoek 65:111-131. [DOI] [PubMed] [Google Scholar]
- 50.Yohalem, D. S., and J. W. Lorbeer. 1997. Distribution of Burkholderia cepacia phenotypes by niche, method of isolation, and pathogenicity to onion. Ann. Appl. Biol. 130:467-477. [Google Scholar]