Abstract
A 613-bp fragment of an essential ketosynthase gene from the biosynthetic pathway of aromatic polyketide antibiotics was sequenced from 99 actinomycetes isolated from soil. Phylogenetic analysis showed that the isolates clustered into clades that correspond to the various classes of aromatic polyketides. Additionally, sequencing of a 120-bp fragment from the γ-variable region of 16S ribosomal DNA (rDNA) and subsequent comparative sequence analysis revealed incongruity between the ketosynthase and 16S rDNA phylogenetic trees, which strongly suggests that there has been horizontal transfer of aromatic polyketide biosynthesis genes. The results show that the ketosynthase tree could be used for DNA fingerprinting of secondary metabolites and for screening interesting aromatic polyketide biosynthesis genes. Furthermore, the movement of the ketosynthase genes suggests that traditional marker molecules like 16S rDNA give misleading information about the biosynthesis potential of aromatic polyketides, and thus only molecules that are directly involved in the biosynthesis of secondary metabolites can be used to gain information about the biodiversity of antibiotic production in different actinomycetes.
Soil actinomycetes, especially those that belong to the genus Streptomyces, have been the focus of intensive research for the past several decades. The interest in Streptomyces arose from the finding that this group of bacteria seems to have the ability to produce a large variety of different bioactive compounds that have a wide spectrum of activity. From the 1950s to the mid-1970s numerous new bioactive molecules were discovered through large screening programs, and these molecules subsequently found their way into various clinical uses ranging from control of infections to cancer treatment (21).
In more recent years, modern high-throughput screening methods have exponentially increased the number of strains screened annually, but the number of novel compounds discovered has not increased in the same proportion. One of the many reasons, presumably the most important one, for this problem is that old molecules (and strains) are being rediscovered with the screening procedures that are in use today (21).
In a previous paper (18) Metsä-Ketelä et al. reported a method that could be used for preliminary classification of strains on the basis of their genetic abilities to produce various compounds belonging to the aromatic polyketide group. This method is based on PCR amplification of a gene fragment that is essential in the biosynthesis pathway of aromatic polyketides and on analysis of the amplified regions by phylogenetic methods. The degenerate primers designed for this purpose amplify a portion of a ketosynthase gene (KSα), which in collaboration with KSβ and an acyl carrier protein condenses small carboxylic acids in a stepwise manner to form a long polyketide chain that is subsequently folded into a range of different aromatic compounds by various ketoreductases, cyclases, and aromatases. Later, the molecule formed is often further modified by diverse oxygenases, methylases, and reductases. Finally, addition of a deoxysugar residue(s) to the molecule gives rise to further diversity within this group of compounds, which include molecules like anthracyclines, naphthaquinone and angucyclines.
In this study we set out to extend our analysis of KSα fragments to investigate if the results for a larger number of samples would supplement our previous results so that different aromatic polyketide classes could be identified from a phylogenetic tree. This would open up the possibility of using the results for DNA fingerprinting of secondary metabolites and as the basis for a search for attractive antibiotic biosynthesis genes that could be used in many applications, the most significant of which can be considered to be combinatorial biosynthesis (9). Moreover, amplified KSα fragments can be used as homologous hybridization probes, which significantly facilitate the cloning of antibiotic biosynthesis gene clusters.
Second, we also wished to address the question of whether a phylogenetic tree calculated from the KSα fragments represents the evolution of the bacterial species or only the molecular evolution of the antibiotic biosynthesis gene(s). To investigate this, we set out to compare the KSα tree to a tree derived from a traditional marker molecule that reflects the evolution of the species; dissimilar phylogenies would suggest that there was independent evolution of aromatic polyketides. The small ribosomal subunit (16S ribosomal DNA [rDNA]) was chosen to represent the evolution of the bacteria (15), since previous reports have indicated that a partial nucleotide sequence of the 16S rDNA molecule containing the γ-variable region has sufficient information content to classify representatives of the genus Streptomyces into different groups (10), albeit with a relatively large error frequency (2%) due to the heterogeneity of Streptomyces 16S rRNA (23). In this paper we describe a phylogenetic analysis of partial 16S rRNA and KSα gene sequences from 12 Streptomyces type strains and 87 soil isolates.
MATERIALS AND METHODS
Bacterial strains and cultivation of bacteria.
The bacterial strains used in this study were isolated from soil samples that were collected from various locations around Europe. The soil samples were diluted in water and cultivated on plates containing ISP4 (Difco, Detroit, Mich.), which is a medium commonly used to isolate Streptomyces and closely related genera from soil. Colonies with distinct morphological features (consistency of colonies, melanin production, sporulation, and color of mycelium, spores, and secondary metabolites) were selected from the plates and maintained on ISP4. Strains were designated by using the following scheme: the first one to three letters indicate the isolation location, the last letter (A to G) indicates the designation of the soil sample from a given location, and the numbers indicate different strains isolated from a given soil sample. The strains used in this study were randomly selected from a larger collection of strains isolated as described above.
DNA template preparation and PCR amplification of 16S rDNA and KSα gene fragments.
The templates for PCRs were generated as follows. Fresh mycelium (20 μl) was scraped from ISP4 plates, and sterile H2O was added to bring the final volume to 200 μl. The samples were incubated in a boiling water bath for 10 min, after which they were placed on ice. One microliter of a boiled solution was used as a template in a PCR. The 16S rDNA was amplified with primers 5′-AGA GTT TGA TCC TGG CTC AG-3′ (sense primer) and 5′ GCG CTT TTT GAG ATT CGC TC-3′ (antisense primer) in a 25-μl reaction mixture that contained 25 pmol of each primer, 1 μl of a preparation containing each deoxynucleoside triphosphate at a concentration of 2.5 mM, 0.8 μl of dimethyl sulfoxide, 2.5 μl of 10× Dynazyme Ext reaction buffer, and 0.7 U of proofreading Dynazyme Ext DNA polymerase (Finnzymes, Espoo, Finland). The PCR was started with a long denaturation phase (99°C for 8 min) before addition of the polymerase to ensure separation of GC-rich DNA strands. A total of 30 cycles were used, and each cycle consisted of denaturation at 96°C for 1 min, annealing at 55°C for 1 min, and extension at 72°C for 1.5 min. An additional 8.5-min extension step was performed after the last cycle to promote complete extension of the products. The amplified area corresponds to bp 1 to 1273 in Streptomyces griseus 16S RNA (12). The KSα fragment was amplified with degenerate primers 5′-TSG CST GCT TGG AYG CSA TC-3′ (sense primer) and 5′-TGG AAN CCG CCG AAB CCG CT-3′ as described previously (18). A PCR product of the correct size (613 bp) was obtained from 80% of the strains tested with the KSα primers.
Cloning and sequencing of the amplified PCR fragments.
The amplified 16S rDNA fragments were purified with a High Pure PCR product purification kit (Roche Molecular, Basel, Switzerland) and were sequenced with the same sense primer that was used for PCR amplifications with an ABI 310 genetic analyzer (Applied Biosystems, Foster City, Calif.) by using Big Dye terminator chemistry, version 2.0, according to the manufacturer's instructions. The KSα fragments were purified, cloned, and sequenced as described previously (18).
Analysis of DNA sequences.
Sequence analysis and multiple alignment were performed with the programs in the Vector NTI Suite program package (version 7.0; Informax Inc., Bethesda, Md.). Phylogenies and bootstrap values were calculated with ClustalX, version 1.81, and phylogenetic trees were drawn with Treeview.
RESULTS
Analysis of the 16S rDNA tree.
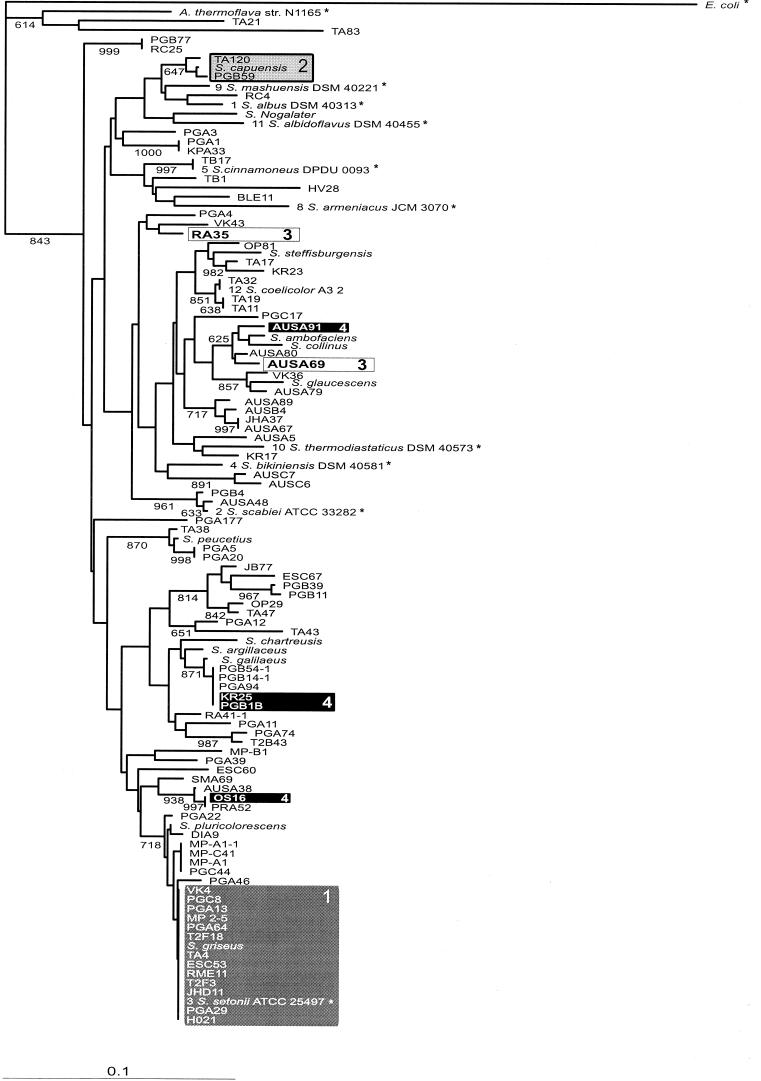
The gene encoding the small ribosomal subunit was amplified from 87 unidentified actinomycete isolates and 12 Streptomyces type strains as described above. Comparison of the sequences with public databases revealed that 85 of the unidentified soil samples belonged to the genus Streptomyces. The remaining two strains, TA21 and TA83, were identified as members of the genera Saccharothrix and Nocardiopsis, respectively. A 194-bp fragment that contains the highly variable γ-region, which accounts for roughly 30% to the variation found in different Streptomyces 16S rRNA sequences (12), was used to construct a multiple alignment and a phylogenetic tree, which is shown in Fig. 1. The area analyzed corresponds to bp 84 to 280 in the S. coelicolor A3 (2) rrnD 16S RNA sequence (GenBank accession number Y00411).
FIG.1.
Phylogenetic tree constructed from partial 16S rDNA sequences. Bootstrap values greater than 60% are shown at the nodes (1,000 = 100%). Scale bar = 10% dissimilarity. Sequences marked with an asterisk were obtained from Ribosomal Database Project II (14). The numbers in front of the Ribosomal Database Project II Streptomyces strains (1 to 12) are the numbers used in the classification system (classes 6 and 7 were omitted as representatives of these classes contained only sequences that had ambiguous characters). The nucleotide sequence accession numbers for the reference strains are as follows: S. albus, X53163; S. thermodiastaticus, Z68101; S. albidoflavus, Z76685; S. coelicolor, Y00411; S. scabies, AB026206; S. setonii, D63872; S. bikiniensis, X79851; S. cinnamoneus, X53171; S. armeniacus, AB018094; S. mashuense, X79323; Amycolatopsis thermoflava, AF052390; and E. coli, AF233451.
In addition, 10 Streptomyces 16S rDNA sequences that represent different Streptomyces groups in Ribosomal Database Project II (16) were added to the analysis, as was an Escherichia coli 16S rDNA sequence that was used as an outgroup in the analysis. One sequence from a closely related gram-positive non-Streptomyces species, which was used to distinguish Streptomyces species from other closely related taxa, was also added to the analysis. Because of the relatively large data set (111 sequences), the distance method (8) was selected as the method used to calculate the phylogenies from the sequences with the program ClustalX.
Bootstrap analysis of the tree (1,000 rounds) revealed that all of the phylogenetic relationships were not fully resolved by using only partial 16S rDNA sequences. This is not very surprising as it is becoming evident that a polyphasic approach that takes into account both genotypic and phenotypic traits is likely to be needed to resolve all of the intraspecies relationships among various Streptomyces strains (1). However, several clearly separable clades were formed within the tree, and the strains found in these branches can be compared to the strains in the KSα tree to investigate the congruence of the two trees.
Analysis of the polyketide synthase (PKS) tree.
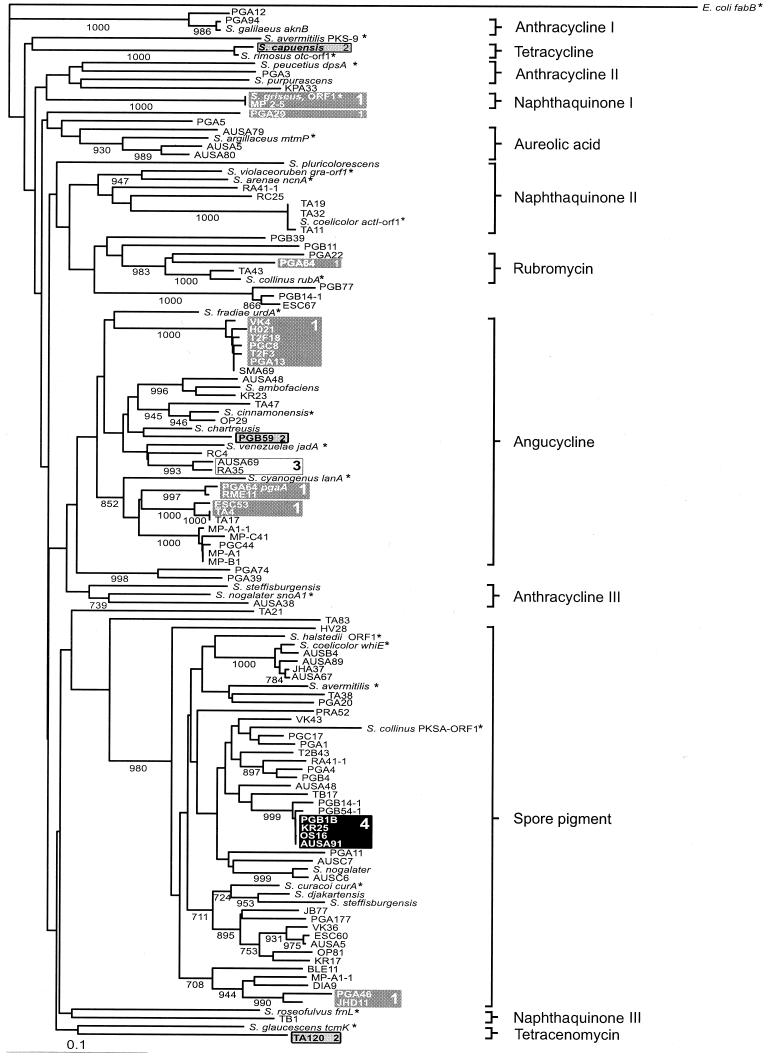
The amino acid sequences encoded by 17 known type II polyketide antibiotic biosynthesis gene fragments and four polyketide spore pigment gene fragments were used as a basis for classification of unknown samples. The sequences obtained from the 87 unknown actinomycete isolates and 12 Streptomyces type strains were also added to the analysis (Fig. 2). Two different sequences were obtained from eight strains, as described previously (18); seven strains contained one antibiotic sequence and one spore pigment sequence, and one strain contained two antibiotic sequences. The Dayhoff (5) PAM350 distance matrix was used to construct a multiple alignment by the distance method, and a phylogenetic tree was constructed by the neighbor method of Saitou and Nei (20) by using 1,000 rounds in a bootstrap analysis. The distantly related fabB gene from the fatty acid biosynthesis pathway of E. coli (11) was used as an outgroup.
FIG.2.
Phylogenetic tree constructed from KSα gene fragments. Bootstrap values greater than 70% are shown at the nodes (1,000 = 100%). Scale bar = 10% dissimilarity. Sequences marked with an asterisk were derived from the GenBank database. The sequences obtained from strains S. galilaeus, S. nogalater, S. coelicolor, S. argillaceus, and S. peucetius in this work were identical to the sequences deposited in the GenBank database. The nucleotide sequence accession numbers for the reference strains are as follows: S. galilaeus, AF257324; S. nogalater, AJ224512; S. coelicolor, X63449 (act) and X55942 (whiE); S. halstedii, L05390; S. avermitilis, AB070937 (spore pigment) and AB070947 (pks-9); S. collinus, AF293354 (PKSA ORF1) and AF293355 (rub); S. curacoi, M33704; S. roseofulvus, L26338; S. glaucescens, M80674; S. violaceoruber, X16144; S. arenae, AF098965; S. argillaceus, X89899; S. peucetius, L35560; S. rimosus, Z25538; S. griseus, X77865; S. fradiae, X87093; S. cinnamonensis, Z11511; S. venezuelae, L33245; and S. cyanogenus, AF080235.
Antibiotic biosynthesis genes occur in separate branches based on the antibiotic produced.
In another study, it was found that an amplified fragment of the KSα gene could be used to separate spore pigment and antibiotic biosynthesis genes and that the antibiotic biosynthesis genes could be placed in separate branches on the basis of the starter unit used in biosynthesis (18). Here, the much larger data set and the existence of many more confirmed antibiotic-producing KSα gene sequences in the GenBank database enabled us to construct a tree that shows the separation of different antibiotic groups in distinct branches in many cases.
The most clearly separated group, which is also the largest group, contains genes involved in the biosynthesis of angucycline antibiotics (Fig. 3A). The clade in Fig. 2 that includes sequences from S. fradiae urdA to Streptomyces sp. strain B1 contains all of the genes that are known to be responsible for the biosynthesis of angucyclines (namely, the gene fragments from S. fradiae, S. venezuelae, and S. cyanogenus). Moreover, the gene clusters of Streptomyces sp. strain PGA64 (GenBank accession number AY034378) and Streptomyces sp. strain H021 (unpublished results) harbor genes that are homologous to jadI from S. venezuelae, which has been shown to be responsible for the cyclization of the fourth angular ring in jadomycin biosynthesis (13). Genes similar to the genes encoding the cyclases have not been identified in other antibiotic-producing gene clusters.
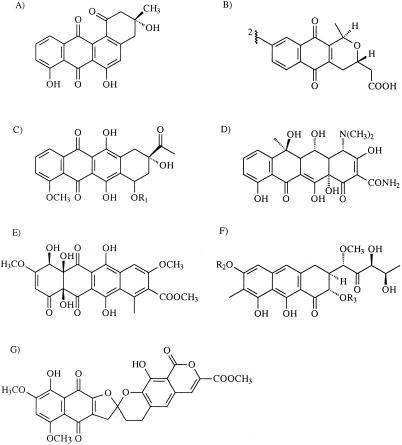
FIG. 3.
Structures of compounds that represent different classes of aromatic polyketides. (A) Angucycline antibiotic rabelomycin; (B) naphthaquinone antibiotic actinorhodin; (C) anthracycline antibiotic daunomycin; (D) tetracycline antibiotic oxytetracycline; (E) tetracenomycin C; (F) aureolic acid antibiotic mithramycin; and (G) rubromycin β. R1, R2, and R3 refer to sugar moieties, which are not derived from the PKS pathway.
Another identifiable group is the group that spans from S. violaceoruber to Streptomyces sp. strain TA11. This group contains three naphthaquinone antibiotic-producing (Fig. 3B) strains, S. violaceoruber, S. arenae, and S. coelicolor. The other two strains that produce naphthaquinone antibiotics, S. griseus and S. roseofulvus, both form their own branches on the tree. However, separation is feasible in the sense that griseusin, which is produced by S. griseus, is formed from a longer polyketide chain (20 carbons) than the other naphthaquinones, which have a polyketide backbone consisting of 18 carbons (26). Similarly, the gene cluster of S. roseofulvus is responsible for the production of two polyketides with different lengths, nanaomycin (C16) and frenolicin (C18) (3).
Anthracycline (Fig. 3C) producers also formed three different branches on the tree; S. galilaeus and Streptomyces sp. strain PGA12 migrated separately from S. peucetius and S. purpurascens. The other anthracycline producers that are represented in the KSα tree, S. nogalater and S. steffisburgensis, formed a third group. It is somewhat surprising to find that the phylogenetic distance between the aclacinomysin producer S. galilaeus and two other anthracycline producers, S. peucetius and S. purpurascens, is as great as the phylogenetic analysis indicates since the biosynthesis pathways of the antibiotics produced by these three organisms proceed through a common intermediate compound, aklavinone (22). In order to see if the distribution of anthracycline producers on various branches was due to the fact that only partial sequences of the KSα gene were used in the analysis, we compared two trees constructed from whole and partial amino acid sequences that were available in the GenBank database. However, the trees derived from whole and partial sequences agreed very well with each other (data not shown), and the early division of anthracycline producers could also be seen on the tree constructed with whole sequences.
Tetracycline, tetracenomycin, aureolic acid, and rubromycin producers (Fig. 3D, E, F, and G, respectively) also were placed on unique branches by the analysis. However, because there is only one known sequence from the producers of each of these compounds, caution should be used in making predictions with these groups of molecules. It is tempting to conclude that the branch that includes the sequences from Streptomyces sp. strain PGA 22 to S. collinus contains sequences that are responsible for the biosynthesis of rubromycins or similar compounds, because in this group Streptomyces sp. strain PGA 64 and S. collinus, from which the genes responsible for rubromycin biosynthesis have recently been cloned (17), are known to produce rubromycins. A variety of sequences also cluster around the aureolic acid producer S. argillaceus, but it is not possible to propose boundaries for this group because there is no information about the compounds produced by the surrounding strains. The oxytetracyline producers S. rimosus and S. capuensis group together, as observed previously (18), but no other strains similar to these organisms were found in the strains analyzed. Finally, no genes homologous to the genes of the tetracenomycin producer S. glaucescens were discovered.
The sequences involved in spore pigment formation are clearly separable from antibiotic biosynthesis genes. The difference in the early parts of the biosynthesis routes of these two classes of compounds is due to the fact that the spore pigment compound, which has eluded chemical identification thus far, is most likely composed of a longer polyketide backbone (C24) than most antibiotics (27). The great distance in the phylogenetic analysis could be attributed to the results for 10 distinct amino acids in the area analyzed, which are highly conserved (93 to 100%) in the spore pigment clade but are variable and dissimilar in the antibiotic class. The chemical and structural nature of these amino acids is also different in many instances in the two classes. Interestingly, the sequences of the two non-Streptomyces strains, TA21 and TA83, which migrated between the spore pigment and antibiotic sequences, contain signature amino acids for both groups, and it is not clear to which group these sequences belong.
Incongruities between the two phylogenetic trees support horizontal transfer of antibiotic biosynthesis genes.
Comparison of the distributions of strains in the two phylogenetic trees shows that there seems to be no correlation between KSα and 16S rDNA sequences. For instance, 13 isolates which have 16S rDNA sequences identical to that of S. griseus (Fig. 1 and 2, boxes 1) have highly diverse antibiotic biosynthesis genes that are distributed in six different antibiotic branches in the KSα tree. Similarly, the strains phylogenetically similar to S. capuensis (boxes 2) are separated by a great phylogenetic distance in the KSα tree. Another example involves strains RA35 and AUSA69 (boxes 3), which have very similar antibiotic biosynthesis genes but different 16S rDNA sequences. In addition to the examples described above, numerous other discrepancies can be found between the two phylogenetic trees. Taken together, these results strongly imply that the evolution of aromatic polyketide antibiotics is independent of the evolution of the bacterial species.
Interestingly, there is also incongruity between the spore pigment clade and the 16S rDNA tree. This is noticeable, for example, with Streptomyces sp. strains OS16, AUSA91, PGB1B, and KR25 (boxes 4), which have identical spore pigment sequences; however, according to the 16S rDNA tree these strains are phylogenetically very distantly related to each other. This is somewhat surprising, as one would expect that these bacteria would gain no evolutionary advantage from the horizontal transfer of genes involved in the biosynthesis of spore pigments unless there is a connection between the different kinds of spore pigments or spores that these strains have and the survival of these strains under various types of environmental conditions.
Finally, it should be noted that the 16S rDNA and PKS tree comparisons were made by using trees that contain sequences that appear in both of the trees (i.e., the same data set was used in both of the trees and the sequences that were obtained from GenBank were omitted), as well as trees in which the paralogous spore pigment and antibiotic biosynthesis genes were analyzed independently. However, the latter trees are not shown here because additional figures would provide no further insight into the matter as the topologies of these trees were extremely similar to those of the trees shown in Fig. 1 and 2.
DISCUSSION
Horizontal transfer of antibiotic biosynthesis genes has been previously studied for streptomycin (6, 24) and β-lactam antibiotics (4). In particular, Egan et al. (7) have shown that two Streptomyces isolates that are phylogenetically related to S. coelicolor have obtained a streptomycin biosynthesis gene(s) that is highly homologous to the genes cloned from S. griseus. Furthermore, molecular genetic analysis of cloned antibiotic biosynthesis gene clusters provides more indirect evidence for the horizontal transfer of antibiotic biosynthesis genes; it has been shown that a 241-bp direct repeat sequence marks the boundaries of the mithramycin biosynthesis gene cluster in S. argillaceus, which could have arisen from the acquisition of the gene cluster through a recombination event (14). In addition, the complete genome sequences of S. avermitilis (19) and S. coelicolor (2) have shown that many of the antibiotic biosynthesis gene clusters reside near the ends of the linear chromosomes of Streptomyces species, where genetic rearrangements are common, and that many transposase genes are adjacent to antibiotic biosynthesis genes.
In this study we collected the largest set of sequences to date for two different marker molecules, traditional 16S rDNA and KSα genes from the aromatic polyketide pathway. The data presented here show that there is no evident correlation between the phylogenetic trees constructed by using these different marker molecules. The results also show that very similar antibiotic biosynthesis genes can be found in distantly related Streptomyces species and that diverse antibiotic genes can be found in closely related species. These results strongly suggest that aromatic polyketide biosynthesis genes are transferred horizontally between different Streptomyces species.
The heterogeneity of Streptomyces 16S rDNA has been estimated to result a 2% error frequency in a phylogenetic tree derived from this marker molecule (23). Additionally, our results indicate that the information content in the γ-region of the molecule is insufficient to resolve all of the intraspecies relationships of different Streptomyces strains, which can be seen from the existence of identical sequences and from the low bootstrap values for some of the nodes. Similar results have been reported recently by other workers (7). Another source of inaccuracy is the possible existence of multiple aromatic polyketide biosynthesis gene clusters in a single organism. Fortunately, it seems that this is not exceedingly common; by using the two complete streptomycete genome sequences of S. coelicolor and S. avermitilis that are available, we determined that the degenerate primers used in this study can anneal to only one aromatic polyketide antibiotic KSα gene and one spore pigment KSα gene fragment for each of the two species (data not shown). Furthermore, for a different data set for 85 Streptomyces isolates, amplified KSα PCR products were sequenced directly with the degenerate sense primer. Forty percent of the products showed multiple KSα fragments in the sequencing reactions; these products were cloned further and screened for several KSα sequences. Two antibiotic biosynthesis gene clusters were found in only 2% of these isolates, and in most cases the multiple signals were due to one antibiotic fragment and one spore pigment fragment (K. Palmu, personal communication). Altogether, the error frequency that arises from the sources mentioned above is well below the level of discrepancy found between the two trees.
The information content of the KSα fragments seems to be sufficient to find a connection to the different antibiotics that the biosynthesis gene products synthesize. Even though it is somewhat surprising that the anthracycline-producing strains form three lineages on the KSα tree, it is significant that there are no inconsistencies in the tree (i.e., different antibiotic groups are not represented in the same clade of the tree). According to our analysis, the angycycline class appears to be the most abundant antibiotic class in Streptomyces. However, in the future it would be interesting to investigate in more detail how well our results coincide with the real distribution of different aromatic polyketide gene clusters and if the degenerate primers have a bias towards certain kinds of antibiotics, which leads to overrepresentation of these classes. The main obstacle to using this method for high-throughput screening of antibiotic biosynthesis genes from newly isolated strains comes from the fact that similar spore pigment genes are amplified at the same time. This compromises the throughput of the method, as the search for all of the clusters in a single organism is laborious.
Nevertheless, we feel that the usefulness of the PKS typing method comes from the fact that since the tree seems to project the different antibiotic metabolites, it can be reasoned that unusual or highly divergent KSα genes would account for more unusual compounds that could have novel characteristics. For example, Streptomyces sp. strains PGB77, PGB14-1, and ESC67 form a clade of their own on the KSα tree that is only weakly related to the strains that synthesize rubromycin (Fig. 2). The apparent movement of antibiotic biosynthesis genes also suggests that typing of antibiotic producers should be done with a representative antibiotic biosynthesis gene, since traditional marker molecules give misleading results. Another advantage comes from the observation that Streptomyces strains harbor interesting antibiotic biosynthesis genes that are not expressed in normal laboratory conditions, like the genes for the biosynthesis of the angucycline antibiotic jadomycin (25). Therefore, the metabolite products synthesized by these gene products cannot be directly observed. However, with the method described here, we can categorize and reach these silent biosynthesis genes, which hopefully will increase the biodiversity of antibiotic biosynthesis genes that are available. This source of new antibiotic biosynthesis genes could then be used in combinatorial biosynthesis for the production of novel “unnatural” natural products.
Acknowledgments
We thank Kaj Räty, Pauli Kallio, and Kaisa Palmu for critical reading of the manuscript and Maria Metsä-Ketelä for revising the language.
This study was supported by TEKES (National Technology Agency of Finland) and by a grant from Turku University Foundation to M.M.-K.
REFERENCES
- 1.Anderson, A. S., and E. M. Wellington. 2001. The taxonomy of Streptomyces and related genera. Int. J. Syst. E vol. Microbiol. 51:797-814. [DOI] [PubMed] [Google Scholar]
- 2.Bentley, S. D., K. F. Chater, A. M. Cerdeno-Tarraga, G. L. Challis, N. R. Thomson, K. D. James, D. E. Harris, M. A. Quail, H. Kieser, D. Harper, A. Bateman, S. Brown, G. Chandra, C. W. Chen, M. Collins, A. Cronin, A. Fraser, A. Goble, J. Hidalgo, T. Hornsby, S. Howarth, C. H. Huang, T. Kieser, L. Larke, L. Murphy, K. Oliver, S. O'Neil, E. Rabbinowitsch, M. A. Rajandream, K. Rutherford, S. Rutter, K. Seeger, D. Saunders, S. Sharp, R. Squares, S. Squares, K. Taylor, T. Warren, A. Wietzorrek, J. Woodward, B. G. Barrell, J. Parkhill, and D. A. Hopwood. 2002. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 417:141-147. [DOI] [PubMed] [Google Scholar]
- 3.Bibb, M. J., D. H. Sherman, S. Õmura, and D. A. Hopwood. 1994. Cloning, sequencing and deduced functions of a cluster of Streptomyces genes probably encoding biosynthesis of the polyketide antibiotic frenolicin. Gene 142:31-39. [DOI] [PubMed] [Google Scholar]
- 4.Buades, C., and A. Moya. 1996. Phylogenetic analysis of the isopenicillin-N-synthetase horizontal gene transfer. J. Mol. Evol. 42:537-542. [DOI] [PubMed] [Google Scholar]
- 5.Dayhoff, M. O., R. M. Schwartz, and B. C. Orcutt. 1978. A model of evolutionary change in proteins, p. 345-352. In M. O. Dayhoff (ed.), Atlas of protein sequence and structure, vol. 5. National Biomedical Research Foundation, Washington, D.C.
- 6.Egan, S., P. Wiener, D. Kallifidas, and E. M. H. Wellington. 1998. Transfer of streptomycin biosynthesis gene clusters within streptomycetes isolated from soil. Appl. Environ. Microbiol. 64:5061-5063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Egan, S., P. Wiener, D. Kallifidas, and E. M. H. Wellington. 2001. Phylogeny of Streptomyces species and evidence for horizontal transfer of entire and partial antibiotic gene clusters. Antonie Leeuwenhoek 79:127-133. [DOI] [PubMed] [Google Scholar]
- 8.Feng, D. F., and R. F. Doolittle. 1996. Progressive alignment of amino acid sequences and construction of phylogenetic trees from them. Methods Enzymol. 266:368-382. [DOI] [PubMed] [Google Scholar]
- 9.Hutchinson, C. R. 1999. Microbial polyketide synthases: more and more prolific. Proc. Natl. Acad. Sci. USA 96:3336-3338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kataoka, M., K. Ueda, T. Kudo, T. Seki, and T. Yoshida. 1997. Application of the variable region in 16S rDNA to create an index for rapid species identification in the genus Streptomyces. FEMS Microbiol. Lett. 151:249-255. [DOI] [PubMed] [Google Scholar]
- 11.Kauppinen, S., M. Siggaard-Andersen, and P. von Wettstein-Knowles. 1988. beta-Ketoacyl-ACP synthase I of Escherichia coli: nucleotide sequence of the fabB gene and identification of the cerulenin binding residue. Carlsberg. Res. Commun. 53:357-370. [DOI] [PubMed] [Google Scholar]
- 12.Kim., E., H. Kim, S.-P. Hong, K. H. Kang, Y. H. Kho, and Y.-H. Park. 1993. Gene organization and primary structure of a ribosomal RNA gene cluster from Streptomyces griseus subsp. griseus. Gene 132:21-31. [DOI] [PubMed] [Google Scholar]
- 13.Kulowski, K., E. Wendt-Pienkowski, L. Han, K. Yang, L. C. Vining, and C. R. Hutchinson. 1999. Functional characterization of the jadI gene as a cyclase forming angucyclinones. J. Am. Chem. Soc. 121:1786-1794. [Google Scholar]
- 14.Lombo, F., A. F. Braña, C. Méndez, and J. A. Salas. 1999. The mithramycin gene cluster of Streptomyces argillaceus contains a positive regulatory gene and two repeated DNA sequences that are located at both ends of the cluster. J. Bacteriol. 181:642-647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ludwig, W., and K. H. Schleifer. 1994. Bacterial phylogeny based on 16S and 23S rRNA sequence analysis. FEMS Microbiol. Rev. 15:155-173. [DOI] [PubMed] [Google Scholar]
- 16.Maidak, B. L., J. R. Cole, T. G. Lilburn, C. T. Parker, Jr., P. R. Saxman, R. J. Farris, G. M. Garrity, G. J. Olsen, T. M. Schmidt, and J. M. Tiedje. 2001. The RDP-II (Ribosomal Database Project). Nucleic Acids Res. 29:173-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martin, R., O. Sterner, M. A. Alvarez, E. de Clercq, J. E. Bailey, and W. Minas. 2001. Collinone, a new recombinant angular polyketide antibiotic made by an engineered Streptomyces strain. J. Antibiot. 54:239-249. [DOI] [PubMed] [Google Scholar]
- 18.Metsä-Ketelä, M., V. Salo, L. Halo, A. Hautala, J. Hakala, P. Mäntsälä, and K. Ylihonko. 1999. An efficient approach for screening minimal PKS genes from Streptomyces. FEMS Microbiol. Lett. 180:1-6. [DOI] [PubMed] [Google Scholar]
- 19.Õmura, S., H. Ikeda, J. Ishikawa, A. Hanamoto, C. Takahashi, M. Shinose, Y. Takahashi, H. Horikawa, H. Nakazawa, T. Osonoe, H. Kikuchi, T. Shiba, Y. Sakaki, and M. Hattori. 2001. Genome sequence of an industrial microorganism, Streptomyces avermitilis: deducing the ability of producing secondary metabolites. Proc. Natl. Acad. Sci. USA 98:12215-12220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol 4:406-425. [DOI] [PubMed] [Google Scholar]
- 21.Strohl, W. R. 1997. Industrial antibiotics: today and the future, p. 1-47. In W. R. Strohl (ed.), Bio/technology of antibiotics, 2nd ed. Marcel Dekker, New York, N.Y.
- 22.Strohl, W. R., M. L. Dickens, V. B. Rajgarhia, A. J. Woo, and N. D. Priestley. 1997. Anthracyclines, p. 577-657. In W. R. Strohl (ed.), Bio/technology of antibiotics, 2nd ed. Marcel Dekker, New York, N.Y.
- 23.Ueda, K., T. Seki, T. Kudo, T. Yoshida, and M. Kataoka. 1999. Two distinct mechanisms cause heterogeneity of 16S rRNA. J. Bacteriol. 181:78-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wiener, P., S. Egan, and E. M. H. Wellington. 1998. Evidence for transfer of antibiotic-resistance genes in soil populations of streptomycetes. Mol. Ecol. 7:1205-1216. [DOI] [PubMed] [Google Scholar]
- 25.Yang, K., L. Haa, and L. C. Vining. 1995. Regulation of jadomycin B production in Streptomyces venezuelae ISP5230: involvement of a repressor gene, jadR2. J. Bacteriol. 177:6111-6117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yu, T.-W., M. J. Bibb, W. P. Revill, and D. A. Hopwood. 1994. Cloning, sequencing, and analysis of the griseusin polyketide synthase gene cluster from Streptomyces griseus. J. Bacteriol. 176:2627-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yu, T.-W., Y. Shen, R. McDaniel, H. G. Floss, C. Khosla, D. A. Hopwood, and B. S. Moore. 1998. Engineered biosynthesis of novel polyketides from Streptomyces spore pigment polyketide synthases. J. Am. Chem. Soc. 120:7749-7759. [Google Scholar]