Abstract
A 16S rRNA-targeted probe was designed and validated in order to quantify the number of uncultured Ruminococcus obeum-like bacteria by fluorescent in situ hybridization (FISH). These bacteria have frequently been found in 16S ribosomal DNA clone libraries prepared from bacterial communities in the human intestine. Thirty-two reference strains from the human intestine, including a phylogenetically related strain and strains of some other Ruminococcus species, were used as negative controls and did not hybridize with the new probe. Microscopic and flow cytometric analyses revealed that a group of morphologically similar bacteria in feces did hybridize with this probe. Moreover, it was found that all hybridizing cells also hybridized with a probe specific for the Clostridium coccoides-Eubacterium rectale group, a group that includes the uncultured R. obeum-like bacteria. Quantification of the uncultured R. obeum-like bacteria and the C. coccoides-E. rectale group by flow cytometry and microscopy revealed that these groups comprised approximately 2.5 and 16% of the total community in fecal samples, respectively. The uncultured R. obeum-like bacteria comprise about 16% of the C. coccoides-E. rectale group. These results indicate that the uncultured R. obeum-like bacteria are numerically important in human feces. Statistical analysis revealed no significant difference between the microscopic and flow cytometric counts and the different feces sampling times, while a significant host-specific effect on the counts was observed. Our data demonstrate that the combination of FISH and flow cytometry is a useful approach for studying the ecology of uncultured bacteria in the human gastrointestinal tract.
The human gastrointestinal (GI) tract harbors a diverse microbial community, which has important metabolic and protective functions in the GI tract (for a review see reference 18). Recent studies have indicated that interactions between the host and the bacterial community are of considerable importance but are very complex and are just starting to be understood (4, 9, 10, 25). Most information about the bacterial community in the human GI tract has been obtained by selective cultivation of microbes from fecal samples. In the past 5 years, culture-independent approaches in which the sequence variability of the 16S rRNA genes has been used have demonstrated that most of the predominant bacteria in human fecal samples have not yet been obtained in culture, illustrating the limitation in our knowledge of these predominant organisms (16, 23, 24). In addition, denaturing gradient gel electrophoresis and temperature gradient gel electrophoresis of fecal 16S ribosomal DNA (rDNA) and rRNA amplicons have been demonstrated to be powerful culture-independent approaches for determining and monitoring the bacterial community in feces (24, 25). Such studies have revealed that the predominant bacterial community in human feces is relatively stable over time, is host specific, and is not significantly altered following consumption of certain probiotic strains (19, 20, 24).
Although the use of 16S rDNA-directed denaturing gradient gel electrophoresis, cloning, and sequencing has provided new insights into the bacterial composition of the human GI tract, these approaches are all based on the use of PCR amplification methods, and hence the results cannot be accurately converted to real numbers of bacteria. Fluorescent in situ hybridization (FISH) performed with 16S rRNA-targeted oligonucleotide probes has been shown to be very powerful for detecting and quantifying uncultured bacteria in environmental samples (for a review see reference 3). FISH analysis of fecal populations has demonstrated that these populations are relatively stable over time (5). The members of various genera, like Bacteroides, Bifidobacterium, Streptococcus, Lactobacillus, Collinsella, Eubacterium, and Clostridium, could be quantified accurately in feces by using the FISH approach (5, 6, 7, 13). In addition, a group of Fusobacterium prausnitzii-like bacteria, which was found to be predominant in several fecal clone libraries (16, 23, 24), was also found to be predominant by using the FISH approach (5, 17).
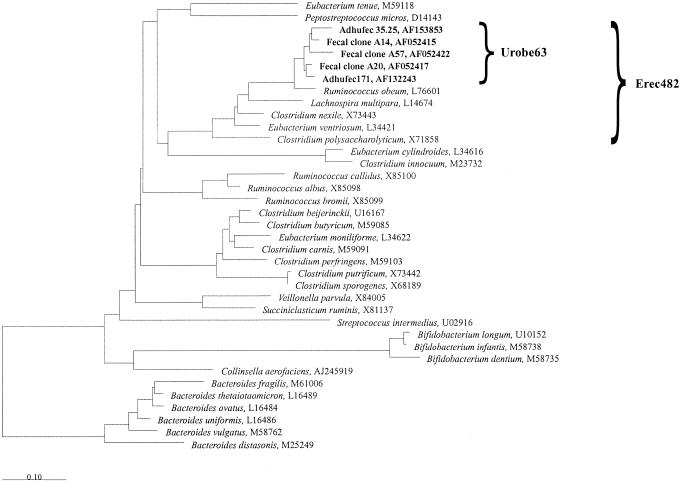
In several studies, a group of closely related uncultured bacteria whose closest cultivable relative is Ruminococcus obeum was found regularly in fecal 16S rDNA clone libraries (16, 24) (Fig. 1). These organisms were also found to be present as determined by fecal temperature gradient gel electrophoresis profiles of dominant and active bacteria and could be important members of the bacterial community in the human GI tract (24). Since the closely related cloned amplicons all have less than 97% sequence similarity to R. obeum, they represent a group of hitherto unknown species. Until now, no quantitative data for these organisms have been obtained, and the morphology of these bacteria remains unknown, since their presence has been determined only on the basis of PCR data.
FIG. 1.
Rooted neighbor-joining tree showing the phylogenetic relationships among the 16S rDNA sequences of the uncultured R. obeum-like bacteria (indicated by boldface type) and some closely and distantly related bacteria. The phylogenetic tree was generated from the tree of life of the ARB software package. The Bacteroides group was used as an outgroup. Accession numbers of the 16S rDNA sequences are indicated. Bar = 10% sequence divergence. The target sequences of the Urobe63 probe and the Erec482 probe are indicated.
In many studies, FISH analysis is combined with microscopic analysis. Although less frequently used, the combination of FISH with flow cytometry has been used successfully to analyze different microbial communities (2, 14, 21, 22). Major advantages of this combination are the multiparametric analysis of samples and the relatively fast and sensitive quantification of populations, even those that make up only about 1% of the total community.
In the present study, we developed a set of two probes to detect and quantify the group of uncultured R. obeum-like bacteria by FISH in combination with direct microscopy and flow cytometry. After validation, the probes were used to quantify the number of hybridized cells in fecal samples from different individuals by microscopic and flow cytometric analysis. In addition, the Eubacterium rectale-Clostridium coccoides group, which includes the uncultured R. obeum-like bacteria, and the total bacteria were quantified. Variations over time, among individuals, and between the approaches were analyzed statistically.
MATERIALS AND METHODS
Design and validation of the oligonucleotide probes.
Sequences of closely related uncultured R. obeum-like bacteria from human fecal samples were obtained from two studies in which fecal 16S rDNA clone libraries were constructed and analyzed (Fig. 1) (16, 24). The identities (and accession numbers) of these sequences are as follows: fecal clone A14 (AF052415), fecal clone A20 (AF052417), fecal clone A57 (AF052422), adhufec171 (AF132243), and adhufec35.25 (AF153853). The sequences were aligned by using the ARB software package (15), and probes targeting these sequences were designed. A newly designed probe was screened for specificity based on comparative analysis of 16S rRNA sequences from the ARB database and the Ribosomal Database Project (RDP) (11) and newly deposited sequences from GenBank by using the ARB and RDP software (Table 1). The CHECK_PROBE analysis function of the RDP and the Probe Match function of ARB were used to screen the 16S and 23S rRNA sequence databases for target sequences for the new probe.
TABLE 1.
Alignment of probe sequences and target sequences having a maximum of two mismatchesa
| Probe or target | Sequence |
|---|---|
| S-∗-Urobe-0063-a-A-20b | 3′ GC-TTG-CCC-TTA-ATG-AAA-TAA |
| S-∗-Urobe-0063-b-A-20b | 3′ GC-TTG-CCC-TTT-ATR-AAR-TAA |
| Target | 5′ CG AAC GGG AAW UAY UUY AUU |
| Fecal clones A14, A20, and A57 | .. ... ... ... ... ... ... |
| adhufec35.25 | .. ... ... ... ... ... ... |
| adhufec171 | NN NNN NNN NN. ... ... ... |
| Ruminococcus obeum | .. ... ... ... CY. ... ... |
| Rumen clone RFN27 | .. ... ... ... ... ... U.A |
N, R, W, and Y are the International Union of Pure and Applied Chemistry codes for ambiguous bases. Dots indicate bases that are identical to bases in the target site. Differences between the probes are indicated by boldface type. Mismatches of target sequences with the probe target site are underlined.
Oligonucleotide Probe Database nomenclature (1). The Urobe63 probe is a 1:1 mixture of probes S-∗-Urobe-0063-a-A-20 and S-∗-Urobe-0063-b-A-20.
Thirty-two reference strains (Table 2) belonging to various phylogenetic groups found in the GI tract, including R. obeum, were used as negative controls to evaluate the specificity of the new probe. This probe, designated Urobe63 (uncultured Ruminococcus obeum-like bacteria), is a 1:1 mixture of the oligonucleotide probes S-*-Urobe-0063-a-A-20 and S-*-Urobe-0063-b-A-20 (Table 1). The Eub338 probe (2) was used as a positive control for hybridization of the negative reference strains. Since the positive target of the Urobe63 probe is a 16S rDNA sequence of an uncultured group of R. obeum-like bacteria, no positive reference strain could be used. This means that the probe specificity could only be determined by FISH analysis by using bacteria from a reference fecal sample as positive controls. Therefore, bacteria from a reference fecal sample were added to the reference strains in order to verify hybridization signals of the Urobe63 probe. The optimal hybridization conditions were determined by performing hybridizations at a constant temperature of 50°C, gradually increasing the formamide concentration in the hybridization buffer, and adjusting the salt concentration in the corresponding washing buffer as described previously (12). In addition, fecal samples were hybridized with the Urobe63 probe and the Erec482 probe (5), which targets the C. coccoides-E. rectale group that includes the uncultured R. obeum-like bacteria (Fig. 1), to determine whether the Urobe63-positive cells appeared to be double labeled. Furthermore, competitive experiments with a probe which targets only the cultured species R. obeum (5′ AAT GAA ARG TTT CCC GTT CG) were performed to verify if cross-reactions occurred with the nontarget organism having the fewest mismatches with the Urobe63 probe.
TABLE 2.
Reference strains used to validate the probe hybridization conditions
| Strain | Origina |
|---|---|
| Bacteroides fragilis | DSM 2151 |
| Bacteroides distasonis | DSM 20701 |
| Bacteroides ovatus | MMB |
| Bacteroides thetaiotaomicron | MMB |
| Bacteroides uniformis | MMB |
| Bacteroides vulgatus | DSM 1447 |
| Bifidobacterium dentium | ATCC 27678 |
| Bifidobacterium infantis | ATCC 15697 |
| Bifidobacterium longum | MMB |
| Clostridium beijerinckii | MMB |
| Clostridium butyricum | MMB |
| Clostridium carnis | DSM 1293 |
| Clostridium innocuum | MMB |
| Clostridium nexile | MMB |
| Clostridium perfringens | MMB |
| Clostridium polysaccharolyticum | DSM 1801 |
| Clostridium putrificum | DSM 1734 |
| Clostridium sporogenes | DSM 795 |
| Collinsella aerofaciens | DSM 13713 |
| Eubacterium cylindroides | MMB |
| Eubacterium moniliforme | MMB |
| Eubacterium tenue | DSM 20695 |
| Eubacterium ventriosum | DSM 3988 |
| Lachnospira multipara | DSM 3073 |
| Peptostreptococcus micros | DSM 20468 |
| Ruminococcus albus | ATCC 27210 |
| Ruminococcus bromii | ATCC 27255 |
| Ruminococcus callidus | ATCC 27760 |
| Ruminococcus obeum | ATCC 29174 |
| Streptococcus intermedius | DSM 20573 |
| Succiniclasticum ruminis | DSM 9236 |
| Veillonella parvula | DSM 20373 |
DSM, Deutsche Sammlung von Mikroorganismen und Zellkulturen, Braunschweig, Germany; MMB, Laboratory for Medical Microbiology, Groningen, The Netherlands; ATCC, American Type Culture Collection, Rockville, Md.
Reference strains, culture conditions, and fixation.
The 32 reference strains used in this study were obtained from various sources, including the Deutsche Sammlung von Mikroorganismen und Zellkulturen, Braunschweig, Germany; the American Type Culture Collection, Rockville, Md.; and the Laboratory for Medical Microbiology, Groningen, The Netherlands (5) (Table 2). The American Type Culture Collection and Deutsche Sammlung von Mikroorganismen und Zellkulturen strains were cultivated as described in the catalogues of these organizations. All Laboratory for Medical Microbiology strains are clinical or human fecal isolates from local and regional public health laboratories that were identified and cultivated by using standard procedures (8). Exponentially grown cells were harvested by centrifugation at 5,000 × g for 10 min, washed with filtered (pore size, 0.2 μm) phosphate-buffered saline (PBS) (8 g of NaCl per liter, 0.2 g of KCl per liter, 1.44 g of Na2HPO4 per liter, 0.24 g of KH2PO4 per liter; pH 7.2), and diluted 1:3 with 4% (wt/vol) paraformaldehyde in PBS. After fixation at 4°C for 16 h, cells were stored in 50% ethanol-PBS until FISH analysis was performed (2).
Fecal sample processing.
Fecal samples were collected from three healthy Dutch male volunteers (ages, 25 to 32 years) and were processed within 30 min. These volunteers provided three samples within a 4-week period and had not been subjected to any feeding trial, specific diet, or antibiotic treatment for the previous 3 years. Additional fecal samples were collected from six volunteers (two men and four women) who were 25 to 40 years old and who originated from different countries (The Netherlands, Germany, People's Republic of China, Portugal, Italy, and Tunisia) but had lived for at least 3 months in The Netherlands. Fecal samples were processed as described previously (5). Briefly, 0.5 g of a fecal sample was resuspended in 4.5 ml of PBS and vortexed with 5 to 10 glass beads for 5 min to homogenize the sample. After centrifugation at 700 × g for 1 min, 1 ml of supernatant was added to 3 ml of 4% paraformaldehyde in PBS and stored overnight at 4°C. After the fixed cells were washed twice with PBS, they were stored in 50% ethanol-PBS at −20°C until they were used (for at least 1 h). Weighed portions of the remains of the fecal samples were lyophilized to determine the dry weights.
FISH analysis of fecal samples by microscopy.
For microscopic analysis, fixed cells were spotted on gelatin-coated glass slides and dried for 20 min at 45°C. Dilution series of fecal samples were prepared in order to determine the optimal cell concentration for counting with the different probes. After the slides were dried, the cells were dehydrated for 2 to 3 min in a graded ethanol series with the ethanol concentration increasing from 50 to 75% and finally in 96% ethanol in H2O. Ten microliters of hybridization buffer (0.9 M NaCl, 20 mM Tris-HCl [pH 7.5], 0.1% [wt/vol] sodium dodecyl sulfate [SDS]) containing 3 ng of Cy3-labeled Urobe63 probe per μl or 5 ng of fluorescein isothiocyanate (FITC)-labeled Erec482 probe per μl was added to each well, and this was followed by incubation at 50°C for 3 h. After hybridization the slides were washed in 50 ml of hybridization buffer without SDS for 10 to 20 min. For the Urobe63 probe 20% (vol/vol) formamide-containing hybridization buffer and a low-salt washing buffer (0.225 M NaCl, 20 mM Tris-HCl [pH 7.5], 10 mM EDTA) were used. For total counts 4′,6-diamidino-2-phenylindole (DAPI) was added to the washing buffer at a final concentration of 100 ng/ml. After the slides were rinsed in water, they were immediately air dried and mounted in Vectashield (Vector Labs, Burlingame, Calif.). Digital images of the slides, viewed with a Leica (Wetzlar, Germany) DMRXA epifluorescence microscope, were taken with a Kodak Megaplus 1.4 charge-coupled device camera. The images were analyzed and fluorescent cells were counted by using Quantimet HR550 image analysis software (Leica). For each analysis 25 microscopic fields were counted.
FISH analysis of fecal samples by flow cytometry.
For each hybridization 50 μl of fixed cells was centrifuged for 3 min at 9,000 × g and resuspended in 20 μl of hybridization buffer (0.9 M NaCl, 20 mM Tris-HCl [pH 8.0], 0.1% [wt/vol] SDS). After addition of 2 μl of the Cy5-labeled probes and/or the probes double labeled (at the 5′ and 3′ ends) with FITC (30 and 50 ng/μl, respectively), the samples were incubated for 16 h at 50°C in the dark. The Urobe63 and Erec482 probes were both labeled with Cy5 for counting. An FITC-labeled Erec482 probe was used to validate the specificity of the Urobe63 probe. After hybridization, 980 μl of prewarmed washing buffer was added, and the samples were incubated at 50°C for 5 min. Hybridization buffer without SDS was used as a washing buffer after hybridization with the Erec482 probe, and low-salt washing buffer consisting of 0.225 M NaCl, 20 mM Tris-HCl (pH 8.0), and 10 mM EDTA was used after hybridization with the Urobe63 probe. The cells were centrifuged at 9,000 × g for 5 min and resuspended in 1 ml of ice-cold PBS (pH 8.4). To avoid losing the signal intensity, the hybridized cells were kept in the dark on ice until flow cytometric analysis was performed. Cells hybridized with the labeled probes were compared to cells which were incubated in hybridization buffer alone in order to optimize the settings for flow cytometric analysis. Afterwards, the unlabeled cells were incubated with 10 μl of a propidium iodide (PI) solution (1 mg/ml) per ml at 37°C for 20 min to determine the total number of bacteria. Different cell concentrations of each sample were analyzed to check for potential clumps. The bacterial concentration was adjusted to keep the count lower than 1,000 events/s in order to avoid coincidence. During analysis of fecal samples, 0.7-μm yellow-green fluorescent beads (for counting PI-labeled cells) or PC fluorescent beads (for counting Cy5-labeled cells) (Polysciences, Inc.) at known concentrations were added according to the manufacturer's instructions in order to determine cell numbers. Samples were analyzed with a Becton Dickinson FACScalibur flow cytometer. An air-cooled argon ion laser (488 nm) and a red diode laser (635 nm) were used for excitation, and the green, red, and far red signals of the bacteria and the beads were collected in the FL1 (515- to 545-nm), FL3 (>600-nm long-pass filter), and FL4 (653- to 669-nm) detectors, respectively. The system threshold was set on forward scatter signals, and all bacterial analyses were performed at the low-flow-rate settings (12 μl/min). Data were collected in list mode as pulse height signals (four decades in logarithmic scale), and 10,000 cells were acquired for further analysis, which was performed by using CellQuest software (Beckton Dickinson) and/or WinMDI software (version 2.8; http//:facs.Scripps.edu/software.html). The whole hybridization and counting analysis was performed three times for each probe and fecal sample.
Statistical analysis.
The analyses performed for the various samples included the following variables: methodology (microscopy or flow cytometry), probe type (DAPI, Erec482 probe, or Urobe63 probe), individual, and time. Therefore, the coefficients of variation (CV) (i.e., standard variation divided by the mean) for time, for the reproducibility of the counts, and for the different individuals were compared. In addition, regression analysis of the mean counts for fecal samples taken at different times was performed for each individual. Since it appeared that time had no significant effect on the counts, these counts were used as replicates in a three-factor analysis of variance test to statistically analyze the effects of the other variables.
RESULTS
Probe design and validation.
Probes targeting the uncultured R. obeum-like bacteria were designed based on comparative analysis of 16S rRNA sequences from the ARB and RDP databases and newly deposited sequences from the GenBank database. Despite the small sequence variation in the hypervariable V1 region for the different sequences of this uncultured group, a combination of two probes was sufficient to discriminate between this uncultured group and the rest of the sequences in all databases (Table 1). These two probes hybridized at the same target site and were used in a 1:1 mixture during all Urobe63 probe hybridizations. Analysis of the 16S rRNA sequence databases showed that the Urobe63 probe has only two mismatches with the targets of the R. obeum sequence and a sequence of a 16S rDNA clone from a rumen sample (Table 1). All other sequences have three or more mismatches with the probe target site. No match was found with the adhufec171 sequence since the first nondiscriminative bases of the Urobe63 probe target were not present in this sequence (16). Furthermore, no match was found with any of the 23S rRNA sequences. Thirty-two reference strains (Table 2), including R. obeum, did not hybridize with the Urobe63 probe, while all of them showed strong hybridization with the Eub338 probe, as determined by microscopic and flow cytometric analyses. For the microscopic analysis, hybridization buffer containing 20% formamide was chosen since a few cells (∼1% of Eub338-positive cells) of the Peptostreptococcus micros culture and about 10 to 20% of Eub338-positive R. obeum cells hybridized with the Urobe63 probe when no formamide was added. For flow cytometric analysis Cy5-labeled probes, which give emission in the far red, were used since no adverse autofluorescence was observed in this detector channel (FL4) when fecal samples or reference strains were analyzed. However, autofluorescence of unlabeled fecal samples was observed in all other detectors (FL1, FL2, and FL3, which detect green, yellow, and red fluorescent signals, respectively) (data not shown). It appeared that formamide could not be added to the hybridization buffer during flow cytometric analysis when Cy5-labeled probes were used. No hybridization signals were detected when formamide was added even when the reference strains were hybridized with the Eub338 probe. However, washing of Urobe63 probe-hybridized cells with the low-salt washing buffer was sufficient to render all negative reference strains unlabeled while specific hybridization in fecal samples was maintained. Only the cells of R. obeum showed some weak positive hybridization with the Urobe63 probe under these conditions, although the signal was more than 10-fold lower than the signal for positively hybridized cells in a fecal sample.
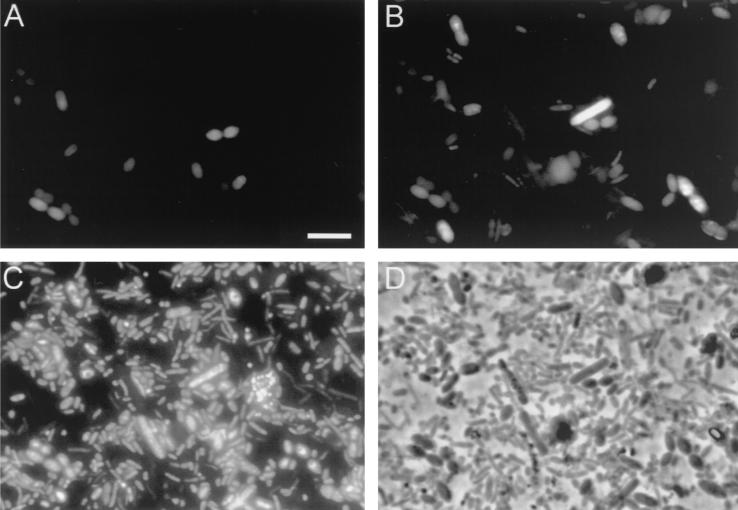
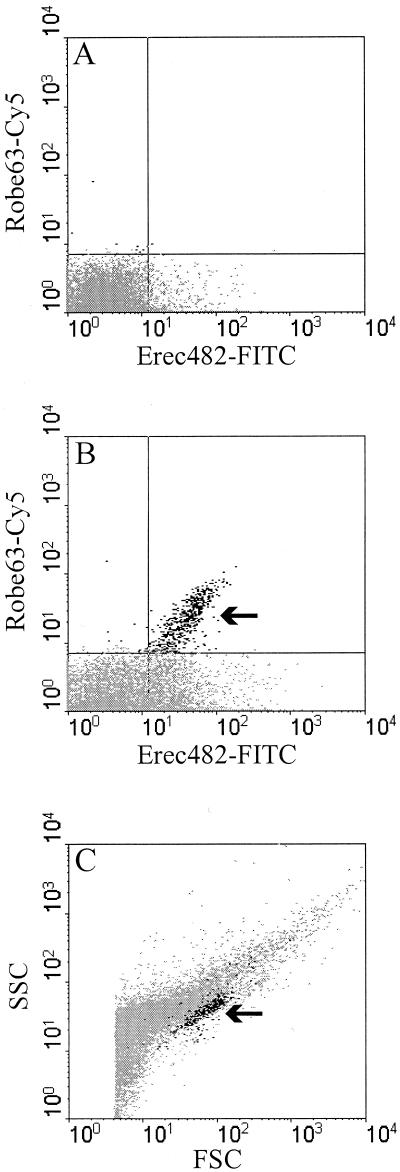
Since there is no isolate that can be used as a positive control strain for the Urobe63 probe, double hybridization of fecal samples was performed with the Urobe63 and Erec482 probes. The latter probe targets the C. coccoides-E. rectale group, to which the uncultured group of R. obeum-like bacteria belongs. Microscopic analysis revealed that all cells that hybridized with the Urobe63 probe also hybridized with the Erec482 probe with similar signal intensities (Fig. 2), indicating that the Urobe63 probe hybridized to the right target group. All Urobe63-positive cells were morphologically similar in all fecal samples examined and were relatively large single or duplococci (Fig. 2A), while the Erec482-positive cells exhibited many different morphotypes (Fig. 2B). The intensities of the Urobe63 probe and Erec482 probe signals varied for different cells, which could have been due to the difference in ribosome content between the cells. As expected, a variety of morphotypes were detected by using DAPI staining and phase-contrast microscopic analysis (Fig. 2C and D). Flow cytometric analysis revealed results similar to those obtained with microscopic examination (Fig. 3). Although the Erec482 probe was double labeled with FITC, it could be used in combination with the Cy5-labeled Urobe63 probe, since the Urobe63-positive cells did not show autofluorescence in the FL1 detector, which detected the FITC-labeled cells. The Cy5-labeled Urobe63-positive cells were also detected as Erec482-positive cells and could clearly be discriminated from unlabeled cells (Fig. 3A and B). When these double-labeled cells were gated, they appeared as very small clusters in the flow cytometric dot blots of the forward-angle light scatter versus side-angle light scatter, indicating that the cells exhibited limited morphological variability (Fig. 3C).
FIG. 2.
Photographs of fecal cells in one microscopic field hybridized with the Cy3-labeled Urobe63 probe (A), the FITC-labeled Erec482 probe (B), and DAPI (C). (D) Phase-contrast photograph. Bar = 5 μm.
FIG. 3.
Flow cytometric analysis of fecal samples, showing the dot plots representing the Urobe63-Cy5 fluorescence intensity and Erec482-FITC fluorescence intensity of unhybridized bacteria (A) and bacteria hybridized with Urobe63 plus Erec482 in feces (B). Cells labeled with both probes (indicated by the arrow in panel B) were marked with a dark color and appeared as small clusters in dot plots of the forward-angle light scatter (FSC) versus side-angle light scatter (SSC) of the total community (C), indicating limited morphological variability among these bacteria.
To ensure that the right target bacteria were detected, cross-hybridization experiments were performed with the Urobe63 probe and the probe targeting R. obeum which showed the fewest mismatches with the Urobe63 probe. No cross-hybridization was found when we used a pure culture of R. obeum, fecal samples, or mixtures of R. obeum and fecal samples. These results and the double-labeling results indicate that the Urobe63 probe specifically detects the uncultured R. obeum-like bacteria.
Comparison between microscopic and flow cytometric analyses.
Microscopic and flow cytometric counts were determined and compared. About 60% of the DAPI-stained cells hybridized with the Eub338 probe, as observed by microscopy, while the percentage was about 80% when Eub338- and PI-positive cells were compared by using flow cytometry. The difference was probably due to dividing cells which contained two or more chromosomes. Such cells are counted by microscopy as two cells when they are DAPI stained, while they are counted as a single cell when they are hybridized with the Eub338 probe. When flow cytometry is used, a cell containing two or more chromosomes is counted as a single cell.
To visualize the difference between the two approaches, the ratio of the mean counts for the probes in each sample was determined in order to quantify the total number of bacteria, the number of uncultured R. obeum bacteria, and the number of members of the C. coccoides-E. rectale group in feces (Table 3). Most ratios varied between 0.64 and 1.67, and only one relatively high ratio (1.93) was found for an Urobe63 count. The mean percentages for the C. coccoides-E. rectale group were 16.9 and 13.5% as determined by microscopy and flow cytometry, respectively. Similarly, the percentages of the uncultured R. obeum group were found to be 2.6 and 2.2% of the total community and 15.6 and 16.7% of the C. coccoides-E. rectale group. This indicated that both groups were numerically important in fecal samples and that the uncultured R. obeum-like bacteria comprised a significant fraction of the C. coccoides-E. rectale group.
TABLE 3.
DAPI and PI total cell counts, Erec482 probe hybridization counts, and Urobe63 probe hybridization counts for fecal samples from three individuals taken over a 4-week period, as determined by microscopy and flow cytometry
| Individual | Time (weeks) | DAPI or PI counts
|
Erec482 counts
|
Urobe63 counts
|
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Microscopy (1011)a | Flow cytometry (1011)a | Ratiob | Microscopy (1010)a | Flow cytometry (1010)a | Ratiob | Microscopy (1010)a | Flow cytometry (1010)a | Ratiob | ||
| A | 0 | 1.87 ± 0.17 | 2.40 ± 0.54 | 0.78 | 3.95 ± 0.63 | 2.80 ± 1.16 | 1.41 | 1.06 ± 0.17 | 0.55 ± 0.36 | 1.93 |
| 2 | 3.27 ± 0.40 | 4.98 ± 0.52 | 0.66 | 4.32 ± 0.77 | 5.69 ± 0.73 | 0.76 | 1.06 ± 0.30 | 1.33 ± 0.31 | 0.79 | |
| 4 | 4.06 ± 0.47 | 5.20 ± 2.60 | 0.78 | 4.90 ± 1.22 | 5.16 ± 0.75 | 0.95 | 1.06 ± 0.25 | 0.72 ± 0.12 | 1.47 | |
| B | 0 | 3.55 ± 0.41 | 4.72 ± 0.71 | 0.75 | 8.35 ± 1.00 | 7.16 ± 1.76 | 1.17 | 0.56 ± 0.07 | 0.88 ± 0.30 | 0.64 |
| 2 | 3.42 ± 0.37 | 3.30 ± 0.59 | 1.04 | 5.93 ± 0.69 | 5.15 ± 1.45 | 1.15 | 0.88 ± 0.20 | 1.31 ± 0.37 | 0.67 | |
| 4 | 3.24 ± 0.33 | 3.15 ± 0.95 | 1.03 | 8.30 ± 1.90 | 4.98 ± 1.51 | 1.67 | 0.71 ± 0.17 | 0.75 ± 0.12 | 0.94 | |
| C | 0 | 2.27 ± 0.41 | 2.06 ± 0.64 | 1.10 | 3.55 ± 0.58 | 4.45 ± 1.91 | 0.80 | 0.71 ± 0.25 | 0.57 ± 0.19 | 1.25 |
| 2 | 2.43 ± 0.25 | 2.41 ± 0.97 | 1.00 | 2.75 ± 0.46 | 2.94 ± 0.91 | 0.94 | 0.38 ± 0.06 | 0.31 ± 0.01 | 1.22 | |
| 4 | 2.23 ± 0.45 | 2.24 ± 1.50 | 1.00 | 2.49 ± 0.62 | 2.96 ± 1.00 | 0.93 | 0.54 ± 0.15 | 0.41 ± 0.16 | 1.30 | |
The values are means ± standard deviations and are expressed in cells per gram (dry weight) of feces.
The ratio was calculated by dividing the mean microscopic count by the mean flow cytometric count.
Regression analysis was performed on the means of the total and specific counts separately as determined by microscopy and flow cytometry for the three samples taken from each individual at different times. None of the 18 analyses resulted in a significant relationship between time and counts (P > 0.05). In addition, the CV for time was found to be comparable to the CV for reproducibility of the counts (data not shown). In only one-half of the counts was the CV for time higher than the CV for reproducibility. These results indicated that the bacterial composition was relatively stable over time. Since no significant differences among the samples taken at different times were found for each individual, the counts were used as replicates in a three-factor analysis of variance test in order to determine the effects of the other variables (Table 4). As expected, a significant difference in the counts was found when DAPI (or PI), the Erec482 probe, or the Urobe63 probe was used (Table 4). In addition, a significant effect of the individual on the different counts was observed. Furthermore, a significant effect on the interaction between the different probes and individuals was observed, which indicated that the different probe counts varied significantly for each individual. The individual differences were most clearly observed when the CV among individuals was calculated for each probe for each sampling time. In 17 of the 18 cases, the CV among individuals was higher than the CV for the assay (data not shown). Furthermore, it was observed that the different counting approaches did not have a significant influence on the outcome of the counts.
TABLE 4.
Three-factor analysis of variance test of the counts obtained by microscopy and flow cytometrya
| Source | df | Mean square | F ratio | P value |
|---|---|---|---|---|
| M | 1 | 2.61 × 1021 | 1.03 | 0.316 |
| P | 2 | 5.05 × 1023 | 199.82 | <0.001b |
| I | 2 | 1.73 × 1022 | 6.85 | 0.003b |
| M × P | 2 | 3.44 × 1021 | 1.36 | 0.269 |
| M × I | 2 | 1.93 × 1021 | 0.76 | 0.474 |
| P × I | 4 | 9.85 × 1021 | 3.90 | 0.010b |
| M × P × I | 4 | 1.96 × 1021 | 0.77 | 0.549 |
| Error | 36 | 2.53 × 1021 |
The degrees of freedom, the mean square, the F ratio, and the P value of the variables methodology (M) (microscopy and flow cytometry), probe (P) (DAPI or PI, Erec482 probe, and Urobe63 probe), and individual (I) are shown.
The variable had a significant effect (P < 0.05).
To determine how common the uncultured R. obeum-like bacteria were in different individuals, the numbers of these bacteria in feces from six additional individuals (adults of both sexes) were determined by microscopic analysis. The percentages of the uncultured R. obeum-like bacteria varied between 1 and 5% of the total bacteria, which is similar to the percentages in feces from the individuals examined previously.
DISCUSSION
In t his paper we describe the development, validation, and application of the Urobe63 probe to detect, examine, and quantify uncultured R. obeum-like bacteria in human fecal samples. We used a microscopic and flow cytometric approach to characterize the specificity of the probe, the morphology of the cells, and the numbers of cells in different fecal samples. None of the reference strains used as negative controls in this study hybridized with the Urobe63 probe, and all Urobe63 probe-labeled fecal cells also hybridized with the Erec482 probe, which is specific for the C. coccoides-E. rectale group (5). These results indicate that the right target group was detected. While many different morphotypes of bacteria hybridized with the Erec482 probe, only one morphotype hybridized with the new Urobe63 probe in all fecal samples. The Urobe63 probe did not hybridize with the type strain of R. obeum, which in all known sequences has the fewest (only two) mismatches at the target site. In addition, no cross-hybridization was observed when the Urobe63 probe was combined with a probe specific for R. obeum. The Urobe63 probe was designed and validated by using currently available 16S rRNA sequences, including several hundred 16S rRNA sequences derived from cultured and uncultured GI tract bacteria. Since various R. obeum-like sequences have been found frequently in fecal clone libraries obtained from different human individuals (16, 24), we concluded from the present data that the new probe can specifically detect a new and numerically important group of intestinal bacteria.
For microscopic analysis and flow cytometric analysis of FISH samples different protocols were required due to the difference in handling procedures. Despite the different approaches our data showed that the counts obtained with the Urobe63 and Erec482 probes by the two methods were similar (Table 3). This indicates that the two approaches should give similar results when the numbers of specific bacteria in environmental samples are quantified. In addition, the 16S rRNA of 60 to 80% of the bacterial cells in feces was accessible, as determined by comparing Eub338 probe and DAPI or PI counts, which is in line with previous observations (5, 19). In general, about 16% of the total bacterial community in the fecal samples belonged the C. coccoides-E. rectale group, which is slightly lower than the percentage reported by Franks and colleagues (5) but similar to values found by Tannock and colleagues (19). The differences in numbers of cells found in a variety of fecal samples may be due to host-specific factors, as observed previously (24, 25). The percentage of the uncultured R. obeum-like bacteria in the samples investigated was on average about 2.5% and varied between 1 and 6%. This indicates that these organisms comprise a significant fraction of the predominant bacterial community in feces, as determined previously by using PCR-based approaches (16, 24). By using statistical analysis, we observed significant differences between the individuals, which are in line with previous observations based on PCR and temperature gradient gel electrophoresis analysis of fecal samples (24). However, the number of individuals used in the statistical test is too small for detailed comparisons.
The results described in this paper reveal that a group of uncultured R. obeum-like bacteria, whose presence was suggested repeatedly by 16S rDNA cloning studies of fecal samples, comprises a significant fraction of the fecal community, as determined by FISH analysis performed with a newly developed specific 16S rRNA-targeted probe. Although the power of 16S rRNA probe hybridization has been demonstrated in many independent studies, its value in studying the ecology of uncultured bacteria requires additional systematic research. The usefulness of FISH combined with flow cytometry was demonstrated, and this technique provides a possible way to sort the R. obeum-like bacteria and other uncultured populations in order to study them in detail. Such studies may ultimately be helpful in refining our knowledge about the ecology of the microorganisms in the human GI tract.
Acknowledgments
We thank the volunteers that provided fecal samples for this study. We thank Chantal Doeswijk for lyophilizing the fecal samples and Patrick Verbaarschot for technical assistance during the flow cytometric analysis. Arjan de Visser was very helpful in the statistical analysis of the data. In addition, we thank Michael Wagner and his coworkers at the Technical University of Munich for their initial help and their advice concerning FISH techniques.
REFERENCES
- 1.Alm, E. W., D. B. Oerther, N. Larsen, D. A. Stahl, and L. Raskin. 1996. The Oligonucleotide Probe Database. Appl. Environ. Microbiol. 62:3557-3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amann, R. I., B. J. Binder, R. J. Olsen, S. W. Chrisholm, R. Devereux, and D. A. Stahl. 1990. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed populations. Appl. Environ. Microbiol. 56:1919-1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amann, R. I., W. Ludwig, and K.-H. Schleifer. 1995. Phylogenetic identification and in situ detection of individual cells without cultivation. Microbiol. Rev. 59:143-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bry, L., P. G. Falk, T. Midtvedt, and J. I. Gordon. 1996. A model of host-microbial interactions in an open mammalian ecosystem. Science 273:1381-1383. [DOI] [PubMed] [Google Scholar]
- 5.Franks, A. H., H. J. M. Harmsen, G. C. Raangs, G. J. Jansen, F. Schut, and G. W. Welling. 1998. Variations of bacterial populations in human feces measured by fluorescent in situ hybridization with group-specific 16S rRNA-targeted oligonucleotide probes. Appl. Environ. Microbiol. 64:3336-3345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Harmsen, H. J. M., P. Elfferich, F. Schut, and G. W. Welling. 1999. A 16S rRNA-targeted probe for the detection of lactobacilli and enterococci in faecal samples by fluorescent in situ hybridization. Microb. Ecol. Health Dis. 11:3-12. [Google Scholar]
- 7.Harmsen, H. J. M., A. C. M. Wildeboer-Veloo, J. Grijpstra, J. Knol, J. E. Degener, and G. W. Welling. 2000. Development of 16S rRNA-based probes for the Coriobacterium group and the Atopobium cluster and their application for enumeration of Coriobacteriaceae in human feces from volunteers of different age groups. Appl. Environ. Microbiol. 66:4523-4527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Holdeman, L. V., E. P. Cato, and W. E. C. Moore. 1977. Anaerobe laboratory manual, 4th ed. Virginia Polytechnic Institute and State University, Blacksburg.
- 9.Hooper, L. V., J. Xu, P. G. Falk, T. Midtvedt, and J. I. Gordon. 1999. A molecular sensor that allows a gut commensal to control its nutrient foundation in a competitive ecosystem. Proc. Natl. Acad. Sci. USA 96:9833-9838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hooper, L. V., M. H. Wong, A. Thelin, L. Hansson. P. G. Falk, and J. I. Gordon. 2000. Molecular analysis of host-microbial relationships in the intestine. Science 291:881-884. [DOI] [PubMed] [Google Scholar]
- 11.Maidak, B. L., J. R. Cole, T. G. Lilburn, C. T. Parker, Jr., P. R. Saxman, R. J. Farris, G. M. Garrity, G. J. Olsen, T. M. Schmidt, and J. M. Tiedje. 2001. The RDP-II (Ribosomal Database Project). Nucleic Acids Res. 29:173-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Manz, W., R. Amann, W. Ludwig, M. Wagner, and K.-H. Schleifer. 1992. Phylogenetic oligodesoxynucleotide probes for the major subclasses of proteobacteria: problems and solutions. Syst. Appl. Microbiol. 15:593-600. [Google Scholar]
- 13.Schwiertz, A., G. Le Blay, and M. Blaut. 2000. Quantification of different Eubacterium spp. in human fecal samples with species-specific 16S rRNA-targeted oligonucleotide probes. Appl. Environ. Microbiol. 66:375-382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Simon, N., N. LeBot, D. Marie, F. Parentsky, and D. Vaulot. 1995. Fluorescent in situ hybridization with rRNA-targeted oligonucleotide probes to identify small phytoplankton by flow cytometry. Appl. Environ. Microbiol. 61:2506-2513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Strunk, O., and W. Ludwig. 1995. ARB—a software environment for sequence data. [Online.] Department of Microbiology, Technical University of Munich, Munich, Germany. http://www.mikro.biologie.tu-Muenchen.de/pub/ARB/documentation/arb.ps.
- 16.Suau, A., R. Bonnet, M. Sutren, J.-J. Godon, G. R. Gibson, M. D. Collins, and J. Doré. 1999. Direct analysis of genes encoding 16S rRNA from complex communities reveals many novel molecular species within the human gut. Appl. Environ. Microbiol. 65:4799-4807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Suau, A., V. Rochet, A. Sghir, G. Gramet, S. Brewaeys, M. Sutren, L. Rigottier-Gois, and J. Doré. 2001. Fusobacterium prausnitzii and related species represent a dominant group within the human fecal flora. Syst. Appl. Microbiol. 24:139-145. [DOI] [PubMed] [Google Scholar]
- 18.Tannock, G. W. 1995. Normal microflora. An introduction to microbes inhabiting the human body. Chapman and Hall, London, United Kingdom.
- 19.Tannock, G. W., K. Munro, H. J. M. Harmsen, G. W. Welling, J. Smart, and P. K. Gopal. 2000. Analysis of the fecal microflora of human subjects consuming a probiotic product containing Lactobacillus rhamnosus DR20. Appl. Environ. Microbiol. 66:2578-2588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vaughan, E. E., G. H. J. Heilig, E. G. Zoetendal, R. Satokari, J. K. Collins, A. D. L. Akkermans, and W. M. de Vos. 1999. Molecular approaches to study probiotic bacteria. Trends Food Sci. Technol. 10(12):400-404. [Google Scholar]
- 21.Wallner, G., R. Amann, and W. Beisker. 1993. Optimizing fluorescent in situ hybridization with rRNA-targeted oligonucleotide probes for flow cytometric identification of microorganisms. Cytometry 14:136-143. [DOI] [PubMed] [Google Scholar]
- 22.Wallner, G., B. Fuchs, S. Spring, W. Beisker, and R. Amann. 1997. Flow sorting of microorganisms for molecular analysis. Appl. Environ. Microbiol. 63:4223-4231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wilson, K. H., and R. H. Blitchington. 1996. Human colonic biota studied by ribosomal DNA sequence analysis. Appl. Environ. Microbiol. 62:2273-2278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zoetendal, E. G., A. D. L. Akkermans, and W. M. de Vos. 1998. Temperature gradient gel electrophoresis analysis of 16S rRNA from human fecal samples reveals stable and host-specific communities of active bacteria. Appl. Environ. Microbiol. 64:3854-3859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zoetendal, E. G., A. D. L. Akkermans, W. M. Akkermans van-Vliet, J. A. G. M. de Visser, and W. M. de Vos. 2001. The host genotype affects the bacterial community in the human gastrointestinal tract. Microb. Ecol. Health Dis. 13:129-134. [Google Scholar]