Abstract
Autoinducer molecules are utilized by gram-negative and gram-positive bacteria to regulate density-dependent gene expression by a mechanism known as quorum sensing. PCR and DNA sequencing results showed that Campylobacter jejuni and Campylobacter coli possessed luxS, which is responsible for autoinducer-2 (AI-2) production. Using a Vibrio harveyi luminescence assay, the production of AI-2 was observed in milk, chicken broth, and brucella broth by C. coli, C. jejuni, Salmonella enterica serovar Typhimurium, and Escherichia coli O157:H7 under different conditions.
Cell-to-cell signaling regulates important microbial processes such as growth, sporulation, toxin production, virulence, antibiotic synthesis, motility, and colonization in a variety of bacteria through alterations in the pattern of gene expression in response to cell density (4, 6, 10, 11). This type of response, known as quorum sensing, may contribute to the enhanced ability of bacteria to survive environmental changes. Quorum sensing was first characterized in the marine bacteria Vibrio harveyi and Vibrio fischeri (13, 14). In V. harveyi, there are two types of density-dependent signaling systems that regulate bioluminescence activity consisting of a sensor and a quorum-sensing molecule called an autoinducer. Signaling system 1 is highly species specific and is composed of sensor 1 and autoinducer-1 (AI-1), a family of related homoserine lactones (3). Signaling system 2 is less species specific and is composed of sensor 2 and autoinducer-2 (AI-2) (16).
There is a growing list of gram-negative bacteria that employ quorum-sensing mechanisms functionally similar to those seen in V. harveyi, and one of these autoinducer systems involves a family of lux genes (1, 7, 17). The luxS gene is required for AI-2 production in signaling system 2 (17), and homologs of luxS have been identified in more than 25 bacterial species, including Salmonella enterica serovar Typhimurium, Escherichia coli, and Helicobacter pylori (8, 17, 18). AI-2 is produced from S-adenosylmethionine in three enzymatic steps that are identical to those in the production of AI-2 in E. coli, Salmonella serovar Typhimurium, V. harveyi, Vibrio cholerae, and Enterococcus faecalis (15). The LuxS protein is the AI-2 synthase in the biosynthetic pathway that cleaves S-ribosylhomocysteine into homocysteine and 4,5-dihydroxy-2,3-pentanedione. Schauder et al. (15) suggested that the pentanedione cyclizes to form a furanone ring, which is the active AI-2 molecule. Crystallography studies of LuxS from H. pylori, Deinococcus radiodurans, and Haemophilus influenzae revealed that it is a zinc metalloenzyme, with the zinc bound to a Cys-His-His triad. The zinc is involved in the cleavage of the ribose ring in the synthesis of AI-2 (12).
It has recently been reported that AI-2 is regulated by bacteria not only in response to cell density but also in response to conditions in the environment (18). Although quorum sensing has been studied extensively under laboratory conditions and quorum-sensing mechanisms in enterohemorrhagic E. coli and Salmonella serovar Typhimurium have been reported (16, 18), relatively little has been published on the roles of quorum sensing in the growth and survival of bacteria in food environments. Gram et al. (9) showed that AI-1 was produced by inoculated members of the family Enterobacteriaceae at concentrations of 106 CFU/ml under conditions that simulated food environments (5°C, reduced oxygen, 4% NaCl) and that the autoinducer could be detected in indigenous members of the Enterobacteriaceae in cold smoked salmon at concentrations of 105 to 106 CFU/g (9). However, AI-1 is generally produced at cell densities of 107 to 108 CFU/ml when bacteria are cultured at higher temperatures in laboratory media (20).
Campylobacter jejuni and other food-borne pathogens possess luxS (12); however, the role of quorum sensing in the production of AI-2 in foods by pathogens of significance to the food industry has not been elucidated. Since quorum sensing involves the transfer of small signaling molecules among bacteria, it is feasible that these autoinducers may be exploited to control bacterial growth, survival, and virulence in foods. Thus, further studies are necessary to elucidate the effect of quorum sensing on bacteria in foods. Furthermore, LuxS has been associated with pathogenicity in enterohemorrhagic E. coli (16) and therefore could possibly be associated with survival and virulence of Campylobacter spp. The objectives of this study were to determine the homology of LuxS in C. jejuni, Campylobacter coli, Salmonella serovar Typhimurium, and E. coli O157:H7 and to determine the production of AI-2 by these organisms in foods.
PCR amplification and sequencing of the Campylobacter luxS gene.
A GenBank search of the C. jejuni sequence (strain NCTC 11168) revealed the presence of a homolog of the luxS gene of V. harveyi (74% identity). Primers were designed according to the luxS homolog (hypothetical protein Cj1198; GenBank accession number AL139077) of C. jejuni NCTC 11168 (CJLUXFR, 5′ AAA AGA ATT CAT GCC ATT ATT AGA CAG CTT TAA AG 3′; CJLUXRE, 5′ AAA AGG ATC CTA AGC ATT CTC GAG TTT TAA TTC 3′) and utilized to amplify the luxS gene in strains of C. jejuni (ATCC 33560) and C. coli (ATCC 33559). Extraction of DNA from isolated colonies on modified charcoal cefoperazone deoxycholate agar (mCCDA; Oxoid, Basingstoke, United Kingdom) plates was achieved using the PrepMan sample preparation reagent (PE Applied Biosystems, Foster City, Calif.) according to the manufacturer's instructions. Amplification reactions were performed in total volumes of 50 μl containing 5 μl of purified template DNA, 10 mM Tris-HCl (pH 8.4), 50 mM KCl, 1.5 mM MgCl2, 200 μM each of the four deoxynucleotide triphosphates, 0.40 μM each of primers CJLUXFR and CJLUXRE, and 1.25 U of Taq DNA polymerase (Promega Core Kit M7660; Promega, Madison, Wis.) (also can obtained from Invitrogen-Life Technologies, Carlsbad, Calif.). Samples were subjected to the following steps: (i) an initial denaturation step of 5 min at 95°C, (ii) 35 amplification cycles, with 1 cycle consisting of 1 min at 94°C, 1 min at 50°C, and 1 min at 72°C, and (iii) a final primer extension step of 7 min at 72°C. These steps were done in a model 9600 or 9700 thermal cycler (PE Applied Biosystems). The PCR products were separated by electrophoresis on 1.5% agarose gels (Gibco-BRL) subsequently stained with ethidium bromide. The amplification products were purified using the Quickstep2 PCR purification kit (catalog no. 92159; Edge Biosystems, Gaithersburg, Md.) and were subsequently sequenced using the Big Dye Terminator kit (catalog no. 4303153; Applied Biosystems, Foster City, Calif.) using an Applied Biosystems 3700 DNA Analyzer. Sequence cleanup was performed with gel filtration columns (catalog no. 42453; Edge Biosystems). In addition to the CJLUXFR and CJLUXRE primers, additional primers for the luxS homolog derived from the C. coli sequence generated in this work were used to confirm the luxS DNA sequence (CCOLILUXSR, 5′ CAT GCC ATT ATT AGA CAG CTT TAA AG 3′; CCOLILUXSF, 5′GAA TGC ATA GCA CAA GTT CCA CAT TGA 3′). Sequence data were imported into Sequencher software (GeneCodes, Ann Arbor, Mich.) for further trimming, assembly, and analysis.
Nucleotide sequence accession numbers.
The C. coli ATCC 33559 and C. jejuni ATCC 33560 luxS homolog GenBank accession numbers are AY078283 and AY122056, respectively.
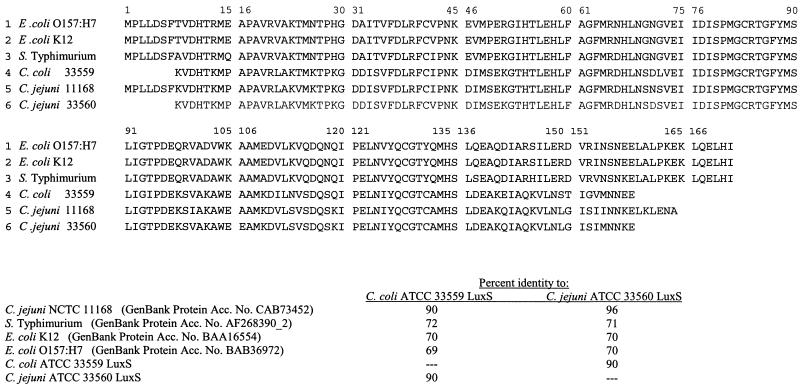
Results from the sequencing of the luxS PCR products indicated that both C. jejuni and C. coli used in this study possessed a luxS gene with homology to luxS of V. harveyi. The C. coli ATCC 33559 LuxS protein displayed 90% identity with the C. jejuni ATCC 33560 and C. jejuni NCTC 11168 LuxS protein and 72, 70, and 69% identity with the Salmonella serovar Typhimurium (GenBank accession no. AF268390_2), E. coli K-12 (GenBank accession no. BAA16554), and E. coli O157:H7 (GenBank accession no. BAB36972) LuxS protein, respectively (Fig. 1). The amino acid substitutions in the C. jejuni and C. coli LuxS protein would not be expected to result in any notable differences in function or binding of the protein in Campylobacter spp. compared to the other bacteria examined as postulated from crystal structural studies and multiple-sequence alignments of the protein (12). The highly conserved amino acid residues in the catalytic site of LuxS are identical in the C. coli (ATCC 33559) and C. jejuni (ATCC 33560) strains sequenced in this study and this region of LuxS in other bacterial genera examined (12).
FIG. 1.
Sequence alignment of LuxS proteins of C. coli (ATCC 33559), C. jejuni (ATCC 33560 and NCTC 11168), E. coli O157:H7, E. coli K-12, and Salmonella serovar Typhimurium. The C. coli ATCC 33559 and C. jejuni ATCC 33560 sequences shown are incomplete. They lack the N-terminal and C-terminal amino acids, since primers for C. jejuni NCTC 11168 were used to amplify luxS in these strains. These incomplete sequences were used to determine percent identities using Blast P (http://www.ncbi.nlm.nih.gov/BLAST/). ClustalW was used for protein alignment: http://dot.imgen.bcm.tmc.edu:9331/multi-align/multi-align.html. Acc. No., accession number.
Autoinducer bioassay.
Cell-free culture supernatants were prepared as previously described by Surette and Bassler (19). Briefly, E. coli O157:H7 933 (U.S. Department of Agriculture Food Safety and Inspection Service; meat isolate) and Salmonella serovar Typhimurium strain LT2 were grown for 18 h in brucella broth (Difco, Detroit, Mich.) at 37°C aerobically, and C. jejuni ATCC 33560 (originally isolated from bovine feces) and C. coli ATCC 33559 (originally isolated from pig feces) were grown in brucella broth at 42°C for 40 h under microaerobic conditions (generated by CampyPak Plus [BBL, Cockeysville, Md.]) in airtight containers. Bacteria were first removed from the growth medium by centrifugation at 16,000 × g for 5 min in a microcentrifuge. The cell-free culture supernatants were subsequently passed through Tuffryn syringe filters (0.2-μm pore size; Pall Gelman, Ann Arbor, Mich.) and stored at −20°C. V. harveyi strains BB120 (has AI-1 and AI-2) and BB152 (has AI-2 but not AI-1) were used as controls. Control strains of V. harveyi were grown overnight at 30°C with aeration in Luria-Bertani (LB; Difco) broth, and 1 ml of cell-free supernatant of each culture was prepared as described above.
The cell-free supernatants from E. coli O157:H7, Salmonella serovar Typhimurium, C. jejuni, C. coli, and V. harveyi were tested for the presence of autoinducers that could induce luminescence in the V. harveyi reporter strain BB170 (has sensor 2 but not sensor 1), capable of sensing AI-2 but not AI-1. In the bioassay, V. harveyi strain BB170 was grown overnight at 30°C with aeration in LB broth and diluted 1:5,000 into autoinducer bioassay (AB) medium (2). Ninety microliters of the diluted bacteria and 10 μl of the cell-free supernatant of each sample were added to individual wells of a 96-well microtiter plate. Ten microliters of cell-free supernatant from strains BB120 and BB152 were used as positive controls, and sterile AB medium (10 μl) was used as a negative control. Luminescence was measured every 20 min using a computer-controlled microplate luminometer (model ML 3000; Dynatech Laboratories, Chantilly, Va.). The microtiter plates were incubated at 30°C and were mixed at 80 rpm for ca. 10 s prior to luminescence readings. Relative light units were expressed as total luminescence per 106 Vibrio harveyi strain BB170 cells per well.
Initial results indicated that both C. jejuni and C. coli produce an AI-2 signaling molecule that caused expression of bioluminescence in V. harveyi. It was also apparent that maximal autoinducer activity occurred during the exponential growth phase, diminishing when the organism entered stationary phase (data not shown). These results parallel the findings of Forsyth and Cover (8), who found that in H. pylori, which is closely related to Campylobacter spp., AI-2 activity was maximal in early to mid-phase exponential growth and declined significantly prior to stationary phase.
Environmental parameters affecting autoinducer production.
Autoinducers are generally thought to accumulate in the growth medium throughout the exponential growth phase, and the expression of target genes is triggered at a threshold level via induction of an autoinducing circuit (9, 16). Environmental conditions, such as the type of food or other sample matrix, incubation temperatures, and atmospheric conditions, may have an effect on the production of autoinducers. Research by Surette and Bassler (18) has shown that the production of AI-2 in Salmonella serovar Typhimurium and E. coli is related not only to high cell densities but also to growth conditions and the presence of glucose and other preferred carbohydrates in the medium. Therefore, we examined the effects of different compounds, including glucose, charcoal, and fetal bovine serum, added to brucella broth on AI-2 production in C. jejuni and C. coli. The bacteria were grown in brucella broth at 42°C for 40 h under microaerobic conditions, and about 2.5 log10 CFU/ml of each organism was then inoculated into 10 ml of brucella broth with 0.5% glucose, brucella broth with 0.5% fetal bovine serum, and brucella broth with 0.5% charcoal. Broths were incubated at 37°C under microaerobic conditions. Portions (3 ml) were withdrawn aseptically from the inoculated broths at 0, 3, 6, 24, and 48 h. Cell-free supernatants were prepared from 1 ml of the withdrawn aliquot as described above. Viable counts were determined in duplicate on mCCDA plates incubated for 48 h at 42°C under microaerobic conditions. Cell-free supernatants (10 μl of each supernatant) prepared at individual sampling times were subjected to the autoinducer bioassay as described above.
The AI-2 activity increased in brucella broth supplemented with glucose or fetal bovine serum, whereas addition of charcoal did not increase AI-2 production over that in brucella broth alone (Table 1). A similar observation was noted by Surette et al. (17) and Surette and Bassler (18), who found that the presence of glucose in LB broth increased autoinducer activity in Salmonella serovar Typhimurium, and the rate of decrease of AI-2 production following growth in a synthetic medium with glucose was affected by the osmolarity of the medium and pH. Therefore, results from previous studies and this study indicate that in addition to cell density, conditions in the environment such as the presence of specific nutrients may affect AI-2 production.
TABLE 1.
AI-2 production of C. jejuni and C. coli in brucella broth with and without supplementsa
| Broth |
C. jejuni
|
C. coli
|
||
|---|---|---|---|---|
| Luminescence | Plate count | Luminescence | Plate count | |
| Brucella broth | 2,000.4 ± 3.4 | 6.8 ± 0.9 | 2,016.2 ± 8.6 | 6.5 ± 0.6 |
| Brucella broth + glucose | 3,075.2 ± 8.5 | 7.5 ± 0.6 | 3,004.5 ± 10.6 | 7.8 ± 0.9 |
| Brucella broth + FBSb | 2,911.3 ± 2.2 | 7.1 ± 0.5 | 2,990.0 ± 4.5 | 7.2 ± 0.6 |
| Brucella broth + charcoal | 2,012.3 ± 3.6 | 7.0 ± 0.8 | 2,025.1 ± 4.7 | 7.1 ± 0.5 |
Cultures were grown at 37°C under microaerobic conditions, and cell-free supernatants were tested by the autoinducer bioassay. Results shown are of samples taken after 24 h of incubation and are the averages ± standard deviations of two separate experiments. Luminescence is shown in luminescence units, and plate counts are log10 CFU per milliliter.
FBS, fetal bovine serum.
Autoinducer production in milk, apple juice, and chicken broth.
The effect of the food environment and the role of quorum sensing in the survival of bacteria within a food matrix have largely been unexplored. To investigate the production of AI-2 in liquid foods, E. coli O157:H7 and Salmonella serovar Typhimurium grown in brucella broth aerobically for 18 h at 37°C and C. jejuni and C. coli grown in brucella broth under microaerobic conditions for 40 h at 42°C were inoculated at levels of ca. 3 to 3.5 log10 CFU/ml individually into 15-ml volumes of brucella broth, ultrahigh-temperature-treated milk (Parmalat brand), pasteurized apple juice (America's Choice brand), and chicken broth (College Inn brand). The samples were incubated at 4, 25, and 37°C for up to 48 h under aerobic conditions. However, samples inoculated with C. jejuni and C. coli and held at 37°C were incubated under microaerobic conditions. Bacteria were inoculated at ca. 6.5 log10 CFU/ml in samples incubated at 4°C, since cells were not expected to grow at that temperature. At each sampling time (0, 3, 6, 24, and 48 h), portions (3 ml) were withdrawn from the inoculated foods, and viable counts were determined in duplicate on MacConkey agar (Difco) and Rainbow Agar Salmonella (Biolog, Hayward, Calif.) for E. coli O157:H7 and Salmonella serovar Typhimurium, respectively, incubated at 37°C for 18 h. Viable counts for Campylobacter spp. were determined by plating in duplicate onto mCCDA and incubating for 48 h at 42°C under microaerobic conditions. Cell-free supernatants were prepared as described above for the autoinducer bioassay to determine the levels of autoinducer production at each sampling time. The experiments were repeated at least twice, with three replicates for each experiment. Analysis of variance was used to determine the effects of storage time and temperature on luminescence values for the four organisms tested in the different matrices.
A comparison of luminescence results of C. jejuni and C. coli showed that AI-2 activity was detected primarily when the bacteria were grown at 37°C for 24 h in milk, chicken broth, and brucella broth (Table 2). Maximal AI-2 activity (ca. 3,000 luminescence units) was evident in brucella broth. The inoculum level for this assay was ca. 3 to 3.5 log10 CFU/ml, which may account for the higher luminescence result than that which was observed in brucella broth when the initial inoculum level was 2.5 log10 CFU/ml (ca. 2,000 luminescence units [Table 1]). There was little or no AI-2 activity in bacteria grown in milk or chicken broth at 4°C; however, there was AI-2 activity in bacteria grown in brucella broth at 4°C at 3 h (ca. 150 luminescence units). The reason for AI-2 activity for bacteria grown at 4°C is not clear, but shifts in intracellular metabolism and stress can alter patterns of AI-2 production in E. coli K-12 (5). At 4 and 25°C, the log10 CFU/ml of C. jejuni and C. coli decreased during the 48-h incubation period. In brucella broth at 25°C at 24 h, luminescence counts were greater than 100, while plate counts decreased to 2.2 log10 CFU/ml (Table 2). The reason for this is unclear, but the results obtained may possibly be due to the inability to culture Campylobacter on mCCDA as a result of cold stress and exposure to air. The bacteria may have been viable and able to produce AI-2. At 37°C, levels of Campylobacter reached >7 log10 CFU/ml by 48 h, and luminescence units were highest at 6 or 24 h. Overall, data were similar for C. jejuni and C. coli.
TABLE 2.
AI-2 production in C. jejuni and C. coli in milk, chicken broth, and brucella broth at 4, 25, and 37°C
| Food or broth | Incubation temp (°C) | Sampling time (h) | Luminescence (luminescence units)a
|
Plate count (log10 CFU/ml)
|
||
|---|---|---|---|---|---|---|
| C. jejuni | C. coli | C. jejuni | C. coli | |||
| Milk | 4 | 0 | 1.20 ± 0.65 A | 1.15 ± 1.10 A | 6.7 | 6.8 |
| 3 | 1.56 ± 0.88 A | 2.50 ± 0.12 A | 6.3 | 6.2 | ||
| 6 | 3.41 ± 1.02 A | 2.11 ± 0.29 A | 5.4 | 5.1 | ||
| 24 | 1.91 ± 0.77 A | 2.01 ± 1.01 A | 3.2 | 3.0 | ||
| 48 | 1.83 ± 0.91 A | 1.13 ± 1.03 A | 2.2 | 2.5 | ||
| 25 | 0 | 1.25 ± 0.77 B | 1.19 ± 0.13 A | 3.9 | 4.0 | |
| 3 | 0.91 ± 0.66 B | 1.00 ± 0.22 A | 3.0 | 3.5 | ||
| 6 | 0.99 ± 0.09 B | 0.89 ± 0.51 A | 2.6 | 3.0 | ||
| 24 | 10.25 ± 1.11 A | 7.25 ± 0.91 A | 2.5 | 2.1 | ||
| 48 | 0.68 ± 0.20 B | 0.69 ± 0.33 A | 1.5 | 1.7 | ||
| 37 | 0 | 2.35 ± 0.99 B | 2.05 ± 0.71 B | 3.5 | 3.1 | |
| 3 | 3.45 ± 1.01 B | 4.01 ± 1.33 B | 3.9 | 3.7 | ||
| 6 | 5.23 ± 2.22 B | 5.43 ± 3.03 B | 4.0 | 4.4 | ||
| 24 | 99.50 ± 1.21 A | 101.50 ± 2.21 A | 6.6 | 6.0 | ||
| 48 | 3.11 ± 1.01 B | 5.21 ± 2.55 B | 7.7 | 7.7 | ||
| Chicken broth | 4 | 0 | 3.05 ± 0.11 A | 2.95 ± 0.35 A | 6.1 | 6.5 |
| 3 | 2.56 ± 0.72 A | 3.16 ± 0.42 A | 6.0 | 6.1 | ||
| 6 | 3.46 ± 0.72 A | 4.06 ± 1.03 A | 5.7 | 5.6 | ||
| 24 | 2.2 ± 0.81 A | 2.11 ± 0.13 A | 3.2 | 3.1 | ||
| 48 | 1.53 ± 0.81 A | 1.61 ± 0.61 A | 2.1 | 2.1 | ||
| 25 | 0 | 0.25 ± 0.03 B | 0.65 ± 0.73 B | 3.9 | 3.9 | |
| 3 | 2.25 ± 1.02 B | 3.05 ± 1.03 AB | 3.2 | 3.4 | ||
| 6 | 14.11 ± 2.03 A | 11.31 ± 1.59 A | 3.5 | 3.4 | ||
| 24 | 6.25 ± 0.11 B | 5.95 ± 0.81 AB | 1.6 | 2.0 | ||
| 48 | 5.21 ± 0.27 B | 5.01 ± 0.12 AB | 1.3 | 1.4 | ||
| 37 | 0 | 1.66 ± 0.11 D | 1.76 ± 0.19 D | 3.5 | 3.5 | |
| 3 | 53.35 ± 7.91 C | 54.45 ± 5.02 C | 4.9 | 4.8 | ||
| 6 | 202.23 ± 9.01 A | 212.03 ± 10.21 A | 6.7 | 6.7 | ||
| 24 | 92.30 ± 3.01 B | 95.30 ± 3.17 B | 7.2 | 7.4 | ||
| 48 | 4.11 ± 0.61 D | 7.11 ± 1.01 D | 7.6 | 8.0 | ||
| Brucella broth | 4 | 0 | 0.22 ± 0.09 C | 0.45 ± 0.11 C | 6.4 | 6.5 |
| 3 | 145.56 ± 7.02 A | 155.00 ± 9.12 A | 5.2 | 5.4 | ||
| 6 | 51.06 ± 1.42 B | 54.06 ± 3.03 B | 5.1 | 4.9 | ||
| 24 | 2.51 ± 0.21 C | 3.51 ± 0.09 C | 2.2 | 2.4 | ||
| 48 | 1.83 ± 0.31 C | 2.33 ± 0.60 C | 1.3 | 1.9 | ||
| 25 | 0 | 0.82 ± 0.33 B | 0.91 ± 0.09 B | 3.9 | 3.9 | |
| 3 | 0.91 ± 0.08 B | 1.01 ± 0.05 B | 3.6 | 3.6 | ||
| 6 | 1.75 ± 0.23 B | 1.95 ± 0.32 B | 2.9 | 2.8 | ||
| 24 | 109.25 ± 5.11 A | 107.00 ± 7.15 A | 2.4 | 2.2 | ||
| 48 | 0.98 ± 0.20 B | 2.98 ± 0.72 B | 2.0 | 1.9 | ||
| 37 | 0 | 1.55 ± 0.10 B | 1.35 ± 0.21 B | 3.7 | 3.7 | |
| 3 | 2.60 ± 0.08 B | 2.70 ± 0.31 B | 4.2 | 4.0 | ||
| 6 | 5.40 ± 0.33 B | 5.40 ± 0.09 B | 4.5 | 4.7 | ||
| 24 | 3029.00 ± 10.66 B | 3,001.05 ± 20.11 A | 6.2 | 6.4 | ||
| 48 | 5.11 ± 0.44 B | 4.65 ± 0.55 B | 8.0 | 7.9 | ||
Results represent the averages ± standard deviations of two experiments with three replicates per experiment. Within each combination of food or broth and temperature, means in the same column with no letter in common are significantly different (P < 0.05) by the Bonferroni least-significant difference separation test.
Similar trends in production of AI-2 were observed for Salmonella serovar Typhimurium (Table 3) and E. coli O157:H7 (Table 4). As with the two Campylobacter spp., the levels of AI-2 had increased by 3 h at 4°C but then returned to levels close to those observed at time zero by 6 h. Luminescence results varied somewhat with incubation time for each combination of food, broth, and temperature; however, the significantly larger luminescence values with the different bacteria at 24 h at 37°C were fairly consistent in milk, chicken broth, and brucella broth when bacterial plate counts were between ca. 7 and 9 log10 CFU/ml, with the exception of C. coli and C. jejuni in chicken broth, where AI-2 levels peaked at 6 h. No notable AI-2 activity was evident in apple juice with any of the organisms examined under any of the conditions tested (data not shown).
TABLE 3.
AI-2 production in Salmonella serovar Typhimurium in milk, chicken broth, and brucella broth at 4, 25, and 37°C
| Food or broth | Incubation temp (°C) | Sampling time (h) | Luminescence (luminescence units)a | Plate count (log10 CFU/ml) |
|---|---|---|---|---|
| Milk | 4 | 0 | 1.42 ± 0.11 B | 6.9 |
| 3 | 106.56 ± 5.92 A | 6.3 | ||
| 6 | 5.96 ± 1.30 B | 6.1 | ||
| 24 | 1.20 ± 0.11 B | 5.7 | ||
| 48 | 0.62 ± 0.29 B | 5.4 | ||
| 25 | 0 | 1.25 ± 0.11 B | 3.8 | |
| 3 | 0.89 ± 0.82 B | 3.9 | ||
| 6 | 0.67 ± 0.30 B | 4.7 | ||
| 24 | 79.25 ± 2.15 A | 7.1 | ||
| 48 | 5.46 ± 1.30 B | 7.9 | ||
| 37 | 0 | 1.54 ± 0.10 B | 3.9 | |
| 3 | 8.92 ± 2.22 B | 4.5 | ||
| 6 | 5.36 ± 0.19 B | 6.5 | ||
| 24 | 920.00 ± 10.22 A | 7.4 | ||
| 48 | 4.65 ± 0.25 B | 9.4 | ||
| Chicken broth | 4 | 0 | 1.70 ± 0.30 B | 7.0 |
| 3 | 101.40 ± 5.20 A | 6.6 | ||
| 6 | 2.45 ± 0.24 B | 5.9 | ||
| 24 | 1.95 ± 0.29 B | 5.3 | ||
| 48 | 1.60 ± 0.30 B | 4.9 | ||
| 25 | 0 | 2.01 ± 0.20 B | 3.5 | |
| 3 | 1.56 ± 0.30 B | 4.1 | ||
| 6 | 1.23 ± 0.31 B | 5.7 | ||
| 24 | 197.50 ± 9.20 A | 7.6 | ||
| 48 | 1.25 ± 0.13 B | 8.9 | ||
| 37 | 0 | 1.21 ± 0.20 D | 3.7 | |
| 3 | 19.69 ± 0.40 C | 4.8 | ||
| 6 | 161.04 ± 4.30 B | 6.1 | ||
| 24 | 1,501.41 ± 12.12 A | 8.9 | ||
| 48 | 6.29 ± 0.19 D | 9.4 | ||
| Brucella broth | 4 | 0 | 1.11 ± 0.50 B | 6.9 |
| 3 | 102.60 ± 7.41 A | 6.4 | ||
| 6 | 4.11 ± 0.23 B | 5.3 | ||
| 24 | 1.29 ± 0.30 B | 5.0 | ||
| 48 | 1.00 ± 0.30 B | 4.2 | ||
| 25 | 0 | 1.11 ± 0.30 B | 3.5 | |
| 3 | 2.69 ± 0.12 B | 4.1 | ||
| 6 | 9.26 ± 0.93 B | 6.9 | ||
| 24 | 546.40 ± 8.30 A | 8.4 | ||
| 48 | 5.19 ± 0.10 B | 8.6 | ||
| 37 | 0 | 0.94 ± 0.40 D | 3.8 | |
| 3 | 42.61 ± 4.06 C | 4.4 | ||
| 6 | 64.30 ± 9.10 B | 6.1 | ||
| 24 | 1,668.40 ± 15.60 A | 8.6 | ||
| 48 | 3.22 ± 0.92 D | 9.7 |
Results represent the average ± standard deviations of two experiments with three replicates per experiment. Within each combination of food or broth and temperature, luminescence means with no letter in common are significantly different (P < 0.05) by the Bonferroni least-significant difference separation test.
TABLE 4.
AI-2 production in E. coli O157:H7 in milk, chicken broth, and brucella broth at 4, 25, and 37°C
| Food or broth | Incubation temp (°C) | Sampling time (h) | Luminescence (luminescence units)a | Plate count (log10 CFU/ml) |
|---|---|---|---|---|
| Milk | 4 | 0 | 1.26 ± 0.31 B | 5.9 |
| 3 | 102.32 ± 11.32 A | 5.6 | ||
| 6 | 3.74 ± 1.02 B | 5.0 | ||
| 24 | 0.97 ± 0.09 B | 4.9 | ||
| 48 | 0.13 ± 0.31 B | 4.5 | ||
| 25 | 0 | 0.50 ± 0.23 B | 3.0 | |
| 3 | 0.25 ± 0.32 B | 4.2 | ||
| 6 | 0.42 ± 0.13 B | 5.7 | ||
| 24 | 55.25 ± 5.11 A | 7.9 | ||
| 48 | 0.34 ± 0.29 B | 8.8 | ||
| 37 | 0 | 1.10 ± 1.01 C | 3.9 | |
| 3 | 12.35 ± 6.01 BC | 4.9 | ||
| 6 | 19.23 ± 5.66 B | 6.4 | ||
| 24 | 822.30 ± 10.01 A | 7.2 | ||
| 48 | 2.11 ± 1.11 C | 9.9 | ||
| Chicken broth | 4 | 0 | 0.85 ± 0.91 B | 6.5 |
| 3 | 303.56 ± 11.02 A | 6.2 | ||
| 6 | 3.74 ± 0.82 B | 4.7 | ||
| 24 | 1.69 ± 0.81 B | 5.6 | ||
| 48 | 1.04 ± 0.33 B | 5.1 | ||
| 25 | 0 | 0.77 ± 0.43 B | 3.1 | |
| 3 | 0.83 ± 0.22 B | 4.0 | ||
| 6 | 1.23 ± 1.20 B | 5.3 | ||
| 24 | 198.25 ± 11.11 A | 7.5 | ||
| 48 | 0.59 ± 0.20 B | 9.1 | ||
| 37 | 0 | 0.66 ± 0.31 D | 3.0 | |
| 3 | 23.35 ± 9.01 C | 4.7 | ||
| 6 | 191.20 ± 2.01 B | 7.1 | ||
| 24 | 1,792.30 ± 20.99 A | 7.2 | ||
| 48 | 5.21 ± 1.11 D | 9.2 | ||
| Brucella broth | 4 | 0 | 0.94 ± 0.39 B | 6.6 |
| 3 | 141.56 ± 9.02 A | 6.3 | ||
| 6 | 8.76 ± 1.02 B | 5.6 | ||
| 24 | 0.66 ± 0.88 B | 5.1 | ||
| 48 | 0.14 ± 0.27 B | 4.9 | ||
| 25 | 0 | 0.84 ± 0.83 B | 3.2 | |
| 3 | 1.75 ± 0.92 B | 4.5 | ||
| 6 | 5.75 ± 1.03 B | 5.9 | ||
| 24 | 479.25 ± 10.11 A | 7.4 | ||
| 48 | 0.67 ± 0.28 B | 8.3 | ||
| 37 | 0 | 0.33 ± 0.41 C | 3.1 | |
| 3 | 33.35 ± 9.71 B | 4.2 | ||
| 6 | 42.23 ± 10.01 B | 6.1 | ||
| 24 | 1,854.30 ± 23.01 A | 7.4 | ||
| 48 | 0.45 ± 0.21 C | 9.0 |
Results represent the averages ± standard deviations of two experiments with three replicates per experiment. Within each combination of food or broth and temperature, luminescence means with no letter in common are significantly different (P < 0.05) by the Bonferroni least-significant difference separation test.
In conclusion, C. jejuni strain ATCC 33560 and C. coli strain ATCC 33559 possess luxS, and the deduced amino acid sequences of LuxS in these two organisms were 90% identical. C. jejuni, C. coli, Salmonella serovar Typhimurium, and E. coli O157:H7 demonstrated AI-2 activity in brucella broth and in liquid foods. Potentially, quorum-sensing systems can serve as targets for the development of novel antibacterial agents and compounds that inhibit the production of signaling molecules in foods. Further research includes examining genes that are regulated by quorum sensing in Campylobacter spp.
Acknowledgments
We thank Bonnie Bassler for supplying the V. harveyi strains utilized in this study, James Kaper for technical advice, Rebecca Tischler for technical assistance in the initial phases of this project, and John Phillips for performing the statistical analyses.
REFERENCES
- 1.Bassler, B. L., E. P. Greenberg, and A. M. Stevens. 1997. Cross-species induction of luminescence in the quorum-sensing bacterium Vibrio harveyi. J. Bacteriol. 179:4043-4045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bassler, B. L., M. Wright, R. E. Showalter, and M. R. Silverman. 1993. Intercellular signaling in Vibrio harveyi: sequence and function of genes regulating expression of luminescence. Mol. Microbiol. 9:773-786. [DOI] [PubMed] [Google Scholar]
- 3.Cao, J., and E. A. Meighen. 1989. Purification and structural identification of an autoinducer for the luminescence system of Vibrio harveyi. J. Biol. Chem. 264:21670-21676. [PubMed] [Google Scholar]
- 4.Davies, D. G., M. R. Parsek, J. P. Pearson, B. H. Iglewski, J. W. Costerton, and E. P. Greenberg. 1998. The involvement of cell-to-cell signals in the development of a bacterial biofilm. Science 280:295-298. [DOI] [PubMed] [Google Scholar]
- 5.DeLisa, M. P., J. J. Valdes, and W. E. Bentley. 2001. Mapping stress-induced changes in autoinducer AI-2 production in chemostat-cultivated Escherichia coli K-12. J. Bacteriol. 183:2918-2928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eberl, L., M. K. Winson, C. Sternberg, G. S. Stewart, G. Christiansen, S. R. Chhabra, B. Bycroft, P. Williams, S. Molin, and M. Givskov. 1996. Involvement of N-acyl-L-homoserine lactone autoinducers in controlling the multicellular behaviour of Serratia liquefaciens. Mol. Microbiol. 20:847-852. [DOI] [PubMed] [Google Scholar]
- 7.Engebrecht, J., N. H. Nealson, and M. Silverman. 1983. Bacterial bioluminescence: isolation and genetic analysis of functions from Vibrio fischeri. Cell 32:773-781. [DOI] [PubMed] [Google Scholar]
- 8.Forsyth, M. H., and T. L. Cover. 2000. Intercellular communication in Helicobacter pylori: luxS is essential for the production of an extracellular signaling molecule. Infect. Immun. 68:3193-3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gram, L., A. B. Christensen, L. Ravn, S. Molen, and M. Givskov. 1999. Production of acetylated homoserine lactones by psychrotrophic members of the Enterobacteriaceae isolated from foods. Appl. Environ. Microbiol. 65:3458-3463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ji, G., R. C. Beavis, and R. P. Novick. 1995. Cell density control of staphylococcal virulence mediated by octapeptide pheromone. Proc. Natl. Acad. Sci. USA 192:12055-12061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kievit, T. R., and B. H. Iglewski. 2000. Bacterial quorum sensing in pathogenic relationships. Infect. Immun. 68:4839-4849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lewis, H. A., E. B. Furlong, B. Laubert, G. A. Eroshkina, Y. Batiyenko, J. M. Adams, M. G. Bergseid, C. D. Marsh, T. S. Peat, W. E. Sanderson, J. M. Sauder, and S. G. Buchanan. 2001. A structural genomics approach to the study of quorum sensing: crystal structures of three LuxS orthologs. Structure 9:527-537. [DOI] [PubMed] [Google Scholar]
- 13.Nealson, K. H., and J. W. Hastings. 1979. Bacterial bioluminescence: its control and ecological significance. Microbiol. Rev. 43:496-518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nealson, K. H., T. Platt, and J. W. Hastings. 1970. Cellular control of the synthesis and activity of the bacterial luminescent system. J. Bacteriol. 104:313-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schauder, S., K. Shokat, M. G. Surette, and B. L. Bassler. 2001. The LuxS family of bacterial autoinducers: biosynthesis of a novel quorum-sensing signal molecule. Mol. Microbiol. 41:463-476. [DOI] [PubMed] [Google Scholar]
- 16.Sperandio, V., J. L. Mellies, W. Nguyen, S. Shin, and J. B. Kaper. 1999. Quorum sensing controls expression of the type III secretion gene transcription and protein secretion in enterohemorrhagic and enteropathogenic E. coli. Proc. Natl. Acad. Sci. USA 96:15196-15201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Surette, M. G., M. B. Miller, and B. L. Bassler. 1999. Quorum sensing in Escherichia coli, Salmonella typhimurium, and Vibrio harveyi: a new family of genes responsible for autoinducer production. Proc. Natl. Acad. Sci. USA 96:1639-1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Surette, M. G., and B. L. Bassler. 1999. Regulation of autoinducer production in Salmonella typhimurium. Mol. Microbiol. 31:585-595. [DOI] [PubMed] [Google Scholar]
- 19.Surette, M. G., and B. L. Bassler. 1998. Quorum sensing in Escherichia coli and Salmonella typhimurium. Proc. Natl. Acad. Sci. USA 95:7046-7050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Swift, S., A. V. Karlyshev, L. Fish, E. L. Durant, M. K. Winson, S. R. Chhabra, P. Williams, S. MacIntyre, and G. S. A. B. Stewart. 1997. Quorum sensing in Aeromonas hydrophila and Aeromonas salmonicida: identification of the LuxRI homologs AhyRI and AsaRI and their cognate N-acylhomoserine lactone signal molecules. J. Bacteriol. 179:5271-5281. [DOI] [PMC free article] [PubMed] [Google Scholar]