Abstract
Glycogen debranching enzyme (GDE) has 4-α-glucanotransferase and amylo-1,6-glucosidase activities in the single polypeptide chain. We analyzed the detailed action profile of GDE from Saccharomyces cerevisiae on amylose and tested whether GDE catalyzes cyclization of amylose. GDE treatment resulted in a rapid reduction of absorbance of iodine-amylose complex and the accumulation of a product that was resistant to an exo-amylase (glucoamylase [GA]) but was degraded by an endo-type α-amylase to glucose and maltose. These results indicated that GDE catalyzed cyclization of amylose to produce cyclic α-1,4 glucan (cycloamylose). The formation of cycloamylose was confirmed by high-performance anion-exchange chromatography, and the size was shown to range from a degree of polymerization of 11 to a degree of polymerization around 50. The minimum size and the size distribution of cycloamylose were different from those of cycloamylose produced by other 4-α-glucanotransferases. GDE also efficiently produced cycloamylose even from the branched glucan substrate, starch, demonstrating its potential for industrial production of cycloamylose.
4-α-Glucanotransferases (EC 2.4.1.25) catalyze the transfer of glucan chain from one α-1,4-glucan molecule to another, as shown in the following equation:
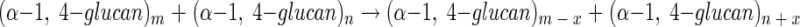 |
(1) |
The enzyme activity is widely distributed in bacteria, yeasts, plants, and mammals and is catalyzed by at least five structurally distinct enzyme groups (22). Studies of the function of these enzymes are important not only for understanding the activity-structure relationship of 4-α-glucanotransferases but also because an increasing number of 4-α-glucanotransferases are known to be able to catalyze a cyclization reaction to produce cycloamylose, as follows:
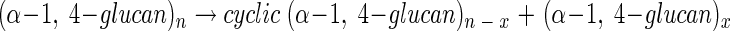 |
(2) |
Cycloamylose can accommodate hydrophobic guest molecules, so changing their solubility, reactivity, or stability, and thus can be used in the food, chemical, and pharmaceutical industries (22). However, it has recently been recognized that the size of cycloamylose produced depends upon the type of 4-α-glucanotransferase employed and that cycloamyloses with different sizes posses different structures and properties (2, 18, 22). Recently, Machida et al. (11) showed that high-molecular-weight cycloamylose can work better than conventional cyclodextrins (cycloamylose with a degree of polymerization [DP] of 6 to 8) as an artificial chaperone to promote refolding of denatured protein.
Glycogen debranching enzyme (GDE) (EC 2.4.1.25/EC 3.2.1.33) is also a 4-α-glucanotransferase but is particularly interesting since it has amylo-1,6-glucosidase activity in the same polypeptide chain (3, 14). The enzyme is distributed in mammals and yeasts and is involved in the degradation of glycogen in vivo. Glycogen phosphorylase (EC 2.4.1.1) degrades glycogen from its nonreducing ends, leaving dextrin with shortened chains. GDE transfers maltosyl or maltotriosyl units from the shortened chain to a nonreducing end of another chain with its transferase activity, leaving a single glucosyl residue attached. Then, GDE removes the glucosyl stub remaining at the branch point of glycogen with its amylo-1,6-glucosidase activity. Thus, GDE has unique structural and enzymatic properties among 4-α-glucanotransferases, and its action on amylose is of particular interest. The aim of this work is to analyze the action of GDE on amylose in detail, and to test the ability of GDE to produce cycloamylose.
MATERIALS AND METHODS
Chemicals.
Baker's yeast (Saccharomyces cerevisiae) was obtained from Kaneka Corporation (Osaka, Japan). Synthetic amyloses (average molecular masses of 5, 10, 30, 70, 110, and 320 kDa) were purchased from Nakano Vinegar Co., Ltd. (Aichi, Japan). Soluble starch (Pinedex no. 100) was purchased from Matsutani Chemical Industry Co., Ltd. (Hyogo, Japan). The standard cycloamylose mixture (DP, 6 to 31) was prepared as described previously (9). All other chemicals were obtained from Wako Pure Chemical Industries Ltd. (Osaka, Japan).
Purification of GDE from S. cerevisiae.
Baker's yeast (700 g) was suspended in 1,400 ml of 10 mM Tris-HCl (pH 7.5) containing 1 mM phenylmethylsulfonyl fluoride, and the cells were disrupted with a continuous operation DYNO-MILL (Shinmaru Enterprises Corporation, Osaka, Japan) using the following conditions: grinding container size, 600 ml; grinding elements, 750 g of 0.25- to 0.5-mm-diameter glass beads; flow rate, 40 ml/min. The lysate was centrifuged (12,000 × g, 30 min), and the supernatant was brought to 30% saturation with ammonium sulfate. The suspension was kept at 4°C for 2 h. The precipitate was removed by centrifugation (16,000 × g, 20 min). Ammonium sulfate was again added to the supernatant to a final concentration of 70% saturation, and the suspension was kept at 4°C overnight. After centrifugation (16,000 × g, 20 min), the precipitate was dissolved in 20 mM Tris-HCl buffer (pH 7.5) and dialyzed against the same buffer. The dialysate was heated at 50°C for 20 min and centrifuged. The supernatant was filtered through a 0.45-μm-pore-size membrane and then loaded onto a Q-Sepharose Fast Flow column (26 by 150 mm; Amersham Pharmacia Biotech, Uppsala, Sweden) and washed with 150 mM NaCl in 20 mM Tris-HCl buffer (pH 7.5). GDE was eluted with 20 mM Tris-HCl buffer (pH 7.5) containing 450 mM NaCl and dialyzed against 20 mM Tris-HCl buffer (pH 7.5). The solution was loaded onto a hydroxyapatite column (15 by 100 mm; Bio-Rad, Hercules, Calif.), and the enzyme was eluted with a linear gradient of 20 to 160 mM phosphate buffer (pH 6.8). Active fractions were pooled and a concentrated Tris buffer containing ammonium sulfate added to give final concentrations of 50 mM Tris-HCl (pH 7.5) and 500 mM ammonium sulfate. The solution was loaded onto a phenyl TOYOPEARL 650 M column (16 by 100 mm; TOSOH, Tokyo, Japan), and the enzyme was eluted with a linear gradient of 500 to 0 mM ammonium sulfate in 50 mM Tris-HCl buffer (pH 7.5). Active fractions were concentrated with an ultrafiltration unit (Centriprep 30; molecular weight cutoff, 30,000; Amicon, Beverly, Mass.) and dialyzed against 20 mM Tris-HCl buffer (pH 7.5).
Other enzymes.
Potato D-enzyme was purified to homogeneity from cell extract of Escherichia coli TG-1 carrying the plasmid pKK233-DPE (23). Thermus aquaticus amylomaltase was purified to homogeneity from cell extract of E. coli MC1061 carrying the plasmid pFQG8 as described previously (24). Isoamylase and pullulanase were purchased from Hayashibara Biochemical Laboratories (Okayama, Japan). Bacterial α-amylase (saccharifying type) was obtained from Nagase Chemtex Co., Ltd. (Kyoto, Japan). Glucoamylase from Rhizopus sp. was purchased from Toyobo Co., Ltd. (Osaka, Japan).
Assay of enzyme activity.
The GDE activity was determined by measuring its amylo-1,6-glucosidase activity using glucosyl-β-cyclodextrin (Glu-CD7) as a substrate. A reaction mixture (120 μl) containing 50 mM sodium acetate buffer (pH 6.0), 15 mM Glu-CD7, and enzyme was incubated at 30°C for 30 min, and the reaction tube was heated at 100°C for 5 min to stop the reaction. Released glucose was measured by the glucose oxidase method (13). One unit of activity is defined as the amount of enzyme that produces 1 μmol of glucose per min under this assay condition. D-enzyme and amylomaltase activities were assayed as described previously (23, 24).
Preparation of amylose solution.
Synthetic amylose (2 mg) was dissolved in 0.2 ml of 1 N NaOH solution and then neutralized by adding 0.4 ml of distilled water, 0.2 ml of 1 M sodium acetate buffer (pH 6.0), and 0.2 ml of 1 N HCl. The solution was used immediately after the neutralization.
Preparation of starch solution.
Soluble starch was purified to remove oligosaccharides as follows. One gram of soluble starch was dissolved in 100 ml of distilled water and 400 ml of ethanol was added, and the mixture was centrifuged (12,000 × g, 10 min). The pellet was dissolved in 10 ml of distilled water and again precipitated by adding 4 volumes of ethanol. The pellet was dissolved in 4 ml of distilled water (final concentration, 10% [wt/vol]).
Determination of the yield of GA-resistant molecules.
A reaction mixture (1 ml) containing 0.5 U of GDE, 0.1% (wt/vol) substrate, and 20 mM sodium acetate buffer (pH 6.0) was incubated at 40°C. To stop reaction, the mixture was heated at 100°C for 5 min. The denatured enzyme was removed by centrifugation. The amount of GA-resistant molecules was determined as follows. Fifty microliters of the reaction mixture was incubated either with GA (1.5 U) only or with an enzyme mixture containing GA (1.5 U), isoamylase (0.05 U), pullulanase (0.1 U), and α-amylase (0.2 U) in 50 mM sodium acetate buffer (pH 5.5) for 4 h at 40°C. Released glucose was measured by the glucose oxidase method (13). The amount of GA-resistant molecules was calculated by subtracting the amount of glucose released by GA only from that released by the enzyme mixture.
HPAEC.
High-performance anion-exchange chromatography (HPAEC) was carried out with a Dionex DX-300 system with a pulsed amperometric detector (model PADII; Dionex Corp., Sunnyvale, Calif.). The column was a Carbopac PA-100 (4 by 250 mm). A sample was injected and eluted with a gradient of sodium nitrate (0 to 2 min, 8 mM; 2 to 13 min, increasing from 22 to 26 mM with the installed gradient program 3; 3 to 35 min, increasing from 26 to 70 mM with the installed gradient program 4; 35 to 50 min, increasing from 70 to 200 mM with the installed gradient program 7; 50 to 52 min, 200 mM) in 150 mM NaOH with a flow rate of 1 ml/min.
Absorbance at 660 nm of amylose-iodine complex.
Twenty-five microliters of reaction mixture containing 0.1% (wt/vol) product was mixed with 1 ml of iodine reagent, and the absorbance at 660 nm was measured using a spectrophotometer (model UV-240; Shimadzu Co., Kyoto, Japan). Iodine reagent was made daily from 100 μl of iodine stock solution (0.26 g of I2 and 2.6 g of KI in 10 ml of water) mixed with 100 μl of 1 M HCl and diluted to 26 ml with distilled water.
Other procedures.
Reducing ends of glucan were quantitated using a modified Park-Johnson method (4). Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was performed on a 5 to 15% gradient gel, and the gel was stained using Coomassie brilliant blue. Time of flight mass spectrometry (TOF-MS) was carried out by a matrix-assisted laser desorption-ionization-TOF MS (Voyager-DE STR; Applied Biosystems, Foster City, Calif.) using 2,5-dihydroxybenzoic acid as the matrix.
RESULTS
Action of GDE on amylose.
GDE from Saccharomyces cerevisiae used in this study was purified as described in the Materials and Methods section. The enzyme was homogeneous as judged by SDS-PAGE (Fig. 1), and the molecular mass (150 kDa) coincided with the value reported by Tabata et al. (19). Furthermore, specific activity of the purified enzyme (2.4 U per mg of protein) was comparable with that reported by Tabata et al. (19). When the purified enzyme acted on maltooligosaccharide with a DP of 4, a series of maltooligosaccharides (DP, 2 or more) were detected by thin-layer chromatography. Glucose, maltose, and maltotriose were not used as substrates. When the enzyme acted on Glu-CD7, only glucose and CD7 were produced (data not shown). The action profiles coincided with those reported by other researchers (19, 20). These results demonstrated that the purity of GDE preparation was sufficient to investigate the GDE action.
FIG. 1.
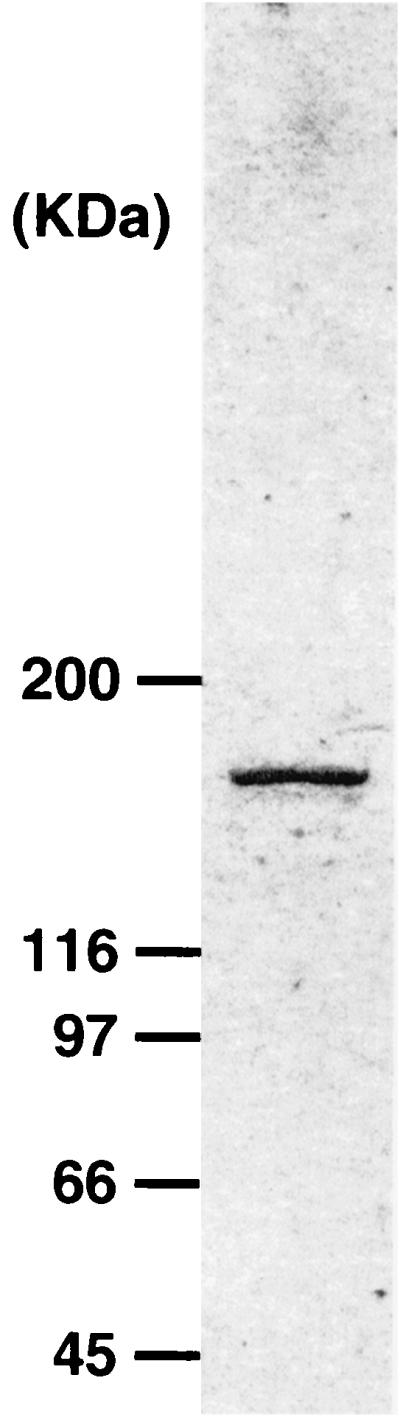
SDS-PAGE of the purified GDE from S. cerevisiae. Numbers on the left are the estimated molecular masses of marker proteins.
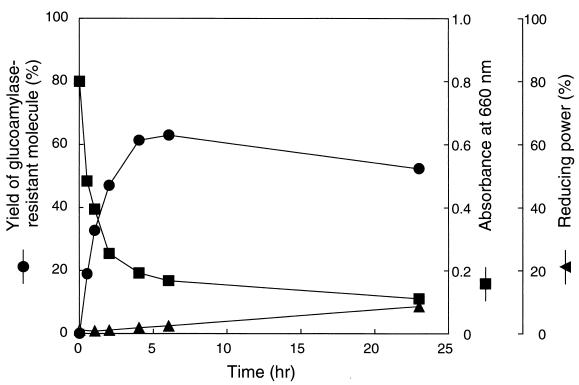
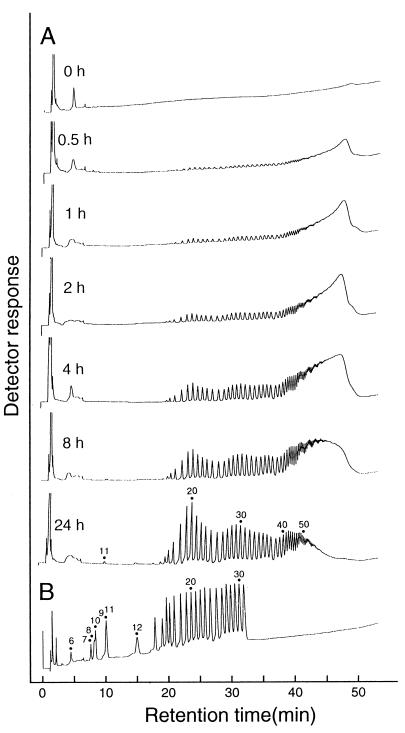
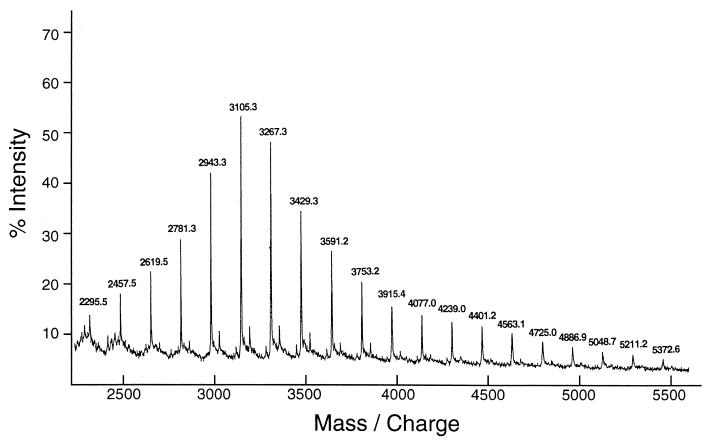
Synthetic amylose (average molecular mass of 30 kDa) was used to investigate GDE action, because it is a linear α-1,4 glucan and free from α-1,6 linkages. Purified GDE from S. cerevisiae was incubated with the amylose. The ability of amylose to form an iodine complex (absorbance at 660 nm) decreased rapidly (Fig. 2), indicating that the amylose chain was shortened by GDE action. Concomitantly, the yield of GA-resistant molecules increased to a maximum level of 63% after 6 h (Fig. 2). GA is an exo-type amylase and hydrolyzes both α-1,4 and α-1,6 linkages in glucans to produce glucose from nonreducing ends. Thus, linear or branched glucans are more or less completely broken down to glucose by GA. However, glucans with cyclic structure are resistant to GA. Therefore, the results strongly suggested that amylose was degraded by a cyclization reaction. To confirm this hypothesis, the GA-resistant molecules from various reaction stages were analyzed by HPAEC (Fig. 3). The GA-resistant molecules from the 0.5-h reaction mixture mainly eluted at 45 to 50 min. In this HPAEC system, glucans with a DP of >60 cannot be separated and are eluted together in this area (45 to 50 min). The sizes of the GA-resistant molecules decreased as the reaction progressed. After 24 h of reaction, however, the elution profile did not significantly change (data not shown). The GA-resistant molecules obtained were separated into many peaks, but the retention time of each peak was the same as that of cycloamylose standards. Furthermore, the GA-resistant molecules were completely degraded to glucose and maltose by α-amylase (endo-type amylase) but were not degraded by isoamylase and pullulanase (data not shown), indicating that the molecules consisted only of α-1,4-glucosidic linkages. A final confirmation of the cyclic nature of GA-resistant molecules was achieved by the determination of their molecular mass by TOF-MS. Many peaks produced from the GA-resistant molecules were detected at equal molecular mass intervals of a glucosyl residue of 162 Da, and their molecular masses agreed with those of the cyclic molecules but did not agree with those of the noncyclic molecules (Fig. 4 and Table 1). Those profiles coincided with those reported previously (23). From these results, we conclude that GDE can catalyze the cyclization of amylose to produce cycloamylose.
FIG. 2.
Action of GDE on amylose with average molecular mass of 30 kDa. A reaction mixture (2 ml) containing 2 mg of synthetic amylose AS-30, 20 mM sodium acetate buffer (pH 6.0), and 1.0 U of GDE was incubated at 40°C. At the indicated time points, 300 μl of the mixture was removed, boiled for 5 min to stop the reaction, centrifuged to remove insoluble materials, and then subjected to analyses.
FIG. 3.
HPAEC analysis of GA-resistant molecules. A 100-μl reaction mixture described in Fig. 2 was treated with a mixture of GA, isoamylase, and pullulanase. The GA-resistant molecules were precipitated by adding 10 volumes of acetone. Cycloamylose with a DP of 6 can be quantitatively precipitated by this treatment. (A) The pellet was dried, redissolved in 7.5 μl of 1 M NaOH, diluted to 50 μl with distilled water, and then subjected to HPAEC analysis. (B) Cycloamylose with DP between 6 and 31 (0.5 nmol each) are shown as standards.
FIG. 4.
TOF-MS analysis of GA-resistant molecules. The GA-resistant molecules were prepared as described in the legend to Fig. 3. After removing glucose with Waters Sep-Pak C18 cartridges, the GA-resistant molecules were subjected to TOF-MS. The number on each peak indicates the molecular mass (in daltons) of the molecules plus 23 Da (sodium ion). Molecules with a molecular mass of less than 2,200 could not be detected because of noises from the matrix under this condition.
TABLE 1.
Experimental and theoretical masses of glucans
| DP | Exptl massa (Da) | Theoretical mass (Da)
|
|
|---|---|---|---|
| Cyclic glucanb | Noncyclic glucanc | ||
| 14 | 2,295.5 | 2,293.0 | 2,311.0 |
| 15 | 2,457.5 | 2,455.1 | 2,473.1 |
| 16 | 2,619.5 | 2,617.2 | 2,635.2 |
| 17 | 2,781.3 | 2,779.4 | 2,797.4 |
| 18 | 2,943.3 | 2,941.5 | 2,959.5 |
| 19 | 3,105.3 | 3,103.7 | 3,121.7 |
| 20 | 3,267.3 | 3,265.8 | 3,283.8 |
| 21 | 3,429.3 | 3,427.9 | 3,445.9 |
| 22 | 3,591.2 | 3,590.1 | 3,608.1 |
| 23 | 3,753.2 | 3,752.2 | 3,770.2 |
| 24 | 3,915.4 | 3,914.4 | 3,932.4 |
| 25 | 4,077.0 | 4,076.5 | 4,094.5 |
| 26 | 4,239.0 | 4,238.6 | 4,256.6 |
| 27 | 4,401.2 | 4,400.8 | 4,418.8 |
| 28 | 4,563.1 | 4,562.9 | 4,580.9 |
| 29 | 4,725.0 | 4,725.1 | 4,743.1 |
| 30 | 4,886.9 | 4,887.2 | 4,905.2 |
| 31 | 5,048.7 | 5,049.3 | 5,067.3 |
| 32 | 5,211.2 | 5,211.5 | 5,229.5 |
| 33 | 5,372.6 | 5,373.6 | 5,391.6 |
Determined by matrix-assisted laser desorption-ionization-TOF-MS.
Calculated as 162.14n + 22.990 Da, where 162.14 is the mass of a glucosyl residue, n is the DP; and 22.990 is the mass of a sodium ion.
Calculated as 162.14n + 18.015 + 22.990 Da, where 162.14 is the mass of a glucosyl residue, n is the DP, 18.015 is the additional mass of a reducing and residue, and 22.990 is the mass of a sodium ion.
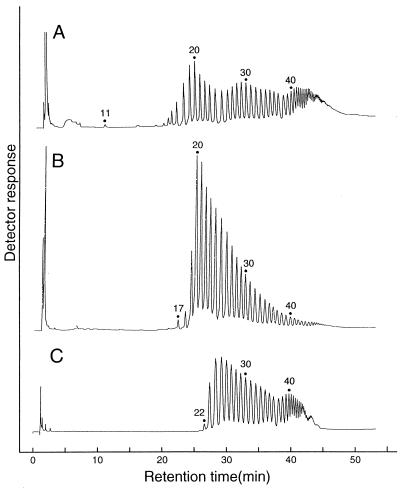
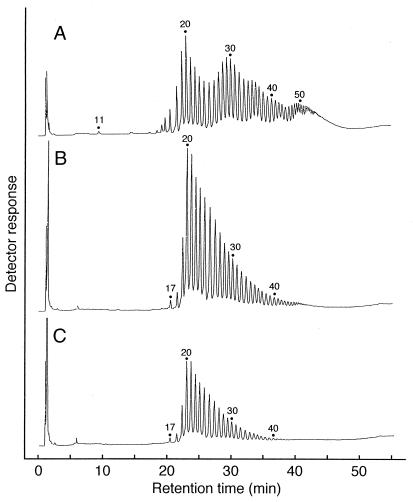
The minimum DP of cycloamylose produced by GDE was considered to be 11 (Fig. 5A) from the HPAEC analysis, although molecules with a DP of 11 to 13 were not detected by the TOF-MS spectrum because of noise derived from matrix. The minimum size is apparently different from those produced by potato D-enzyme (Fig. 5B) and T. aquaticus amylomaltase (Fig. 5C). The elution profile of cycloamylose produced by GDE was also unique, showing characteristic trimodal distribution (Fig. 5A).
FIG. 5.
HPAEC analysis of cycloamylose produced by GDE (A), D-enzyme (B), or T. aquaticus amylomaltase (C). The cycloamyloses produced by GDE were prepared as described in the legend to Fig. 3. Cycloamyloses were prepared from amylose with an average molecular mass of 30 kDa by potato D-enzyme and T. aquaticus amylomaltase as described previously (23, 24).
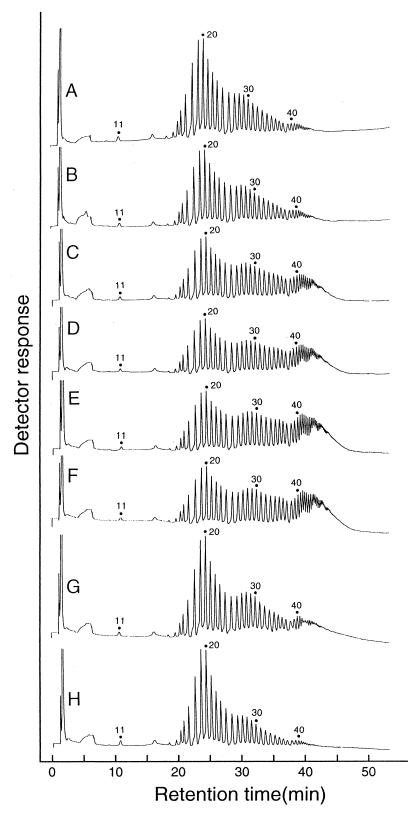
Cycloamylose production from various substrates.
GDE was incubated with 0.1% (wt/vol) amyloses with various molecular sizes (molecular weight, 5,000 to 320,000), amylopectin, and debranched starch (short chain amylose) (Fig. 6). In all cases, the yield of cycloamylose was around 50 to 60% after 24-h reactions, and the cycloamylose showed characteristic trimodal distribution.
FIG. 6.
HPAEC analysis of cycloamyloses derived from various substrates. A reaction mixture (100 μl) containing 0.1% (wt/vol) substrate, 0.05 U of GDE, and 20 mM sodium acetate buffer (pH 6.0) was incubated at 40°C. After 24 h, the mixture was heated at 100°C for 5 min to stop the reaction. Cycloamylose was prepared as described in the legend to Fig. 2. The substrates were amyloses with average molecular masses of 5 kDa (A), 10 kDa (B), 30 kDa (C), 70 kDa (D), 110 kDa (E), and 320 kDa (F); amylopectin (G); and debranched starch (short-chain amylose) (H).
Because starch is economically preferred to amylose as a substrate, the production of cycloamylose from starch was investigated in further detail. Potato D-enzyme and GDE were incubated with soluble starch. The yield of cycloamylose with D-enzyme was only 15%. Addition of isoamylase and pullulanase to hydrolyze the α-1,6 linkages of starch increased the yield to 40%. On the other hand, GDE produced cycloamylose with a yield of 60% (after 6 h) without any other enzymes. The HPAEC analysis indicated that cycloamyloses produced with D-enzyme and GDE had different size distributions (Fig. 7).
FIG. 7.
HPAEC analysis of cycloamylose produced by GDE (A), D-enzyme plus isoamylase (B), or D-enzyme (C) from soluble starch. A reaction mixture (2.5 ml) containing 0.1% (wt/vol) soluble starch, 0.625 U of GDE, and 10 mM sodium acetate buffer (pH 6.0) was incubated at 40°C. As controls, 875 U of D-enzyme or enzyme mixture (875 U of D-enzyme, 4 U of isoamylase) was used in place of GDE. The cycloamyloses were prepared from 24-h reaction mixture as described in the legend to Fig. 2.
Cycloamylose production by GDE in the presence of glucose.
The first process for the purification of cycloamylose from the reaction mixture is to degrade linear glucan into glucose by exo-type amylase. However, when cyclodextrin glucanotransferases (CGTase), D-enzyme, and amylomaltase are incubated with cycloamylose in the presence of glucose, these enzymes catalyze the linearization of cycloamylose (coupling reaction) (22). The coupling reaction would reduce the yield of cycloamylose, so, these 4-α-glucanotransferases must be completely inactivated before the purification process. On the other hand, it has been reported that GDE cannot transfer glucan to D-glucose or maltose as acceptors (20). This report suggested that GDE would not catalyze linearization of cycloamylose in the presence of glucose or maltose and that the inactivation step could be omitted in the process of cycloamylose production.
GDE and D-enzyme were incubated with amylose (average molecular mass of 30 kDa) with or without glucose. As expected, D-enzyme could not produce cycloamylose in the presence of a low concentration of glucose. On the other hand, glucose did not affect the cycloamylose production by GDE at the concentrations tested (0.01 and 0.1%).
DISCUSSION
Takaha and Smith (22) classified 4-α-glucanotransferases into five types (types I to V) based on their primary structures and substrate specificities. Characteristics of these enzymes are shown in Table 2. Cyclization reactions catalyzed by enzymes belonging to types I, II, and V have been already described. Type I enzyme is well known for the synthesis of cycloamylose with a DP between 6 and 8 (α-, β-, and γ-cyclodextrins) and is commonly referred to as CGTase. Previously, we have demonstrated that CGTase can produce cycloamylose with higher DP (on the order of several hundred), especially in the initial reaction stages (27). Here we have demonstrated that the type III enzyme, GDE, also catalyzes cyclization, in spite of its unique structural and enzymatic properties among 4-α-glucanotransferases. This result may suggest that 4-α-glucanotransferases generally catalyze the cyclization reaction. We anticipate that further research on the type IV enzymes and the noncategorized glucanotransferases may well reveal cyclization activities.
TABLE 2.
Characteristics of activities of each type of 4-α-glucanotransferase
| Type | Source | Smallest donor | Smallest acceptor | Smallest transferred unit | Disproportionated products | Cyclization reaction | Smallest cycloamylose (DP) | Reference(s) |
|---|---|---|---|---|---|---|---|---|
| I | Bacillus ohbensis | G3b | G | G | G, G2, G3, Gn | + | 6 | 8 |
| Bacillus macerans | G3 | G | G | G, G2, G3, Gn | + | 6 | 8 | |
| Bacillus megaterium | G2 | G | G | G, G2, G3, Gn | + | 6 | 8 | |
| II | Potato | G3 | G | G2 | G, G3, Gn | + | 17 | 7, 23 |
| Escherichia coli K-12 | G2 | G | G | G, G2, G3, Gn | + | 17 | Unpublished data | |
| Thermus aquaticus | G2 | G | G | G, G2, G3, Gn | + | 22 | 24 | |
| III | Rabbit muscle | NT | G | G2 | NT | NT | 1 | |
| Yeast | G4 | G3 | G2 | G2, G3, Gn | + | 11 | 20, this work | |
| IV | Thermotoga maritima | G4 | G2 | G2 | G2, G3, Gn | NT | 10 | |
| V | Thermococcus litoralis | G2 | G | G | G, G2, G3, Gn | + | 16 | 5, 6 |
| Pyrococcus kodakaraensis | G2 | G | G | G, G2, G3, Gn | + | 16 | 21 | |
| Othera | Streptococcus mitis | G2 | G | G | G, G2, G3, Gn | NT | 28 | |
| Streptococcus bovis | G3 | G | G | G, G2, G3, Gn | NT | 29 | ||
| Bacillus subtilis | G4 | G4 | G2 | G2, G3, Gn | NT | 15 | ||
| Streptococcus mutans | G2 | G | G | G, G2, G3, Gn | NT | 12 |
These enzymes could not be categorized because of the lack of structural information.
G, G2, G3, G4 and Gn are glucose, maltose, maltotriose, maltotetraose, and maltooligosaccharides with a DP of 5 and larger, respectively.
The most distinctive difference among cycloamylose produced by each 4-α-glucanotransferase is the DP of the smallest molecule (Table 2). CGTases (type I) produce a molecule with a DP of 6, while amylomaltases and D-enzyme (type II) produce molecules with DP between 17 and 22 as the smallest cycloamyloses. Recently, the tertiary structure of T. aquaticus amylomaltase has been obtained by X-ray crystallographic studies and compared with that of CGTase (16, 17). It was suggested that the enzyme structures of CGTase and amylomaltase, respectively, determine the minimum size of cycloamylose. GDE produces cycloamylose with a DP of 11 as the smallest molecule. GDE may have structural features to create this size of cycloamylose. The other difference is the size-distribution of the cycloamylose (Fig. 5). GDE may have a structural motif that promotes production of cycloamylose with a DP around 30. The X-ray analysis of GDE may bring a novel insight into the cyclization mechanism of 4-α-glucanotransferases.
Cycloamylose with a high DP is potentially useful for various industries. We propose two processes for cycloamylose production. One process uses type II enzyme (D-enzyme/amylomaltase) as a catalyst, and amylose as a substrate. The enzymes efficiently produce cycloamylose, especially when high-molecular-weight amylose is used as a substrate. For example, the yield of cycloamylose from 320-kDa amylose reached more than 95% (23, 24). Furthermore, the average size of cycloamylose can be controlled by changing the reaction time or amount of enzyme. Type I (CGTase), III (GDE), and V (glucanotransferases from hyper-thermophilic archaea) enzymes are not as suitable as type II in this process, for the following reasons. The main products of CGTases are cyclodextrins with a DP between 6 and 8 (25, 26), and the yield of cycloamylose with a high DP would be lower than that by type II. The cycloamylose yield by the type V enzymes was also relatively low; 50% (6) to 67% (21). This is because these enzymes have relatively high hydrolytic activities compared with type II enzymes (6, 21). GDE is also not suitable for this process because of its low cycloamylose yield (around 60%). This is probably due to hydrolytic activity of GDE as indicated in Fig. 2.
The other process uses GDE as a catalyst and starch as substrate. The yield of cycloamylose is around 60%. Starch is much preferred to amylose as a substrate from the viewpoint of availability and cost. Furthermore, GDE does not catalyze the coupling reaction to degrade cycloamylose even in the presence of glucose. These properties would make the process simple. However, the size distribution cannot be controlled. Thus, each process has its own merits and demerits, and we have to select the process with consideration to the cost and desired quality of the cycloamylose product.
Acknowledgments
This work was supported by a grant for the development of the next generation of bioreactor systems from the Society for Techno-Innovation of Agriculture, Forestry and Fisheries (STAFF), Tokyo, Japan, to Ezaki Glico Co., Ltd.
REFERENCES
- 1.Brown, D. H., and B. Illingworth. 1962. The properties of an oligo-1,4-1,4-glucantransferase from animal tissues. Proc. Natl. Acad. Sci. USA 48:1783-1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gessler, K., I. Uson, T. Takaha, N. Krauss, S. M. Smith, S. Okada, G. M. Sheldrick, and W. Saenger. 1999. V-amylose at atomic resolution: X-ray structure of a cycloamylose with 26 glucose residues (cyclomaltohexaicosaose). Proc. Natl. Acad. Sci. USA 96:4246-4251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gillard, B. K., and T. E. Nelson. 1977. Amylo-1,6-glucosidase/4-α-glucanotransferase: use of reversible substrate model inhibitors to study the binding and active sites of rabbit muscle debranching enzyme. Biochemistry 16:3978-3987. [DOI] [PubMed] [Google Scholar]
- 4.Hizukuri, S., K. Shirasaka, and B. O. Juliano. 1983. Phosphorus and amylose branching in rice starch granules. Starch/Stärke 35:348-350. [Google Scholar]
- 5.Imamura, H., S. Fushinobu, M. Yamamoto, T. Kumasaka, T. Wakagi, and H. Matsuzawa. 2001. Reaction mechanism and crystal structure of 4-α-glucanotransferase from a hyperthermophilic archaeon, Thermococcus litoralis. J. Appl. Glycosci. 48:171-175. [Google Scholar]
- 6.Jeon, B. S., H. Taguchi, H. Sakai, T. Ohshima, T. Wakagi, and H. Matsuzawa. 1997. 4-α-Glucanotransferase from the hyperthermophilic archaeon Thermococcus litoralis: enzyme purification and characterization, and gene cloning, sequencing and expression in Escherichia coli. Eur. J. Biochem. 248:171-178. [DOI] [PubMed] [Google Scholar]
- 7.Jones, G., and W. J. Whelan. 1969. The action pattern of D-enzyme, a transmaltodextrinylase from potato. Carbohydr. Res. 9:483-490. [Google Scholar]
- 8.Kitahata, S. 1995. Cyclomaltodextrin glucanotransferase, p. 6-17. In The Amylase Research Society of Japan (ed.), Enzyme chemistry and molecular biology of amylases and related enzymes. CRC Press, Boca Raton, Fla.
- 9.Koizumi, K., H. Sanbe, Y. Kubota, Y. Terada, and T. Takaha. 1999. Isolation and characterization of cyclic α-(1→4)-glucans having degrees of polymerization 9-31 and their quantitative analysis by high-performance anion-exchange chromatography with pulsed amperometric detection. J. Chromatogr. A 852:407-416. [DOI] [PubMed] [Google Scholar]
- 10.Liebl, W., R. Feil, J. Gabelsberger, J. Kellermann, and K. H. Schleifer. 1992. Purification and characterization of a novel thermostable 4-α-glucanotransferase of Thermotoga maritima cloned in Escherichia coli. Eur. J. Biochem. 207:81-88. [DOI] [PubMed] [Google Scholar]
- 11.Machida, S., S. Ogawa, S. Xiaohua, T. Takaha, K. Fujii, and K. Hayashi. 2000. Cycloamylose as an efficient artificial chaperone for protein refolding. FEBS Lett. 486:131-135. [DOI] [PubMed] [Google Scholar]
- 12.Medda, S., and E. E. Smith. 1984. A radioisotope method for assays of amylomaltase and D-enzyme. Anal. Biochem. 138:354-359. [DOI] [PubMed] [Google Scholar]
- 13.Miwa, I., J. Okudo, K. Maeda, and G. Okuda. 1972. Mutarotase effect on colorimetric determination of blood glucose with D-glucose oxidase. Clin. Chim. Acta 37:538-540. [DOI] [PubMed] [Google Scholar]
- 14.Nakayama, A., K. Yamamoto, and S. Tabata. 2001. Identification of the catalytic residues of bifunctional glycogen debranching enzyme. J. Biol. Chem. 276:28824-28828. [DOI] [PubMed] [Google Scholar]
- 15.Pazur, J. H., and S. Okada. 1968. The isolation and mode of action of a bacterial glucanotransferase. J. Biol. Chem. 243:4732-4738. [PubMed] [Google Scholar]
- 16.Przylas, I., Y. Terada, K. Fujii, T. Takaha, W. Saenger, and N. Strater. 2000. X-ray structure of acarbose bound to amylomaltase from Thermus aquaticus implications for the synthesis of large cyclic glucans. Eur. J. Biochem. 267:6903-6913. [DOI] [PubMed] [Google Scholar]
- 17.Przylas, I., K. Tomoo, Y. Terada, T. Takaha, K. Fujii, W. Saenger, and N. Strater. 2000. Crystal structure of amylomaltase from Thermus aquaticus, a glycosyltransferase catalysing the production of large cyclic glucans. J. Mol. Biol. 296:873-886. [DOI] [PubMed] [Google Scholar]
- 18.Saenger, W., J. Jacob, K. Gessler, T. Steiner, D. Hoffmann, H. Sanbe, K. Koizumi, S. M. Smith, and T. Takaha. 1999. Structures of the common cyclodextrins and their larger analogues: beyond the doughnut. Chem. Rev. 98:1787-1802. [DOI] [PubMed] [Google Scholar]
- 19.Tabata, S., and S. Hizukuri. 1992. Properties of yeast debranching enzyme and its specificity toward branched cyclodextrins. Eur. J. Biochem. 206:345-348. [DOI] [PubMed] [Google Scholar]
- 20.Tabata, S., and T. Ide. 1988. Electorochemical detection of reducing carbohydrates produced by the transferase action of yeast debranching enzyme on maltosaccharides. Carbohydr. Res. 176:245-251. [Google Scholar]
- 21.Tachibana, Y., T. Takaha, S. Fujiwara, M. Takagi, and T. Imanaka. 2000. Acceptor specificity of 4-α-glucanotransferase from Pyrococcus kodakaraensis KOD1, and synthesis of cycloamylose. J. Biosci. Bioeng. 90:406-409. [DOI] [PubMed] [Google Scholar]
- 22.Takaha, T., and S. M. Smith. 1999. The functions of 4-α-glucanotransferases and their use for the production of cyclic glucans. Biotechnol. Genet. Eng. Rev. 16:257-280. [DOI] [PubMed] [Google Scholar]
- 23.Takaha, T., M. Yanase, H. Takata, S. Okada, and S. M. Smith. 1996. Potato D-enzyme catalyzes the cyclization of amylose to produce cycloamylose, a novel cyclic glucan. J. Biol. Chem. 271:2902-2908. [DOI] [PubMed] [Google Scholar]
- 24.Terada, Y., K. Fujii, T. Takaha, and S. Okada. 1999. Thermus aquaticus ATCC 33923 amylomaltase gene cloning and expression and enzyme characterization: production of cycloamylose. Appl. Environ. Microbiol. 65:910-915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Terada, Y., H. Sanbe, T. Takaha, S. Kitahata, K. Koizumi, and S. Okada. 2001. Comparative study of the cyclization reactions of three bacterial cyclomaltodextrin glucanotransferases. Appl. Environ. Microbiol. 67:1453-1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Terada, Y., H. Sanbe, T. Takaha, S. Okada, S. Kitahata, and K. Koizumi. 2001. Reaction of CGTase on amylose. J. Appl. Glycosci. 48:177-186. [Google Scholar]
- 27.Terada, Y., M. Yanase, H. Takata, T. Takaha, and S. Okada. 1997. Cyclodextrins are not the major cyclic α-1,4-glucans produced by the initial action of cyclodextrin glucanotransferase on amylose. J. Biol. Chem. 272:15729-15733. [DOI] [PubMed] [Google Scholar]
- 28.Walker, G. J. 1966. Metabolism of the reserve polysaccharide of Streptococcus mitis. Biochem. J. 101:861-872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Walker, G. J. 1965. A transglucosylase of Streptococcus bovis. Biochem. J. 94:299-308. [DOI] [PMC free article] [PubMed] [Google Scholar]