Abstract
Chlamydiae are important pathogens of humans and animals but diagnosis of chlamydial infections is still hampered by inadequate detection methods. Fluorescence in situ hybridization (FISH) using rRNA-targeted oligonucleotide probes is widely used for the investigation of uncultured bacteria in complex microbial communities and has recently also been shown to be a valuable tool for the rapid detection of various bacterial pathogens in clinical specimens. Here we report on the development and evaluation of a hierarchic probe set for the specific detection and differentiation of chlamydiae, particularly C. pneumoniae, C. trachomatis, C. psittaci, and the recently described chlamydia-like bacteria comprising the novel genera Neochlamydia and Parachlamydia. The specificity of the nine newly developed probes was successfully demonstrated by in situ hybridization of experimentally infected amoebae and HeLa 229 cells, including HeLa 229 cells coinfected with C. pneumoniae and C. trachomatis. FISH reliably stained chlamydial inclusions as early as 12 h postinfection. The sensitivity of FISH was further confirmed by combination with direct fluorescence antibody staining. In contrast to previously established detection methods for chlamydiae, FISH was not susceptible to false-positive results and allows the detection of all recognized chlamydiae in one single step.
Chlamydiae cause a wide variety of diseases in humans and animals, including infections of the eye and the respiratory and the genital tracts (40, 43). More recently they were also associated with cardiovascular disease, atherosclerosis, and intrinsic asthma (4, 5, 34, 39). Chlamydiae are characterized by a complicated, obligate intracellular developmental cycle which is unique among the prokaryotes. The metabolically active, obligate intracellular chlamydial reticulate bodies (RB) multiply within their eukaryotic host cells and subsequently differentiate to inactive elementary bodies (EB), which are released and are able to infect new host cells and start a new developmental cycle (40, 43).
Until recently, the genus Chlamydia comprised only the four species Chlamydia pneumoniae, Chlamydia trachomatis, Chlamydia psittaci, and Chlamydia pecorum. This genus was considered the single genus within the family Chlamydiaceae, the only family within the order Chlamydiales. However, the recent description of several novel chlamydia-related bacteria (2, 15, 22, 29, 36) and an encompassing phylogenetic analysis of rRNA and other gene sequences (the major outer membrane protein, GroEL chaperonin, 3-deoxy-d-manno-octulosonic acid [KDO] transferase, small cysteine-rich lipoprotein and the 60-kDa cysteine-rich protein) changed our view on chlamydial diversity and taxonomy, and led to the reclassification of the Chlamydiales (6, 13, 19). Everett and coworkers subdivided the family Chlamydiaceae into the two genera Chlamydia (containing C. trachomatis, C. muridarum, and C. suis) and Chlamydophila (containing C. pneumoniae, C. pecorum, C. psittaci, C. felis, C. abortus, and C. caviae) and established the novel families Parachlamydiaceae, Simkaniaceae, and Waddliaceae, comprising the newly discovered Chlamydia-related organisms (13, 36). Since this new classification is not accepted by all members of the scientific community, we use the neutral genus abbreviation “C.” when possible, leaving the interpretation as Chlamydia or Chlamydophila up to the reader.
While C. trachomatis, C. psittaci, and C. pneumoniae are considered as clinically relevant pathogens in humans, it is still unclear whether the novel chlamydia-related bacteria, which were found as endosymbionts in free-living amoebae (2, 15, 22), as contaminants of a tissue culture (29), within an aborted bovine fetus (36), or in a wastewater treatment plant (21), have any clinical significance.
Diagnosis of chlamydial infections can be attempted by various detection methods, including culture, antigen detection, serology and nucleic acid amplification. However, in spite of the multiplicity of available test methods, the detection of chlamydiae from clinical specimens remains a major challenge, at least for routine laboratories. Especially, laboratory diagnosis of the fastidious C. pneumoniae is currently hampered by a lack of standardized and validated assays leading to a considerable interlaboratory variation of test results (18). This is one of the major reasons why the role of C. pneumoniae in respiratory tract disease as well as in atherosclerosis is still unclear (9, 38). Therefore, both standardizing of available test assays (10) and development of new molecular methods are urgently needed to obtain reliable tools for sensitive and specific detection of C. pneumoniae.
Fluorescence in situ hybridization (FISH) using fluorescently labeled oligonucleotide probes complementary to unique target sites on the rRNA has been shown to be a useful method for the detection and identification of microorganisms without cultivation (1a, 16, 23). FISH is therefore widely used for the analysis of composition and structure of complex microbial communities (26, 27, 41, 42). More recently, this technique was also shown to be a valuable tool for the rapid and specific detection of pathogens from blood cultures as well as for detection of slowly growing organisms including Helicobacter pylori and Legionella pneumophila (24, 25, 37).
In this study a hierarchical set of oligonucleotide probes for the detection of C. pneumoniae and all other known members of the Chlamydiales with FISH was constructed. The described set comprises nine new oligonucleotide probes complementary to order-, genus-, and species-specific sequence stretches on the 16S rRNA of the target organisms and is supplemented by two oligonucleotide probes published previously (2, 22). Analysis of the developmental cycle of C. pneumoniae in HeLa 229 cells using the FISH technique and comparison of the results with transmission electron microscopy revealed that 16S rRNA staining might be a suitable marker for viability and metabolic activity of chlamydiae. The specificity of the probes was demonstrated using cell cultures as well as amoebae infected with the different target organisms. The discriminatory power of the probes was further demonstrated using cell cultures, experimentally coinfected with different chlamydial species. Finally, it was shown that FISH can be combined with the direct immunofluorescence antibody (DFA) technique, allowing the concurrent demonstration of chlamydial rRNA and antigen within a single infected cell.
MATERIALS AND METHODS
Chlamydial strains and sample preparation.
Sample preparation for FISH and transmission electron microscopy was performed by experimental chlamydial infection of semiconfluent HeLa 229 cell monolayers (CCL2.1; American Type Culture Collection, Manassas, Va.) on glass coverslips in 24-well plates, according to standard procedures (10). Semipurified EB were harvested from C. pneumoniae strain TW 183 (Washington Research Foundation, Seattle), C. trachomatis strain UW-31/CX serovar K (ATCC VR 887) and a wild-type C. psittaci strain, isolated from a patient with ornithosis (12). Chlamydial strains were maintained on cycloheximide-treated HeLa 229 cell monolayers (CCL2.1; American Type Culture Collection) in six-well culture plates by standard procedures. C. pneumoniae TW183 was purified from Mycoplasma sp. by pulmonary passage in SCID mice, and cell monolayers were monitored routinely for Mycoplasma contamination by 4′,6′-diamidino-2-phenylindole (DAPI) staining and PCR, following procedures described previously (11). Cultures with at least 80% infected host cells were harvested after 72 h and homogenized with glass beads. Cell suspensions were centrifuged for 10 min at 4°C and 1,600 × g to remove cellular debris. Supernatants obtained were stored in aliquots at −70°C. The numbers of inclusion-forming units from stored aliquots were determined by quantitative chlamydial cell culture inoculation of confluent HeLa 229 cell monolayers with serial 10-fold dilutions, and staining was performed by genus-specific fluorescein isothiocyanate (FITC)-labeled monoclonal antibodies (Pathfinder; Kallestad, Chaska, Minn.) after an incubation period of 48 h for C. trachomatis and C. psittaci and after 72 h for C. pneumoniae. Stored aliquots were thawed and adjusted to the number of target cells to reach a multiplicity of infection of 1 that was used throughout the study. For simultaneous demonstration of C. trachomatis and C. pneumoniae within one sample by FISH, cells were first infected with C. pneumoniae and superinfected with C. trachomatis 24 h later to achieve inclusions of comparable size. Experimentally infected cell monolayers were fixed within the multiwell plates with 2% paraformaldehyde for 20 min at 4°C at various time points spanning the complete chlamydial developmental cycle. Subsequently, coverslips were washed with phosphate-buffered saline (10 mM sodium phosphate, 130 mM sodium chloride, pH 7.2) and stored at −20°C in 50% ethanol-phosphate-buffered saline. Prior to FISH coverslips were dehydrated with increasing concentrations of ethanol (50, 80, and 96%). For electron microscopy infected cells were fixed in a solution of 4% paraformaldehyde and 2.5% glutaraldehyde in 0.1 M phosphate-cacodylate buffer for 2 h. After dehydration in an ascending series of ethanol, the samples were embedded in Epon 812 resin (Fluka, Neu Ulm, Germany). Sections of 70 nm were stained with 1% lead citrate before examination in a Zeiss EM 10 electron microscope.
A representative member of the novel family Parachlamydiaceae, the obligate intracellular endosymbiont of Acanthamoeba sp. strain UWE25, included in this study was maintained in Acanthamoeba sp. strain UWC1 and fixed for FISH as described elsewhere (15).
Oligonucleotide probes and FISH.
In addition to previously published oligonucleotide probes, nine new probes with hierarchical specificity ranging from order to species level (Table 1) were designed using the ARB program package (software available at http://www.arb-home.de) and a comprehensive database comprising more than 15,000 16S rRNA sequences (including more than 150 published and unpublished rRNA sequences of members of the Chlamydiales). In order to ensure probe specificity, all publicly available 16S rRNA sequences included in the ARB database were checked for the presence of the probe target sites. Oligonucleotide probes were synthesized and directly 5′ labeled with 5(6)-carboxyfluorescein-N-hydroxysuccinimide ester (FLUOS), or the hydrophilic sulfoindocyanine fluorescent dyes Cy3 and Cy5 (Thermo Hybaid, Ulm, Germany). Newly designed probes were deposited at the oligonucleotide probe database probeBase (www.probebase.net).
TABLE 1.
Oligonucleotide probe sequences specific for the Chlamydiales and formamide concentration in the hybridization buffer required for specific FISH
| Probea | Target organisms | Sequence (5′-3′) | % FA | Reference |
|---|---|---|---|---|
| S-O-Chls-0523-a-A-18b | Chlamydiales | CCTCCGTATTACCGCAGC | 25 | This study |
| S-F-Chlae-0574-a-A-18 | Chlamydiaceae | CTTTCCGCCTACACGCCC | 20 | This study |
| S-G-Chla-0232-A-18 | Chlamydia | TAGCTGATATCACATAGA | 10 | This study |
| S-G-Chlph-0583-a-A-18 | Chlamydophila | CTAACTTTCCTTTCCGCC | 25 | This study |
| S-S-Ct-0623-a-A-18 | C. trachomatis | ATTAGATGCCGACTCGGG | 30 | This study |
| S-S-Cpn-0214-a-A-18c | C. pneumoniae | CTCTTCCTCAACCGAAAG | 30 | This study |
| S-S-Cpn-0974-a-A-18d | C. pneumoniae | AAGTCCAGGTAAGGTCCT | 30 | This study |
| S-S-Cps-1414-a-A-18 | “C. psittaci” groupe | AAGGCAAAACCAACTCCC | 30 | This study |
| S-S-Cps-1353-a-A-18 | “C. psittaci” groupe | GGCGTTATAGCTGACACG | 30 | This study |
| Bn9658 | Subgroup of the Parachlamydiaceae | TCCGTTTTCTCCGCCTAC | 10f | Amann et al. (2) |
| S-*-ParaC-0658-a-A-18 | Subgroup of the Parachlamydiaceae | TCCATTTTCTCCGTCTAC | 10f | Horn et al. (22) |
Probe designation according to Alm et al. (1); boldface type indicates the probe name as used in the text.
To be used in combination with competitor (5′-CCTCCGTATTACCGCGGC-3′).
To be used in combination with competitors (5′-CTCTCCCTCAACCGAAAG-3′ and 5′-CTCTTCCCCAACCGAAAG-3′).
To be used in combination with competitor (5′-AAACCCAGGTAAGGTCCT-3′).
C. psittaci, including C. felis, C. abortus, and C. caviae.
Formamide concentration when probes Bn9658 and S-*-ParaC-0658-a-A-18 are used simultaneously.
In situ hybridization of HeLa 229 cells and amoebae was performed using the hybridization and washing buffers described elsewhere (3, 32). The samples were rapidly air dried and examined with a Zeiss LSM 510 or a Leica TCS SP confocal laser scanning microscope. The optimal hybridization stringency was determined for each probe by increasing the formamide concentrations in the hybridization buffer in increments of 5 to 10% and by lowering the salt concentration accordingly in the wash buffers (32). The highest formamide concentration which still resulted in an intense fluorescence signal with the probe target organism was subsequently used for specificity control hybridizations with nontarget organisms of the Chlamydiales (for all probes developed, the nontarget organisms with the lowest number of mismatches were members of this order). The bacterial probe EUB338 (5′-GCTGCCTCCCGTAGGAGT-3′), targeting most but not all members of the domain Bacteria (8), was used as a positive control in all hybridization experiments. In addition, probe EUK516 (5′-ACCAGACTTGCCCTCC-3′) complementary to a unique target site on the 18S rRNA of many eukaryotes was used to counterstain the host cells.
Combination of FISH and immunofluorescence.
HeLa 229 cells fixed 48 h after infection with C. pneumoniae were used for FISH according to the protocol described above. In a second step the hybridized sample was subjected to direct fluorescence antibody staining (DFA) with FITC-labeled antibodies directed against chlamydial lipopolysaccharide (LPS) (Progen, Heidelberg, Germany). In order to prevent probe dissociation this second step was performed at room temperature in a 0.9 M sodium chloride solution. In addition, DNA was stained by DAPI in the DFA buffer.
RESULTS
Design and evaluation of oligonucleotide probes.
The newly developed hierarchical set of 16S rRNA-targeted oligonucleotide probes comprises nine species-, genus-, family-, and order-specific probes for members of the order Chlamydiales and is supplemented by two previously published probes for the Chlamydia-related Parachlamydiaceae (Fig. 1, Table 1). Application of this Chlamydiales probe set for FISH allows the specific identification of those chlamydial species that are recognized to cause human disease (including C. trachomatis, C. pneumoniae, and C. psittaci) and the simultaneous detection of chlamydia-like bacteria that have not been attributed to human and animal disease until now.
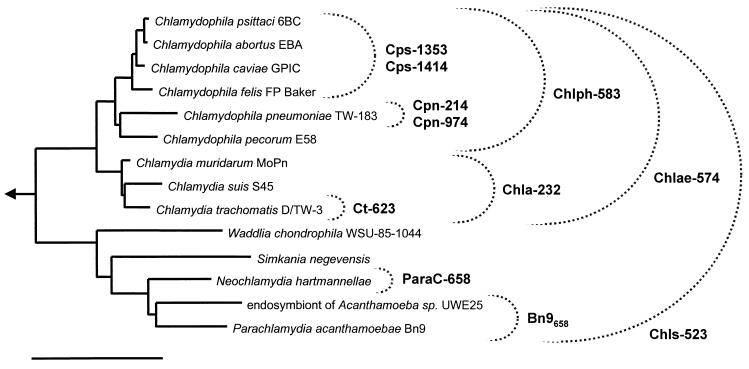
FIG. 1.
16S rRNA-based neighbor-joining tree showing the phylogenetic relations of representative members of the Chlamydiales. The specificities of oligonucleotide probes designed and tested for FISH are indicated. The taxonomy based on the reclassification of the Chlamydiales by Everett et al. (13) was used. Arrow, to outgroup; bar, 10% estimated evolutionary distance.
In order to ensure probe specificity, the optimal hybridization conditions for each probe were determined with the respective target species (Table 1). Using these conditions, each probe was subsequently tested with suitable nontarget chlamydiae (which showed the least number of mismatches with the analyzed probes). In order to enhance probe specificity, unlabeled competitor oligonucleotides corresponding to the respective target sites on the 16S rRNA of the nontarget organisms were used together with probes Cpn-974, Cpn-214, and Chls-523, respectively (Table 1). In all experiments performed (Table 2), the probes specifically hybridized with their respective target organisms and did not give positive signals with the tested nontarget organisms (Table 2). In addition, no detectable signals were obtained when the chlamydia-specific probes were applied on environmental samples (activated sludge from a municipal and an industrial wastewater treatment plant) known to contain a high bacterial diversity (27, 42).
TABLE 2.
FISH experiments demonstrating the specificity of the Chlamydiales probe set
| Probea | Intended specificity | Signal demonstrated withc:
|
|||
|---|---|---|---|---|---|
| C. pneumoniae TW 183 | C. psittaci | C. trachomatis UW-31/CX | “Parachlamydia sp.” UWE25 | ||
| Chls-523 | Chlamydiales | + | + | + | + |
| Chlae-574 | Chlamydiaceae | + | + | + | − |
| Chla-232 | Chlamydia | − | − | + | − |
| Chlph-583 | Chlamydophila | + | + | − | − |
| Ct-623 | C. trachomatis | − | − | + | − |
| Cpn-214 | C. pneumoniae | + | − | − | − |
| Cpn-974 | C. pneumoniae | + | − | − | − |
| Cps-1414 | “C. psittaci” groupb | − | + | − | − |
| Cps-1353 | “C. psittaci” groupb | − | + | − | − |
FISH was performed at the determined optimal hybridization conditions given in Table 1.
C. psittaci including C. felis, C. abortus, and C. caviae.
Symbols: +, positive hybridization signal; −, no fluorescence signal.
Since the newly designed oligonucleotide probes are complementary to different target sites on the 16S rRNA of their target organisms, they can be used simultaneously within a single FISH experiment (with only one exception: probes Chlae-574 and Chla-583 have overlapping target regions). This was exemplarily demonstrated for the identification of C. pneumoniae in HeLa 229 cells (Fig. 2), and the Parachlamydia-related strain UWE25 in amoebae (Fig. 3).
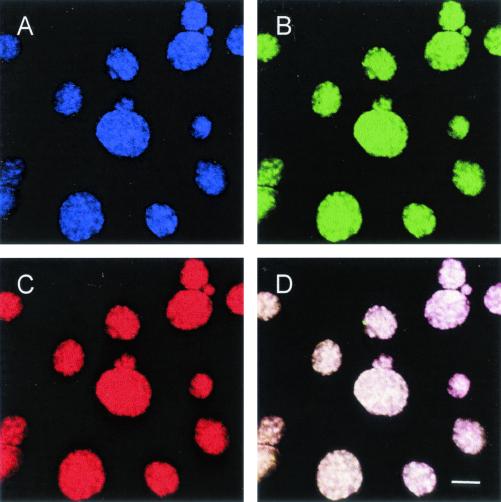
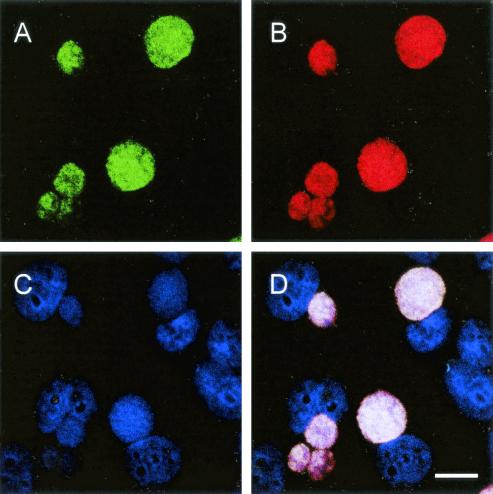
FIG. 2.
In situ identification of C. pneumoniae in HeLa 229 cells by FISH using Cy5-labeled probe Cpn-974, specific for C. pneumoniae (A) (showing an infra-red fluorescence, assigned to blue color); FLUOS-labeled probe Chlae-574, which targets all members of the family Chlamydiaceae (B) (green); and Cy3-labeled probe Chls-523, which hybridizes to all chlamydiae and chlamydia-like bacteria (C) (red). (D) Due to the overlap of colors, the chlamydial inclusions, which bound all three probes, appear white in the composite image. Bar, 10 μm.
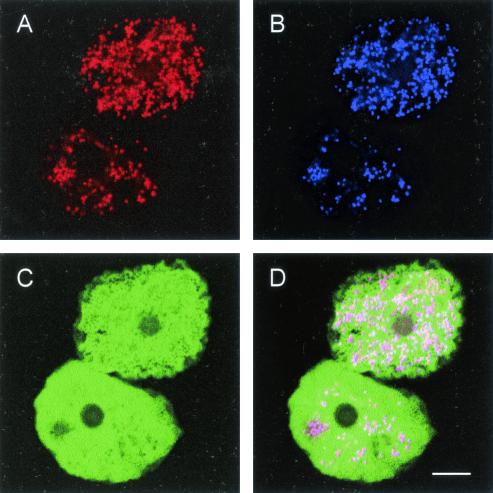
FIG. 3.
In situ identification of the chlamydia-like bacteria UWE25 within free-living amoebae by FISH using Cy3-labeled probe Bn9658 specific for a subgroup of the Parachlamydiaceae (A) (red) and Cy5-labeled probe Chls-523, which targets all chlamydiae (B) (blue). The FLUOS-labeled probe EUK516, which hybridizes to most eukaryotes (C) (green), was used to stain the amoebal host cells. (D) The chlamydiae appear purple in the overlap image. Bar, 10 μm.
Demonstration of coinfection of cell cultures with C. pneumoniae and C. trachomatis.
The simultaneous application of different oligonucleotide probes as well as their discriminatory capacity was also demonstrated in HeLa 229 cell cultures experimentally coinfected with both C. trachomatis and C. pneumoniae (Fig. 4). While C. pneumoniae inclusions were exclusively stained with the C. pneumoniae-specific probe Cpn-214, inclusions of C. trachomatis were specifically recognized by the C. trachomatis-specific probe Ct-623, demonstrating that approximately 20% of HeLa 229 cells were double infected, containing inclusions of both species.
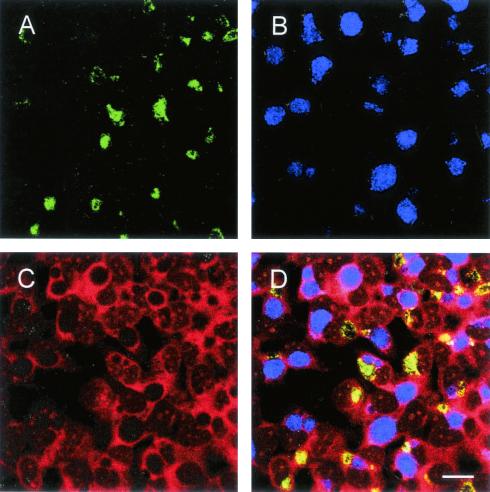
FIG. 4.
Coinfection of HeLa 229 cells with C. trachomatis and C. pneumoniae. The simultaneous application of FLUOS-labeled probe Ct-623 specific for C. trachomatis (A) (green) and Cy5-labeled probe Cpn-214 specific for C. pneumoniae (B) (blue) allows the differentiation between the two chlamydia species. The simultaneous use of the Cy3-labeled eukaryotic probe EUK516 (C) (red) clearly demonstrated that a single HeLa 229 cell can be infected with both chlamydiae (D). Bar, 10 μm.
Colocalization of chlamydial rRNA and chlamydial antigen.
A combination of FISH with immunofluorescence staining was achieved by performance of the immunofluorescence step under high salt concentration (0.9 M NaCl) subsequent to the standard FISH protocol, thus preventing the dissociation of the oligonucleotide probes during antibody binding. The combination of these two techniques was successfully applied for the detection of C. pneumoniae in HeLa 229 cells, using the C. pneumoniae-specific probe Cpn-974 and a monoclonal FITC-conjugated antibody directed against the chlamydial LPS, and demonstrated a close colocalization of rRNA-derived FISH signals and chlamydial antigen (Fig. 5).
FIG. 5.
Combination of FISH and DFA. The performance of DFA subsequent to FISH allows the identification of chlamydiae (by the rRNA-targeted oligonucleotide probes) simultaneously with the detection of chlamydial antigen (by antichlamydia antibodies). (A) Fluorescent signal derived from DFA using fluorescein-labeled antichlamydia-LPS antibodies. (B) Fluorescent signal derived from FISH with C. pneumoniae-specific probe Cpn-214 labeled with Cy3. (C) Fluorescent signal derived from DAPI staining of chlamydiae and nuclei of the HeLa 229 host cells. (D) Composite image. Bar, 10 μm.
Characterization of the C. pneumoniae developmental cycle by FISH.
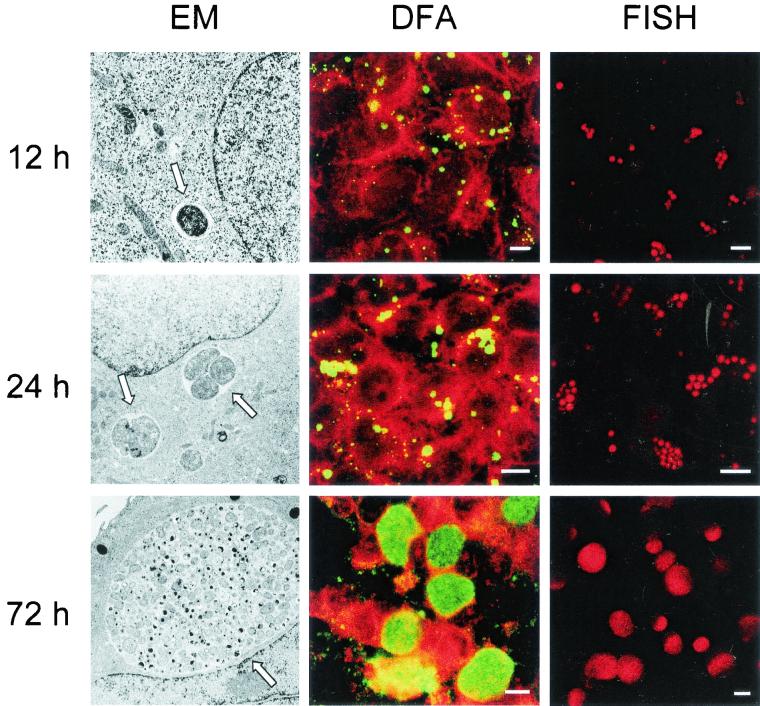
FISH and the newly developed oligonucleotide probes were successfully used to track the developmental cycle of C. pneumoniae in HeLa 229 cells. FISH using the C. pneumoniae-specific probe Cpn-214 was conducted on HeLa 229 cells infected with C. pneumoniae at 4, 6, and 12 h and subsequently every 12 h until 6 days after infection. For comparison, samples from each time point were also analyzed by electron microscopy and stained by immunofluorescence using a monoclonal antibody directed against chlamydial LPS. While intracellular elementary bodies were detected by electron microscopy already 6 h after infection, FISH was able to visualize C. pneumoniae from 12 h postinfection, when the chlamydial inclusions reached 1 to 3 μm in size, until the end of the observation period (Fig. 6). This finding is consistent with our observation that the hybridization of purified elementary bodies using fluorescently labeled oligonucleotide probes did not result in detectable fluorescent signals (data not shown), indicating that only metabolically active chlamydial forms are detected by FISH. Also DFA failed to detect the chlamydiae earlier than 12 h postinfection.
FIG. 6.
Visualization of the developmental cycle of C. pneumoniae by FISH (using Cy3-labeled probe Cpn-214; FISH column) and comparison with electron microscopy (EM column) and DFA staining (using fluorescein-labeled antichlamydia antibodies; DFA column). Three different points of time postinfection are shown. Arrows, chlamydial inclusions. Bars, 10 μm.
DISCUSSION
Despite the remarkable clinical significance of chlamydial infections, there is still a lack of satisfying approaches for laboratory diagnosis of chlamydiae. Especially for C. pneumoniae and C. psittaci one has still to rely on methods which (i) are not well standardized (nucleic acid amplification-based tests), (ii) have insufficient discriminatory power (serology-based methods), or (iii) are time-consuming and technically demanding and carry the risk of laboratory infection (culture). Consequently, there is an urgent need for alternative diagnostic approaches (10). FISH using rRNA-targeted oligonucleotide probes has become a useful diagnostic tool for detection and identification of bacteria that are slowly growing or difficult to cultivate such as L. pneumophila (17, 24) and H. pylori (37). In addition, rapid identification of bacteria in blood cultures (25, 30) and sputum samples (20) was achieved by FISH. Although Meijer and coworkers recently described the application of a digoxigenin-labeled oligonucleotide probe (fully complementary to a 16S rRNA region of 26 nucleotides in length present in most members of the Chlamydiales) for ISH of atherosclerotic tissue (33), no chlamydia-specific oligonucleotide probes with proven suitability for FISH are currently available. We therefore constructed and evaluated a comprehensive and hierarchical set of 16S rRNA-targeted oligonucleotide probes for the specific FISH detection of C. pneumoniae, C. psittaci, C. trachomatis, and all other recognized members of the Chlamydiales, including the so-called environmental chlamydiae (21). The last of these groups encompasses more than five novel evolutionary lineages of chlamydia-like bacteria which were discovered during the past 4 years (2, 15, 23, 31, 36). In addition, there is increasing evidence for an even greater diversity of chlamydiae in the environment (21, 35). While the clinical significance of these only recently detected Chlamydia-related bacteria is still unclear, some of them have been associated with respiratory disease in humans (7, 14, 28, 35; R. J. Birtles, T. J. Rowbotham, C. Storey, T. J. Marrie, and D. Raoult, Letter, Lancet 349:925-926, 1997). Using FISH and the new probe set, a new tool becomes available facilitating a rapid detection of all known chlamydiae and chlamydia-like bacteria, which might help to clarify their role in disease of humans.
The simultaneous application of multiple probes with hierarchical specificity labeled with different dyes increases the reliability of the identification, since nontarget organisms incidentally possessing the target site for one of the probes would not hybridize with the other probes and would thus be characterized by unexpected hybridization patterns. Furthermore, the application of multiple probes labeled with different dyes minimizes the risk of false-positive signals caused by unspecific binding of the fluorescent dyes to nontarget organisms or structures of the host cells. For the new Chlamydiales probe set, the successful application of multiple probes was, for example, demonstrated for the identification of C. pneumoniae in experimentally infected HeLa 229 cells by using the C. pneumoniae-specific probe Cpn-974 and the taxonomically superordinated probes Chlae-574, specific for the family Chlamydiaceae, and Chls-523, which targets all known members of the order Chlamydiales, including the recently discovered chlamydia-like bacteria (Fig. 2).
The newly developed oligonucleotide probes also allowed us to clearly differentiate between C. psittaci and C. pneumoniae. To our knowledge there is no monoclonal antibody available that enables specific staining of C. psittaci grown from human as well as from veterinary samples. Unambiguous identification of the highly infectious C. psittaci is, however, important for patient management as well as for laboratory safety considerations and is currently based mainly on PCR methods or genotyping (12). FISH using the C. psittaci-specific 16S rRNA-targeted oligonucleotide probes might offer a rapid alternative for reliable identification of C. psittaci. However, it should be noted that the C. psittaci-specific probes designed in this study target all chlamydiae formerly referred to as Chlamydia psittaci, including the recently described species Chlamydophila felis, Chlamydophila abortus, and Chlamydophila caviae.
In order to visualize the specificity and discriminatory power of the newly designed FISH probes, HeLa 229 cells were simultaneously infected with C. pneumoniae and C. trachomatis. The subsequent application of FISH using C. pneumoniae and C. trachomatis-specific oligonucleotide probes was able to clearly discriminate C. pneumoniae-derived inclusions from C. trachomatis-derived inclusions and demonstrated for the first time that the same HeLa 229 cells can be infected simultaneously with both chlamydia species. The coexistence of related pathogens in the same host cell might facilitate lateral gene transfer events between them. However, fusion of C. pneumoniae inclusions with C. trachomatis inclusions was never observed in the double-infected cells.
The applicability of FISH for basic chlamydia research as well as for diagnostic purposes was further demonstrated by the analysis of the complete developmental cycle of C. pneumoniae, which revealed that chlamydiae can be detected by FISH as early as 12 h after infection. It was, however, not possible to stain purified EB by FISH. This could result either from a low ribosome content of the EB during this metabolically inactive stage of the developmental cycle or from the inaccessibility of the EB for oligonucleotide probes due to their rigid cell wall that is highly cross-linked by disulfide bonds.
A previous study revealed a similar sensitivity of FISH and culture-based methods for the detection of Pseudomonas aeruginosa from sputa of children with cystic fibrosis (20). However, the sensitivity of FISH for the detection of chlamydiae in patient samples has still to be investigated in detail in further studies. While the restriction of FISH to detect only the metabolically active forms of chlamydiae might be a disadvantage in terms of the sensitivity of FISH in clinical diagnostics, FISH might be able to circumvent several problems of PCR-based diagnostic methods. These are for example illustrated by the highly discrepant results for PCR detection of C. pneumoniae in atheromatous lesions by different study groups spanning positivity rates from 0 to 100%, which have contributed to the current confusion about the role of C. pneumoniae in development of atherosclerosis (4). The possibility to apply FISH directly on the specimens (in situ), thereby detecting only intact and viable cells (in contrast to PCR, which is susceptible to false-positive results due to the presence of dead bacteria or extracellular DNA), and the encompassing coverage of all chlamydiae render FISH a promising tool for the investigation of blood vessels for the presence of metabolically active C. pneumoniae or chlamydia-like bacteria. In addition, FISH might help to improve the laboratory diagnosis of, in particular, C. pneumoniae and C. psittaci infections.
Acknowledgments
This work was supported by a grant of the Sonderforschungsbereich (grant 451) to R.M. and A.E.; a grant of the University of Ulm, Forschungsförderung, to S.P.; and a grant from the BMB+F (German ministry for education and science) within the framework of the CAPNETZ program to M.W. and M.H.
We gratefully acknowledge the excellent technical assistance of Ulrike Simnacher and Sonja Weiss. We also acknowledge the expert technical support and advice of the department of electron microscopy, namely, Rainer Bleher and Monika Hagedorn.
S.P. and A.E. contributed equally to this work.
REFERENCES
- 1.Alm, E. W., D. B. Oerther, N. Larsen, D. A. Stahl, and L. Raskin. 1996. The oligonucleotide probe database. Appl. Environ. Microbiol. 62:3557-3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1a.Amann, R., N. Springer, W. Ludwig, H. D. Görtz, and K. H. Schleifer. 1991. Identification in situ and phylogeny of uncultured bacterial endosymbionts. Nature 351:161-164. [DOI] [PubMed] [Google Scholar]
- 2.Amann, R., N. Springer, W. Schönhuber, W. Ludwig, E. Schmid, K. Müller, and R. Michel. 1997. Obligate intracellular bacterial parasites of acanthamoebae related to Chlamydia spp. Appl. Environ. Microbiol. 63:115-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amann, R., W. Ludwig, and K.-H. Schleifer. 1995. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 59:143-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Apfalter, P., F. Blasi, J. Boman, C. A. Gaydos, M. Kundi, M. Maass, A. Makristathis, A. Meijer, R. Nadrchal, K. Persson, M. L. Rotter, C. Y. Tong, G. Stanek, and A. M. Hirschl. 2001. Multicenter comparison trial of DNA extraction methods and PCR assays for detection of Chlamydia pneumoniae in endarterectomy specimens. J. Clin. Microbiol. 39:519-524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brinke, A., T. J. Van Dissel, P. J. Sterk, A. H. Zwinderman, K. F. Rabe, and E. H. Bel. 2001. Persistent airflow limitation in adult-onset nonatopic asthma is associated with serologic evidence of Chlamydia pneumoniae infection. J. Allergy Clin. Immunol. 107:449-54. [DOI] [PubMed] [Google Scholar]
- 6.Bush, R. M., and K. D. Everett. 2001. Molecular evolution of the Chlamydiaceae. Int. J. Syst. E vol. Microbiol. 51:203-220. [DOI] [PubMed] [Google Scholar]
- 7.Corsaro, D., D. Venditti, A. L. Faou, P. Guglielmetti, and M. Valassina. 2001. A new chlamydia-like 16S rDNA sequence from a clinical sample. Microbiology 147:515-516. [DOI] [PubMed] [Google Scholar]
- 8.Daims, H., K.-H. Schleifer, and M. Wagner. 1999. Probe EUB338 is insufficient for the detection of all Bacteria: development and evaluation of a more comprehensive probe set. Syst. Appl. Microbiol. 22:438-448. [DOI] [PubMed] [Google Scholar]
- 9.Danesh, J., P. Whincup, M. Walker, L. Lennon, A. Thomson, P. Appleby, Y. Wong, M. Bernardes-Silva, and M. Ward. 2000. Chlamydia pneumoniae IgG titres and coronary heart disease: prospective study and meta-analysis. BMJ 321:208-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dowell, S. F., R. W. Peeling, J. Boman, G. M. Carlone, B. S. Fields, J. Guarner, M. R. Hammerschlag, L. A. Jackson, C. C. Kuo, M. Maass, T. O. Messmer, D. F. Talkington, M. L. Tondella, and S. R. Zaki. 2001. Standardizing Chlamydia pneumoniae assays: recommendations from the Centers for Disease Control and Prevention (USA) and the Laboratory Centre for Disease Control (Canada). Clin. Infect. Dis. 33:492-503. [DOI] [PubMed] [Google Scholar]
- 11.Essig, A., M. Heinemann, R. Schweitzer, U. Simnacher, and R. Marre. 2000. Decontamination of a mycoplasma-infected Chlamydia pneumoniae strain by pulmonary passage in SCID mice. Int. J. Med. Microbiol. 290:289-92. [DOI] [PubMed] [Google Scholar]
- 12.Essig, A., P. Zucs, M. Susa, G. Wasenauer, U. Mamat, M. Hetzel, U. Vogel, S. Wieshammer, H. Brade, and R. Marre. 1995. Diagnosis of ornithosis by cell culture and polymerase chain reaction in a patient with chronic pneumonia. Clin. Infect. Dis. 21:1495-1497. [DOI] [PubMed] [Google Scholar]
- 13.Everett, K. D., R. M. Bush, and A. A. Andersen. 1999. Emended description of the order Chlamydiales, proposal of Parachlamydiaceae fam. nov. and Simkaniaceae fam. nov., each containing one monotypic genus, revised taxonomy of the family Chlamydiaceae, including a new genus and five new species, and standards for the identification of organisms. Int. J. Syst. Bacteriol. 49:415-440. [DOI] [PubMed] [Google Scholar]
- 14.Friedman, M. G., A. Galil, S. Greenberg, and S. Kahane. 1999. Seroprevalence of IgG antibodies to the chlamydia-like microorganism ‘Simkania Z' by ELISA. Epidemiol. Infect. 122:117-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fritsche, T. R., M. Horn, M. Wagner, R. P. Herwig, K.-H. Schleifer, and R. K. Gautom. 2000. Phylogenetic diversity among geographically dispersed Chlamydiales endosymbionts recovered from clinical and environmental isolates of Acanthamoeba spp. Appl. Environ. Microbiol. 66:2613-2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fritsche, T. R., M. Horn, S. Seyedirashti, R. K. Gautom, K.-H. Schleifer, and M. Wagner. 1999. In situ detection of novel bacterial endosymbionts of Acanthamoeba spp. phylogenetically related to members of the Rickettsiales. Appl. Environ. Microbiol. 65:206-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grimm, D., H. Merkert, W. Ludwig, K.-H. Schleifer, J. Hacker, and B. C. Brand. 1998. Specific detection of Legionella pneumophila: construction of a new 16S rRNA-targeted oligonucleotide probe. Appl. Environ. Microbiol. 64:2686-2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hammerschlag, M. R. 2000. Chlamydia pneumoniae and the lung. Eur. Respir. J. 16:799-800. [DOI] [PubMed] [Google Scholar]
- 19.Herrmann, B., B. Pettersson, K. D. Everett, N. E. Mikkelsen, and L. A. Kirsebom. 2000. Characterization of the rnpB gene and RNase P RNA in the order Chlamydiales. Int. J. Syst. E vol. Microbiol. 50:149-58. [DOI] [PubMed] [Google Scholar]
- 20.Hogardt, M., K. Trebesius, A. M. Geiger, M. Hornef, J. Rosenecker, and J. Heesemann. 2000. Specific and rapid detection by fluorescent in situ hybridization of bacteria in clinical samples obtained from cystic fibrosis patients. J. Clin. Microbiol. 38:818-825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Horn, M., and M. Wagner. 2001. Evidence for additional genus-level diversity of Chlamydiales in the environment. FEMS Microbiol. Lett. 204:71-74. [DOI] [PubMed] [Google Scholar]
- 22.Horn, M., M. Wagner, K.-D. Müller, E. N. Schmid, T. R. Fritsche, K. H. Schleifer, and R. Michel. 2000. Neochlamydia hartmannellae gen. nov., sp. nov. (Parachlamydiaceae), an endoparasite of the amoeba Hartmannella vermiformis. Microbiology 146:1231-1239. [DOI] [PubMed] [Google Scholar]
- 23.Horn, M., T. R. Fritsche, R. K. Gautom, K.-H. Schleifer, and M. Wagner. 1999. Novel bacterial endosymbionts of Acanthamoeba spp. related to the Paramecium caudatum symbiont Caedibacter caryophilus. Environ. Microbiol. 1:357-367. [DOI] [PubMed] [Google Scholar]
- 24.Hu, J., A. P. Limaye, M. Horn, S. Juretschko, R. Gautom, and T. R. Fritsche. 2002. Direct detection of legionellae in respiratory tract specimens by using fluorescence in situ hybridization, p. 221-224. In R. Marre, Y. Abu Kwaik, C. Bartlett, N. P. Cianciotto, B. S. Fields, M. Frosch, J. Hacker, and P. C. Lück (ed.), Legionella. ASM Press, Washington, D.C.
- 25.Jansen, G. J., M. Mooibroek, J. Idema, H. J. Harmsen, G. W. Welling, and J. E. Degener. 2000. Rapid identification of bacteria in blood cultures by using fluorescently labeled oligonucleotide probes. J. Clin. Microbiol. 38:814-817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Juretschko, S., G. Timmermann, M. Schmid, K.-H. Schleifer, A. Pommerening-Röser, H.-P. Koops, and M. Wagner. 1998. Combined molecular and conventional analyses of nitrifying bacterium diversity in activated sludge: Nitrosococcus mobilis and Nitrospira-like bacteria as dominant population. Appl. Environ. Microbiol. 64:3042-3051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Juretschko, S., A. Loy, A. Lehner, and M. Wagner. 2002. The microbial community composition of a nitrifying-denitrifying activated sludge from an industrial sewage treatment plant analyzed by the full-cycle rRNA approach. Syst. Appl. Microbiol. 25:84-99. [DOI] [PubMed] [Google Scholar]
- 28.Kahane, S., D. Greenberg, M. G. Friedman, H. Haikin, and R. Dagan. 1998. High prevalence of “Simkania Z,” a novel Chlamydia-like bacterium, in infants with acute bronchiolitis. J. Infect. Dis. 177:1425-1429. [DOI] [PubMed] [Google Scholar]
- 29.Kahane, S., K. D. Everett, N. Kimmel, and M. G. Friedman. 1999. Simkania negevensis strain ZT: growth, antigenic and genome characteristics. Int. J. Syst. Bacteriol. 49:815-820. [DOI] [PubMed] [Google Scholar]
- 30.Kempf, V. A., K. Trebesius, and I. B. Autenrieth. 2000. Fluorescent In situ hybridization allows rapid identification of microorganisms in blood cultures. J. Clin. Microbiol. 38:830-838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lieberman, D., S. Kahane, D. Lieberman, and M. G. Friedman. 1997. Pneumonia with serological evidence of acute infection with the Chlamydia-like microorganism ‘Z.’ Am. J. Respir. Crit. Care Med. 156:578-582. [DOI] [PubMed] [Google Scholar]
- 32.Manz, W., R. Amann, W. Ludwig, M. Wagner, and K.-H. Schleifer. 1992. Phylogenetic oligodeoxynucleotide probes for the major subclasses of Proteobacteria: problems and solutions. Syst. Appl. Microbiol. 1:593-600. [Google Scholar]
- 33.Meijer, A., P. J. M. Roholl, S. K. Gielis Proper, Y. F. Meulenberg, and J. M. Ossewaarde. 2000. Chlamydia pneumoniae in vitro and in vivo: a critical evaluation of in situ detection methods. J. Clin. Pathol. 53:904-910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mendell, M. A., D. Carrington, D. Strachan, P. Patel, N. Molineaux, J. Levi, T. Toosey, A. J. Camm, and T. C. Northfield. 1995. Chlamydia pneumoniae: risk factors for seropositivity and association with coronary heart disease. J. Infect. 30:121-128. [DOI] [PubMed] [Google Scholar]
- 35.Ossewaarde, J. M., and A. Meijer. 1999. Molecular evidence for the existence of additional members of the order Chlamydiales. Microbiology 145:411-417. [DOI] [PubMed] [Google Scholar]
- 36.Rurangirwa, F. R., P. M. Dilbeck, T. B. Crawford, T. C. McGuire, and T. F. McElwain. 1999. Analysis of the 16S rRNA gene of microorganism WSU 86-1044 from an aborted bovine foetus reveals that it is a member of the order Chlamydiales: proposal of Waddliaceae fam. nov., Waddlia chondrophila gen. nov., sp. nov. Int. J. Syst. Bacteriol. 49:577-581. [DOI] [PubMed] [Google Scholar]
- 37.Rüssmann, H., V. A. J. Kempf, S. Koletzko, J. Heesemann, and I. B. Autenrieth. 2001. Comparison of fluorescent in situ hybridization and conventional culturing for detection of Helicobacter pylori in gastric biopsy specimens. J. Clin. Microbiol. 39:304-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Saikku, P. 1999. Epidemiology of Chlamydia pneumoniae in atherosclerosis. Am. Heart J. 138:S500-S503. [DOI] [PubMed] [Google Scholar]
- 39.Saikku, P., M. Leinonen, K. Mattila, M. R. Ekman, M. S. Nieminen, P. H. Makela, J. K. Huttunen, and V. Valtonen. 1988. Serological evidence of an association of a novel Chlamydia, TWAR, with chronic coronary heart disease and acute myocardial infarction. Lancet ii:983-986. [DOI] [PubMed] [Google Scholar]
- 40.Schachter, J., and W. E. Stamm. 1999. Chlamydia, p. 669-677. In P. R. Murray, E. J. Baron, M. A. Pfaller, F. C. Tenover, and R. H. Yolken/ASM (ed.), Manual of clinical microbiology, 7th ed. Press, Washington, D.C.
- 41.Schmid, M., U. Twachtmann, M. Klein, M. Strous, S. Juretschko, M. S. M. Jetten, J. W. Metzger, K.-H. Schleifer, and M. Wagner. 2000. Molecular evidence for genus level diversity of bacteria capable of catalyzing anaerobic ammonium oxidation. Syst. Appl. Microbiol. 23:93-106. [DOI] [PubMed] [Google Scholar]
- 42.Snaidr, J., R. Amann, I. Huber, W. Ludwig, and K.-H. Schleifer. 1997. Phylogenetic analysis and in situ identification of bacteria in activated sludge. Appl. Environ. Microbiol. 63:2884-2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stephens, R. S. (ed.). 1999. Chlamydia: intracellular biology, pathogenesis, and immunity. American Society for Microbiology, Washington, D.C.