Abstract
We investigated the diversity, distribution, and phenotypes of uncultivated Chloroflexaceae-related bacteria in photosynthetic microbial mats of an alkaline hot spring (Mushroom Spring, Yellowstone National Park). By applying a directed PCR approach, molecular cloning, and sequence analysis of 16S rRNA genes, an unexpectedly large phylogenetic diversity among these bacteria was detected. Oligonucleotide probes were designed to target 16S rRNAs from organisms affiliated with the genus Chloroflexus or with the type C cluster, a group of previously discovered Chloroflexaceae relatives of this mat community. The application of peroxidase-labeled probes in conjunction with tyramide signal amplification enabled the identification of these organisms within the microbial mats by fluorescence in situ hybridization (FISH) and the investigation of their morphology, abundance, and small-scale distribution. FISH was combined with oxygen microelectrode measurements, microscope spectrometry, and microautoradiography to examine their microenvironment, pigmentation, and carbon source usage. Abundant type C-related, filamentous bacteria were found to flourish within the cyanobacterium-dominated, highly oxygenated top layers and to predominate numerically in deeper orange-colored zones of the investigated microbial mats, correlating with the distribution of bacteriochlorophyll a. Chloroflexus sp. filaments were rare at 60°C but were more abundant at 70°C, where they were confined to the upper millimeter of the mat. Both type C organisms and Chloroflexus spp. were observed to assimilate radiolabeled acetate under in situ conditions.
Bacteriochlorophyll (Bchl)-containing, filamentous bacteria are conspicuous inhabitants of hot springs, in which they may form macroscopically visible, benthic microbial mats (reviewed in references 9 and 11). Commonly they are found in association with cyanobacteria, but they may be the principal mat component where elevated concentrations of sulfide inhibit cyanobacterial growth (10, 20, 52). The first cultivated and most extensively studied representative of these thermophilic filaments is Chloroflexus aurantiacus, a bacterium capable of anoxygenic photoautotrophy, photoheterotrophy, and aerobic chemoorganotrophy (37). For several years following the discovery of C. aurantiacus, this organism was considered the predominant filamentous component of hot spring microbial mats in North America and elsewhere (6, 14, 38). Greater diversity among the phototrophic filamentous bacteria concealed by a common morphology was suggested, however, by the observation of deep orange mat layers containing abundant filaments and high concentrations of Bchl a but lacking Bchl c, the principal photosynthetic pigment of C. aurantiacus (8, 9). Using immune serum specific to cultivated C. aurantiacus, Tayne et al. (50) demonstrated that the mat community in Octopus Spring contained nonreacting and, hence, presumably unrelated filamentous microorganisms.
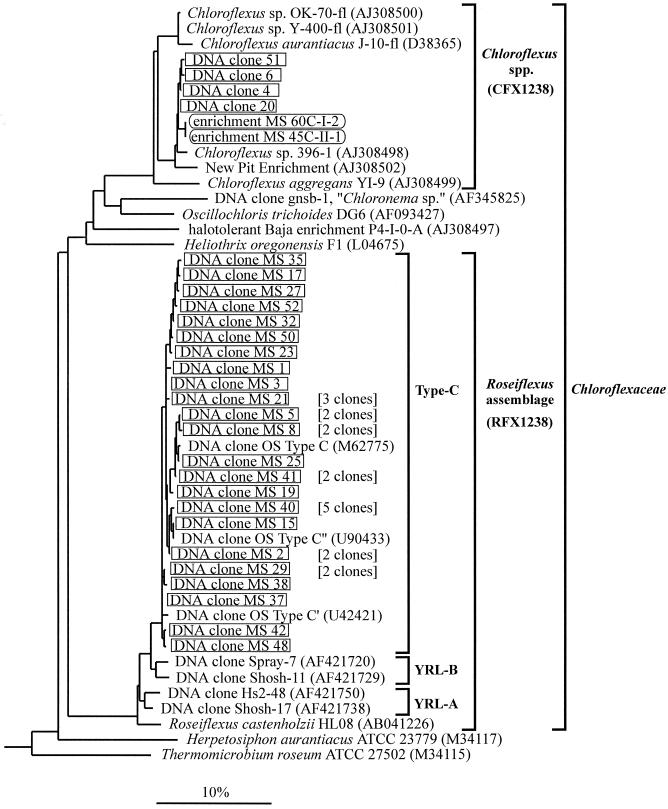
To date, several additional species of filamentous phototrophic bacteria have been cultivated from hot springs, including Chloroflexus aggregans (21), Heliothrix oregonensis (39), and Roseiflexus castenholzii (22). Together with their morphological counterparts in nonthermal waters (Oscillochloris spp. [24] and Chloronema spp. [19]) and hypersaline environments (33), they constitute a phylogenetic lineage within the green nonsulfur bacterial kingdom with the proposed family name Chloroflexaceae (Fig. 1) (37).
FIG. 1.
Phylogenetic affiliations of Chloroflexaceae relatives detected in Mushroom Spring. Sequences from DNA clones and enrichments are boxed. For sequences detected several times in the same mat sample, only single representatives are shown and the respective numbers of identical clones are in parentheses. Accession numbers are indicated for sequences retrieved from public databases. Thirty-five sequences from organisms with various phylogenetic affiliations (not shown) were used to root the tree. The scale bar indicates 10% estimated sequence divergence. The FISH probes CFX1238 and RFX1238 target 16S rRNAs from organisms in the Chloroflexus sp. and Roseiflexus assemblage clusters, as indicated by brackets. The PCR primers used target 16S rRNA genes from organisms in the present family Chloroflexaceae (33). Clusters YRL-A and YRL-B were described previously (7).
Through our previous molecular studies of microbial mats in alkaline hot springs in Yellowstone National Park, we have discovered 16S rRNA genes from organisms that are distantly related phylogenetically to R. castenholzii and that we have termed type C (Fig. 1) (56). The detection of highly similar, yet different, type C-like gene sequences with differential temperature distributions suggested the existence of several temperature-adapted, genetically unique populations (17, 54). These bacteria may cooccur with Chloroflexus spp., as indicated by rRNA distribution patterns revealed through probe hybridization studies (dot blots) (44) and by the fact that bacteria closely related to C. aurantiacus could be cultivated from the same habitats (46). Recently, related 16S rRNA gene sequences from low-temperature Yellowstone hot springs and small geyser splash zones containing thick mats exhibiting distinct red layers (clusters YRL-A and YRL-B in Fig. 1) have been reported (7).
In the present study, we have investigated the phylogenetic and phenotypic diversity of filamentous phototrophs in 60 to 70°C regions of Mushroom Spring in Yellowstone National Park. First, we reexamined the phylogenetic diversity of Chloroflexaceae relatives, targeting their 16S rRNA genes specifically by applying a recently developed PCR protocol (33). Based on the new nucleotide sequence information, we developed and applied rRNA-directed fluorescent in situ hybridization (FISH) probes for the identification of type C organisms and the genus Chloroflexus to analyze their morphology and small-scale distribution in the microbial mat. By combining FISH and microautoradiography, we monitored the uptake of potential carbon sources by type C organisms and Chloroflexus spp. in situ. Corresponding microscope spectrophotometry permitted inferences about pigmentation.
MATERIALS AND METHODS
Bacterial cultures.
C. aurantiacus Y-400-fl (DSM 637) and C. aggregans YI-9 (DSM 9486) were obtained from the Deutsche Sammlung von Mikroorganismen und Zellkulturen (Braunschweig, Germany) and cultivated as described in the Deutsche Sammlung von Mikroorganismen und Zellkulturen catalogue. Frozen cells of R. castenholzii HL08 (DSM 13941) were obtained from S. Hanada, National Institute of Bioscience and Human Technology, Tsukuba, Japan (22). Enrichments from microbial mat material were obtained in water collected from the source pool of Mushroom Spring modified by the addition of nutrients as in medium D (12) to 0.1× strength, yeast extract (0.1 g/liter), NH4Cl (0.2 g/liter), glycylglycine (1.0 g/liter), Na2S · 9H2O (0.2 g/liter), 3-(3,4-dichlorophenyl)-1,1-dimethylurea (1.2 mg/liter), and resazurin (0.0001%, wt/vol). The medium was boiled for 2 min to drive out dissolved oxygen, and 10-ml aliquots were transferred into 20-ml screw cap tubes with rubber septae under a stream of nitrogen. The tubes were immediately closed and autoclaved. Aliquots from a series of mat samples that had been homogenized and diluted to extinction in growth medium were injected. Tubes were incubated at 45 and 55°C and received light from a fluorescent lamp and a tungsten lamp (approximately 25 μE m−2 s−1). Upon visible growth, aliquots of enrichment cultures were transferred to fresh medium anaerobically by using syringes.
Microbial mat sample collection and preparation.
Microbial mat samples were collected from 60 and 70°C sites in the effluent channel of Mushroom Spring, an alkaline (pH 8.3) hot spring about 0.2 km northeast of Great Fountain Geyser in the Lower Geyser Basin in Yellowstone National Park, Wyoming (5, 41). Mushroom Spring is located 0.5 km from the more extensively studied Octopus Spring. The effluent waters of these two springs have very similar compositions, and the microbial mats have similar appearances (41). Microbial mat cores were removed from the mat with a cork borer (core diameter, 8 mm; thickness, 3 mm). Samples destined for DNA analyses or FISH were collected on 24 August 1999, frozen on dry ice, and stored at −80°C.
Radiolabeling experiments were performed on 7 October 2000. Mat cores sampled from a 60°C site were transferred to 7-ml screw cap glass vials containing 4 ml of water from the collection site. Each mat core received 2 μCi of sodium [1-14C]acetate (54 mCi/mmol) or 4 μCi of sodium [14C]bicarbonate (50 mCi/mmol) (NEN Life Science Products). Control samples were killed by adding formaldehyde (4%). Initial experiments had demonstrated diffusion of radiolabeled compounds to greater depths within the mat cores under dark conditions (our unpublished results). Therefore, to enhance diffusion into the mat cores, vials were wrapped with aluminum foil before addition of radiolabel and then placed in the effluent channel at the site of sample collection for 1 h. Subsequently, the aluminum foil was removed and vials were incubated at ambient light intensity and hot spring temperature by placing them in the effluent channel for 3.5 h (8:00 to 11:30 a.m.). To stop biological activity, samples were frozen on dry ice and stored at −80°C.
DNA extraction.
Mat cores were homogenized by gentle grinding with pipette tips. Community DNAs were extracted from microbial mats by use of a previously published protocol (30) modified by omission of the DNA purification step. Briefly, Na-phosphate buffer, sodium dodecyl sulfate, and zirconium beads (diameter, 0.1 mm) were added to the homogenized mat material, and cells were lysed by bead beating. Microscopic observation indicated complete cell lysis. Cell debris was removed by centrifugation, and, after ammonium acetate precipitation, nucleic acids were precipitated from the supernatant by addition of isopropanol.
PCR, molecular cloning, and sequence analysis.
PCR primers specific for 16S rRNA genes from Chloroflexaceae (CCR-344-F, ACGGGAGGCAGCAGCAAG; CCR-1338-R, ACGCGGTTACTAGCAACT) and appropriate PCR conditions have been described previously (33). During the course of the present study, amplification products were obtained with this PCR from a culture of R. castenholzii HL08, which became available only recently (22) (data not shown). For molecular cloning of PCR products the TOPO TA cloning kit (Invitrogen) was used, and plasmid DNA was extracted by using the Turbo Miniprep kit (Qiagen). Sequence analyses were performed by using the ABI Big Dye Terminator Cycle Sequencing Ready Reaction kit (Applied Biosystems) and an ABI Prism 310 capillary sequencer (Applied Biosystems) (33).
Phylogeny reconstruction.
Phylogenetic trees were reconstructed by using the software package ARB (available at http://www.mikro.biologie.tu-muenchen.de) as described previously (33). To detect chimeric DNA molecules that may be generated by PCR, we additionally determined the phylogenetic affiliations of the 5′ ends (nucleotides 400 to 700 [Escherichia coli numbering]) and 3′ ends (nucleotides 1000 to 1300) of newly determined sequences in separate analyses (25).
Rarefaction analysis.
Rarefaction curves were computed based on the analytical approximation algorithm of Hurlbert (23) by applying the freeware program aRarefactWin (available at http://www.uga.edu/∼strata/AnRareReadme.html). Asymptotic maxima (Emax) approached by the rarefaction curves were estimated by fitting two-parameter hyperbolas as described previously (35). Conservative estimates of Emax were calculated, taking into account the possibility of PCR artifact generation at high frequency. By randomly dropping 33% of unique sequences from the data set, new frequency distributions were generated which were then used to compute the corresponding rarefaction curves and values for Emax. The procedure was replicated 10 times to assess stochastic effects.
FISH.
Oligonucleotide probes targeting 16S rRNAs from organisms in clusters Chloroflexus spp. and Roseiflexus assemblage (Fig. 1) were designed by using the PROBE DESIGN and PROBE MATCH options of the phylogeny software package ARB (available at http://www.mikro.biologie.tu-muenchen.de) and the BLAST program (1) at the National Center for Biotechnology Information (Washington, D.C.). Probes specific for Chloroflexus spp. or the Roseiflexus assemblage were CFX1238 (CGCATTGTCGTGGCCATT) and RFX1238 (CGCATTGTCGGCGCCATT), respectively. In addition, probe EUB338 (GCTGCCTCCCGTAGGAGT), matching 16S rRNAs from most bacteria (2), was used. Oligonucleotides labeled with the fluorescent dye cyanine Cy3 or fluorescein or with horseradish peroxidase (HRP) were synthesized commercially (Interactiva, Ulm, Germany).
Prior to FISH, bacterial cells from cultures were fixed in 4% paraformaldehyde in phosphate-buffered saline (PBS) (130 mM NaCl, 10 mM sodium phosphate [pH 7.2]) for 1 h at 4°C. Fixed cells and microbial mat cores were washed in PBS and stored in a 1:1 mixture of PBS and 96% ethanol at −20°C (2). Mat samples were homogenized by gentle grinding with pipette tips. Cell suspensions were pipetted onto gelatin-coated microscope slides, air dried at 35°C, and dehydrated in an ethanol series (50, 70, and 96%, 3 min each). In situ hybridizations with Cy3-, fluorescein-, and HRP-labeled probes were performed as described previously (3, 29), with the incubation temperature for both hybridization and washing steps lowered to 35°C (47). For fluorescent detection of hybridized HRP-labeled probes, the tyramide signal amplification kit (NEN Life Science Products) with fluorescein-tyramide was used as described by Schönhuber et al. (48). Subsequently, slides were mounted in Vectashield (Vector Laboratories). Conditions for hybridizations with HRP-labeled probes were optimized empirically by performing experiments under increasingly stringent hybridization and washing conditions. Fluorescence was detected with an epifluorescence microscope (Axioskop; Zeiss) equipped with Zeiss filter sets 10 (excitation filter BP 450-490, beamsplitter FT 510, and emission filter BP 515-565) and 15 (BP 546/12, FT 580, and LP 590). Microscopic images were recorded with a digital camera (Spot RT; Diagnostic Instruments), and fluorescence intensities of at least 50 filaments were measured by using the NIH-Image software package version 1.62 (National Institutes of Health, Bethesda, Md.). From the resulting probe dissociation curves (data not shown), optimum formamide concentrations for maximum hybridization specificity at high signal strength were deduced to be 50, 40, and 40% for probes EUB338, CFX1238, and RFX1238, respectively (32). For equivalent stringencies of corresponding washing steps, the NaCl concentrations in washing buffer were 0.019 and 0.037 M, respectively (48). These hybridization conditions were used in subsequent experiments. No fluorescence was detected in control experiments performed with no probe added, thereby excluding fluorescence generation by potentially present endogenous peroxidases.
Combination of FISH and microautoradiography.
Radiolabeled microbial mat material was fluorescently hybridized on microscope slides as described above. For control experiments, nonradioactive mat material was processed in parallel. In a darkroom equipped with a Safelight lamp (filter no. 2; Kodak), microscope slides were subsequently coated with a photographic emulsion solution (50% photographic emulsion NTB2 [Kodak], 0.2% gelatin, 0.02% CrKSO4) (36), air dried for 30 min, and incubated in the dark at 4°C for 1 to 30 days. Slides were developed using developer D-19 (Kodak) at a 50% concentration according to the manufacturer's recommendation and examined by dark-field and epifluorescence microscopy. Digital images were recorded with a Spot RT camera (Diagnostic Instruments) and overlaid by using the software package MetaVue (version 4.6; Universal Imaging).
Vertical distribution analysis.
Prior to cryosectioning, ethanol-fixed mat cores were embedded in OCT compound (Tissue Tek) at room temperature overnight. Cores were frozen and sectioned (50 μm) in a cryomicrotome (2800 Frigcut; Reichert Scientific Instruments) at −17°C. Combining adjacent slices, 100-μm-thick horizontal mat sections (i.e., cut parallel to the mat surface) were obtained and collected in 1.5-ml tubes. This mat material was homogenized, and aliquots were spread on microscope slides and subjected to FISH with HRP-labeled probe RFX1238 or CFX1238. For at least 25 randomly chosen microscopic fields per mat section and probe, lengths of filaments were measured by using an ocular micrometer grid (18). Scattered filaments were considered exclusively, while those enclosed in optically dense aggregates were omitted from these analyses. Proportional abundances were determined by comparing lengths of probe-stained filaments to lengths of filaments visible by phase-contrast microscopy.
Spectrometry.
For microscope spectrometry, vertical cryosections of OCT-embedded microbial mat cores (cut perpendicular to the mat surface; thickness, 50 μm) were air dried onto microscope slides. Spectral absorbance characteristics were measured by using a microscope (Olympus BX50 WI) connected to a Hamamatsu PMA-11 spectrometer (250 to 950 nm) (Hamamatsu Photonics, Hamamatsu City, Japan) via a fiber optic cable mounted on the video port of the microscope (26). The halogen lamp of the microscope was used with the heat filter removed from the light path. In vivo absorbance spectra of cell suspensions from cultures were measured with a Lambda-2 spectrometer (260 to 1,100 nm) (Perkin-Elmer).
Microsensor measurements.
Microprofiles of oxygen were measured in situ under ambient irradiance with Clark-type oxygen microsensors equipped with a guard cathode (42) and connected to a sensitive picoamperemeter (Unisense, Århus, Denmark). The microsensors had a low stirring sensitivity (< 2%) and a fast response time (t90 < 0.4 s). They were positioned with a manually operated micromanipulator (Märzhäuser, Wetzlar, Germany) mounted on a heavy stand, and sensor signals were recorded on a stripchart recorder (Servogor 110; Spectronic, Leeds, United Kingdom). All equipment was run via a 12-V car battery equipped with a 12-V/220-V DC/AC converter. Linear calibration was done from readings in the overlying spring water and from readings either in the anoxic parts of the mats or in spring water made anoxic by addition of sodium dithionite. The oxygen content of the spring water was determined by Winkler titration. The in situ measurements were made under an incident saturating irradiance of 1,800 to 2,500 μE m−2 s−1, as monitored with a quantum irradiance meter (QSL-101; Biospherical Instruments) during the measurements.
Nucleotide sequence accession numbers.
The EMBL accession numbers for the 16S rRNA gene sequences reported in this study are AJ421643 to AJ421669, AJ421694, and AJ421695.
RESULTS
Enrichments.
Enrichments were obtained in tubes that had received at least 0.1 mg of microbial mat sample. Two enrichments, obtained from 60 and 45°C sites, incubated at 55 and 45°C and designated 60C-I-2 and 45C-II-1, respectively, contained abundant green filamentous organisms of indeterminate length and 0.8 to 1.0 μm in diameter. Cells exhibited spectral absorbance maxima at 742, 795, and 870 nm, indicating the contents of Bchl c and Bchl a (data not shown). Segments of 16S rRNA genes were PCR amplified by applying primers specific for the Chloroflexaceae (Fig. 1) (33) and sequenced. The 16S rRNA gene sequences recovered from these enrichments were identical. Phylogenetic analysis indicated a close relationship to Chloroflexus sp. strain 396-1 (sequence difference, 1.0%), cultivated from a siliceous hot spring in Yellowstone National Park (28) (Fig. 1).
Diversity analysis.
To analyze the phylogenetic diversity of Chloroflexaceae relatives in the microbial mat at 60°C, segments of their 16S rRNA genes were amplified from community DNA by applying a directed PCR approach (33). The PCR product was cloned, and 42 plasmid inserts were sequenced (sequence lengths, 860 to 865 nucleotides). Separate phylogenetic analyses of the 5′ ends and 3′ ends of the sequences revealed three chimeric DNA molecules, composed of sequence stretches with different phylogenetic affiliations (fragment from type C cluster in combination with fragment from Chloroflexus cluster) (Fig. 1) and likely generated by PCR (25). Among the remaining 39 clones, 27 different sequences were found, indicating that sequence diversity was high (Fig. 1). Many of these sequences differed by only one or a few nucleotides. Four clone sequences were highly similar to 16S rRNA gene sequences from the above-described enrichments and Chloroflexus sp. strain 396-1 (maximum difference among these sequences, 1.6% [14 nucleotides]) (Fig. 1). Twenty-three clone sequences were highly similar to sequences from type C-like organisms that previously had been detected in Mushroom Spring (41) and in nearby Octopus Spring (17, 56) (maximum difference among these sequences, 2.5% [22 nucleotides]) (Fig. 1). One of the new sequences (clone MS 15) was identical in the overlapping 213-nucleotide stretch to sequence type C" previously reported by Ferris and Ward (17). Type C-like organisms constitute a monophyletic cluster, excluding related bacteria from other hot springs in Yellowstone National Park (clusters YRL-A and YRL-B in Fig. 1) (7) and their closest cultivated relative, R. castenholzii, a filamentous phototrophic bacterium recently isolated from a hot spring in Japan (22). This result is moderately supported by the respective internodal branch lengths in the phylogenetic tree (Fig. 1) and bootstrap analysis (7). Sequences of 16S rRNA genes of type C-like organisms and R. castenholzii differ by 5.9 to 8.0%.
We used rarefaction analysis to calculate the average number of different sequences to be expected in a series of subsamples with increasing numbers of clones investigated (33). This analysis indicated that the investigation of additional plasmid inserts very likely would have led to the discovery of a significant number of additional 16S rRNA gene sequences (data not shown). By extrapolating the rarefaction curve, the total number of 16S rRNA gene sequences from Chloroflexaceae relatives probably present in the microbial mat sample was estimated to be 82. To calculate a more conservative estimate of this number, we postulated a high PCR error rate and accordingly decreased the number of different sequences in our data set from 27 to 18 by randomly excluding 33% of unique sequences. In comparison, frequencies of PCR artifact generation described in the literature for 16S rRNA gene amplifications range from 7 to 32%, including nucleotide misincorporations, chimeras, and heteroduplex formations (40, 49, 53). In replicate computations, depending on the sequences dropped and the respective resulting frequency distribution, the number of different 16S rRNA gene sequences from Chloroflexaceae relatives present in the microbial mat sample was conservatively estimated to be 30 to 39 (data not shown). We also note that 8 of the 27 genotypes were observed in replicate. Since artifactual sequences were singletons in a previous study (49), this further increases our confidence in the reality of much of the detected diversity. In addition, many identical sequences were found in another clone library that was generated by using a different set of PCR primers (unpublished data).
FISH.
Initially, we used Cy3-labeled and fluorescein-labeled oligonucleotide probes (EUB338) for in situ hybridizations. When applied to microbial mat samples from Mushroom Spring, however, these probes did not produce sufficiently strong fluorescent hybridization signals (data not shown). Strong autofluorescence of cyanobacteria, dimly detectable even with the green emission filter (Fig. 2), interfered with the unequivocal identification of hybridized cells. To obtain more intense hybridization signals in subsequent experiments, we used HRP-labeled oligonucleotide probes in combination with a tyramide signal amplification system using fluorescein-tyramide (47). Due to the precipitation of multiple fluorescent dye molecules per probe molecule, the application of this technique produced significantly brighter hybridization signals than the use of fluorescently monolabeled probes.
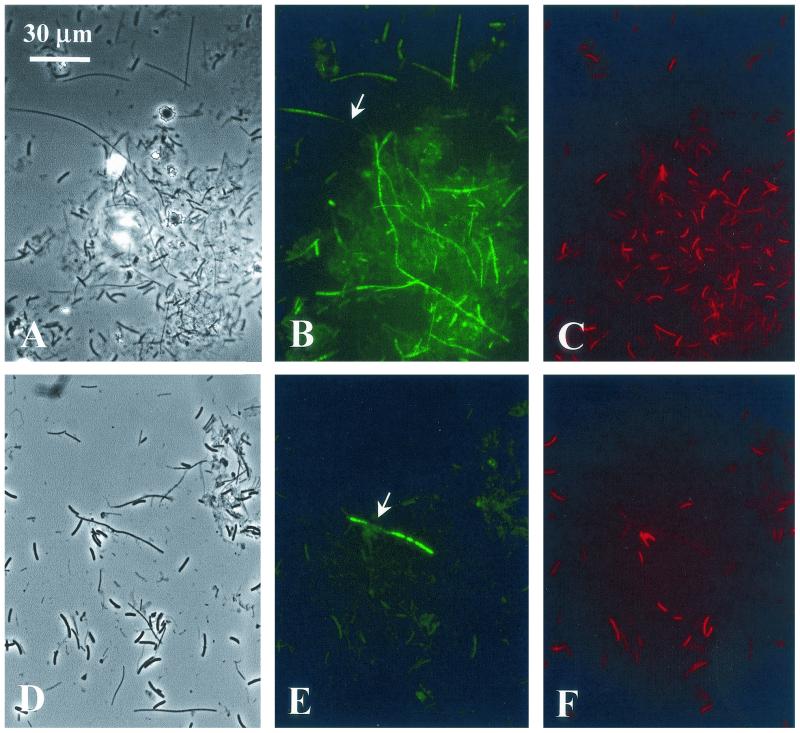
FIG. 2.
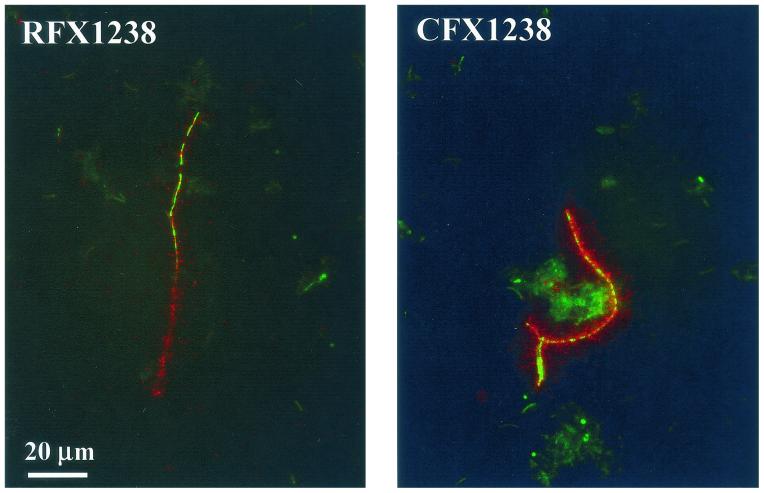
FISH identification of type C organisms and Chloroflexus spp. in a microbial mat sampled from a 60°C site in the Mushroom Spring effluent channel. (A to C) Hybridization with probe RFX1238. (D to F) Hybridization with probe CFX1238. Phase-contrast (A and D) and epifluorescence (B and E [fluorescein fluorescence] and C and F [cyanobacterial autofluorescence]) photomicrographs of identical microscopic fields are shown. Arrows point to heterogeneous FISH staining of individual filaments. The bar applies to all panels.
A series of hybridization experiments at increasing stringency resulted in probe dissociation curves (data not shown). The intensities of the hybridization signals obtained with probes CFX1238, RFX1238, and EUB338 strongly dropped at formamide concentrations in the hybridization buffer of higher than 40, 40, and 50%, respectively. Aside from different maximum fluorescence intensities, virtually identical dissociation curves were obtained when probes were hybridized to cultivated bacterial cells with matching 16S rRNAs or to filaments in microbial mat samples (data not shown). Formamide concentrations for half-maximal probe binding for cultures and natural samples differed by only 1% (RFX1238) and 6% (CFX1238), indicating that stained organisms in the mats also exhibited matching rRNAs. In comparison, even a single lateral mismatch between an 18-nucleotide probe and bacterial rRNA commonly decreases the formamide concentration for half-maximal FISH probe binding by approximately 13 to 17% (13, 32). Furthermore, application of probe RFX1238 to cultures of C. aurantiacus and C. aggregans and of probe CFX1238 to R. castenholzii did not yield any measurable hybridization signals even at the lowest stringency tested (20% formamide) (data not shown), indicating that rRNAs with two mismatches were safely discriminated against in subsequent experiments performed at 40 and 50% formamide.
Phylogenetic probes RFX1238 and CFX1238 both stained filamentous organisms 0.7 to 1.0 μm in width and indeterminate in length (Fig. 2).
Distribution of Chloroflexaceae relatives.
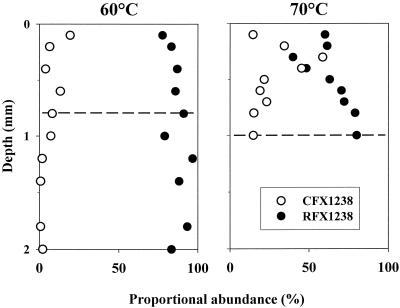
In the microbial mat sampled from the 60°C site, 78 to 97% of filaments visible in phase contrast were FISH stained with probe RFX1238 (identifying type C and related bacteria within the Roseiflexus assemblage), and 0 to 19% were stained with probe CFX1238 (identifying Chloroflexus spp.) (Fig. 3). While Chloroflexus spp. were found exclusively in the top 1 mm, coinciding with the occurrence of cyanobacteria, type C was the dominant filament at all depths and appeared to be almost the only component of deeper, orange-colored mat layers. At 70°C, type C was also predominant, making up 40 to 80% of the filamentous biomass depending on the vertical position within the mat. Here, however, 59% of the filaments were stained with probe CFX1238 at a subsurface maximum (0.4 to 0.6 mm), indicating that Chloroflexus spp. accounted for a relatively larger proportion of filaments than at the 60°C site. Type C filaments comprised the majority of filaments above and below this layer. Together, the two probes RFX1238 or CFX1238 stained 75 to 99% of all filaments visible in phase contrast.
FIG. 3.
Proportional abundances of filaments stained with FISH probes CFX1238 and RFX1238, depending on depth within the microbial mats sampled from 60 and 70°C sites. The dashed lines indicate the greatest depths at which cyanobacteria were microscopically observed.
Pigment distribution.
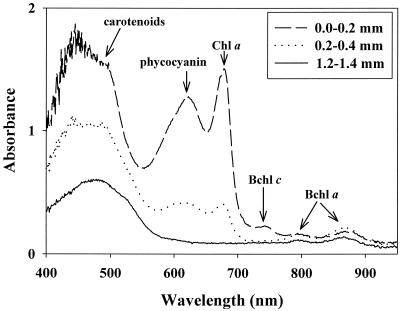
The absorbance of photosynthetic pigments and their vertical distribution within the microbial mat at 60°C were analyzed by microscope spectrometry (Fig. 4). At the mat surface (0- to 0.2-mm depth), strong absorbance peaks at characteristic wavelengths indicated large amounts of the cyanobacterial pigments chlorophyll a (wavelength, 678 nm) and phycocyanin (622 nm). Additional peaks indicated the presence of Bchl c (744 nm) and Bchl a (794 and 869 nm) in the same mat layer. At greater depths (e.g., 0.2 to 0.4 mm), the contents of cyanobacterial pigments and Bchl c diminished. In deep, orange mat layers (i.e., below 0.9 mm), only Bchl a was detected. The broad absorbance peak at 400 to 500 nm detected throughout the mat is due to the presence of carotenoids and includes additional absorbance maxima of the various chlorophylls. Overall, cyanobacterial pigments were detected at higher concentrations than Bchls, probably due to both larger biomasses of cyanobacteria in upper mat layers and low specific pigment contents of filaments in deep layers. These observations were confirmed by measurements of mat samples with fiber optic scalar irradiance microprobes (M. Kühl, unpublished data).
FIG. 4.
Absorbance spectra obtained by microscope spectrometry of a thin section of a microbial mat from a 60°C site in Mushroom Spring cut perpendicular to the mat surface. Absorbance was calculated for 0.2-mm depth intervals at three depths as indicated. Absorbance peaks: phycocyanin, 622 nm; chlorophyll a, 678 nm; Bchl c, 744 nm; Bchl a, 794 and 869 nm.
Oxygen concentration profiles.
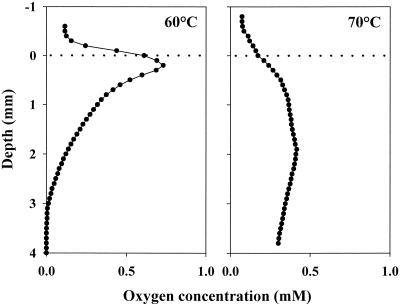
Figure 5 shows representative oxygen concentration profiles measured under strong solar irradiation in microbial mats at 60 and 70°C sites in Mushroom Spring. At both of these sites, the maximum oxygen concentrations within the mats due to cyanobacterial photosynthesis exceeded the concentration of air-saturated water (148 and 127 μM at 60 and 70°C, respectively). At 60°C, the highest concentration was found at 0.2 mm below the mat surface and oxygen penetrated to a 3-mm depth. At 70°C, the oxygen profile did not show a pronounced peak. Here, the oxygen concentration was several times higher than that in the overlying water at all depths within the mat and was maximal at a depth of 1.9 mm. At 70°C, the mat is more gelatinous and translucent to light. Concomitant oxygen production at greater depth and lower volumetric oxygen consumption rates due to lower cell densities cause the observed oxygen penetration to greater depth at this temperature.
FIG. 5.
Oxygen concentration profiles in microbial mats at 60 and 70°C sites in Mushroom Spring. The measurements were made in situ under an ambient-light irradiance of 1,800 to 2,500 μE m−2 s−1. The dotted lines indicate positions of mat surfaces.
Microautoradiography.
We combined FISH and microautoradiography on a single microscope slide to simultaneously investigate the phylogenetic affiliation and substrate uptake of Chloroflexaceae-related bacteria in microbial mats (Fig. 6). The best results were obtained when cells were FISH stained before being coated with an autoradiographic emulsion film, similar to the protocol of Lee et al. (27). Alternative experiments, with FISH performed after autoradiography development (36), were less satisfactory due to swelling and frequent detachment of the autoradiographic film. The optimum exposure time for autoradiography, enabling unequivocal substrate uptake detection but avoiding complete coverage of filaments with silver grains and concomitant masking of the fluorescent hybridization signal, was determined empirically and varied from 7 to 16 days.
FIG. 6.
Photomicrographs of filamentous organisms in a Mushroom Spring microbial mat sample incubated with 14C-labeled acetate at in situ temperature (60°C) and irradiance. Epifluorescence and dark-field images of identical microscopic fields have been overlaid. Filaments were simultaneously stained by FISH, indicating phylogenetic affiliation with type C organisms or Chloroflexus spp. (probes RFX1238 and CFX1238, respectively, shown in green), and by microautoradiography, revealing substrate uptake (silver grains, shown in red). The bar applies to both panels.
In microbial mat samples that had been incubated with sodium [1-14C]acetate, approximately 1 to 3% of the filaments exhibited radioactivity as determined by the high density of silver grains appearing adjacent to them (Fig. 6). Nonradiolabeled or formaldehyde-killed cells in control experiments did not accumulate any silver grains, indicating that chemography and acetate adsorption did not occur. Organisms from both phylogenetic groupings investigated, type C and Chloroflexus spp., apparently had taken up radiolabeled acetate during the incubation experiments (Fig. 6). Acetate uptake was observed in samples incubated in the light and in the dark. In contrast, assimilation of CO2 could not be detected in any sample.
DISCUSSION
Diversity of Chloroflexaceae-related bacteria.
From a single 150-μl sample drawn from a Mushroom Spring microbial mat, we recovered 27 different 16S rRNA gene sequences from Chloroflexaceae-related bacteria (Fig. 1). This exceeds the sequence diversity detected in this hot spring mat and in a similar mat in Octopus Spring through previous extensive molecular studies (15-17, 41, 55, 56) by almost one order of magnitude (three different type C-like sequences were recovered previously). Furthermore, rarefaction analysis suggested that the recovered sequences may represent only a fraction of the extant gene diversity. There are many sequences that differ at only a few nucleotide positions, and it cannot be excluded at present that some of the sequence diversity detected might be due to PCR misincorporations. However, even if the highest PCR error rate reported in the literature (7 to 32%) (40, 49, 53) is postulated, the previously detected sequence diversity among 16S rRNA gene sequences from Chloroflexaceae relatives in these hot springs is exceeded at least severalfold. Sequences from bacteria affiliated with the present genus Chloroflexus (Fig. 1) were recovered from this habitat for the first time.
This discovery of greater diversity among Chloroflexaceae-related bacteria (Fig. 1) to some extent may be due to the use of a directed PCR approach (33), which enabled a more focused study of this phylogenetic group compared to previous investigations targeting all bacteria. In addition, early cloning studies may have missed particular genotypes, simply because they were restricted to the analysis of a smaller number of clones (55, 56). Diversity studies based on denaturing gradient gel electrophoresis (DGGE) generally are believed to detect the numerically dominant genotypes (31) and hence may miss less abundant genotypes. It is also possible, however, that a large portion of extant sequence diversity may have remained undetected in previous DGGE studies on these microbial mats (15-17, 41) due to insufficient electrophoretic separation of DNA molecules with highly similar sequences. We are currently investigating this potential limitation.
Clusters of highly similar yet distinct 16S rRNA gene sequences have been observed in molecular studies of microbial diversity in numerous habitats, including hot springs (54). Unique distributions of similar 16S rRNA genes from hot spring cyanobacteria and type C-like bacteria previously suggested evolutionary divergence of closely related microorganisms into ecologically distinct populations (54). At present, no information is available about the spatial distribution of the large sequence diversity detected in this study, and hence it cannot be excluded that heterogeneities of 16S rRNA genes within genomes of individual bacteria to some extent may contribute to this diversity (34). However, it seems likely that a considerable diversity of Chloroflexaceae-related bacteria in hot springs in the past had remained undetected through both cultivation-based and molecular investigations. Numerous closely related populations may exist within a small spatial scale, and each may be adapted to particular conditions in the hot spring mat environment (e.g., with respect to carbon sources, light conditions, etc.).
In a recent survey of various hot springs in Yellowstone National Park, covering a temperature range of 35 to 53°C, Boomer et al. discovered two previously unknown phylogenetic clusters of bacteria related to R. castenholzii (YRL-A and YRL-B in Fig. 1), and assumed that these were filamentous phototrophs since they were recovered from Bchl-rich mat layers (7). Interestingly, they observed some site-specific phylogenetic clustering and did not detect any type C-like organisms in their samples (7). It will be interesting to unravel the environmental conditions that govern the differential compositions of these microbial mats, which are still little understood. It is possible that temperature adaptation explains some of the diversity within the Roseiflexus assemblage, since the three major clades (YRL-A, YRL-B, and type C) contain clones obtained from samples collected at 47 to 53°C and 35 to 41°C (7) and at 60 to 70°C (this study), respectively. While Chloroflexus spp. have repeatedly been cultivated from these and similar hot springs (references 28, 37, 38, and 46 and this study), the isolation of new strains of numerically more relevant mat-forming bacteria would enable in-depth studies of their physiological diversity and hence would be particularly enlightening.
Application of the FISH technique to cyanobacterial mats.
Due to their content of photosynthetic pigments, phototrophic organisms in hot spring microbial mats commonly exhibit massive autofluorescence that may interfere with fluorescence-based identification techniques such as FISH. In the present study, this problem could be overcome by use of HRP-labeled oligonucleotide probes in conjunction with fluorescein-tyramide signal amplification as previously described by Schönhuber et al. (47, 48). Probe-mediated fluorescence could be unequivocally distinguished from autofluorescence due to very strong signal intensity and differential color (Fig. 2). Comparison of probe dissociation curves obtained with cultures and microbial mat samples provided evidence for highly specific phylogenetic identification of type C-like organisms and Chloroflexus spp. with probes RFX1238 and CFX1238, respectively (data not shown).
The majority (75 to 99%) of filaments in the microbial mats were stained by probes RFX1238 or CFX1238, indicating that they are phylogenetically affiliated with either type C or Chloroflexus spp. While filaments with nonmatching rRNAs may exist, the observation of incompletely stained, individual filaments (Fig. 2) suggested that unstained filaments may instead be due to variable concentrations of cellular rRNA or to technical limitations of the FISH method. Similarly, using HRP-labeled oligonucleotide probes and tyramide signal amplification for detection, Schönhuber et al. previously observed irregular FISH staining of cells within pure cultures of gram-positive bacteria and assumed that this problem may be caused by heterogeneous cell wall permeability and concomitant probe accessibility (47).
Radiolabeled, FISH-stained microbial mat samples were further processed for microautoradiographic detection of substrate uptake. While, to our knowledge, microautoradiography had not been previously combined with the use of HRP-labeled probes and tyramide signal amplification-based hybridization detection, we did not experience any specific difficulties.
Phenotype of uncultivated Chloroflexaceae relatives.
In the past, type C-like 16S rRNA gene sequences had repeatedly been discovered through molecular surveys of microbial mats from alkaline hot springs (Fig. 1) (15-17, 41, 54, 56). Our FISH results showed that these sequences originated from abundant bacteria with a filamentous morphology (Fig. 3 and 4). These filaments predominate the microbial mats at 60 and 70°C sites in Mushroom Spring (Fig. 3). In turn, Chloroflexus spp. were indicated through our present and previous (44) probe hybridization studies to represent only a minor component of the natural microbial mats, even though several related strains could be readily cultivated from the studied habitat. Therefore, the majority of morphologically similar filaments in hot springs, which, in the past, collectively have been referred to as Chloroflexus spp. (8, 9, 14) or Chloroflexus-like organisms (41), not only are remarkably diverse (see above) but may be drastically unrelated to Chloroflexus spp. and instead form a separate phylogenetic lineage affiliated with the recently described R. castenholzii (Fig. 1) (22).
The vertical distribution of FISH-identified Chloroflexus spp. and type C organisms (Fig. 3) within the mats correlated with the detection of Bchl c and Bchl a, respectively (Fig. 4). This distribution is consistent with the hypothesis that Chloroflexus spp. in the microbial mat, like their cultivated relatives, produce both of these Bchls and that type C organisms are Bchl a-producing phototrophs, similar to their relative R. castenholzii (22).
Both phylogenetic groups investigated, type C organisms and Chloroflexus spp., were detected at high numbers within the upper 1 mm of the microbial mats (Fig. 3). Here, they cooccured with abundant cyanobacteria and obviously tolerated very high concentrations of oxygen during the day (Fig. 5). This result is in agreement with earlier light and electron microscopic observations of filamentous organisms intermingled with cyanobacteria close to the mat surface (6, 11, 41), PCR-DGGE detection of 16S rRNA genes from both type C organisms and cyanobacteria within the top mat layer (41), and dot blot hybridization experiments which measured the highest concentrations of type C-like rRNA in the upper 1-mm layer of the analyzed (48 to 51°C) mat (44). Decreasing concentrations of type C-like rRNA with depth in the mat (44) and the fact that Chloroflexus spp. were not found at all outside the cyanobacterial layer (Fig. 3) suggested a preference of both of these groups of organisms for a life close to the mat surface. In the laboratory, cultures of both C. aurantiacus and R. castenholzii are able to grow under atmospheric oxygen tension (11, 22). While cultivated strains of C. aurantiacus require anoxic conditions for the synthesis of Bchls (11), which in these microbial mats prevail at night (41), pigment production by R. castenholzii HL08 may occur under both aerobic and anaerobic conditions (22).
Through combined FISH and microautoradiography, we observed uptake of acetate, but not bicarbonate, by both Chloroflexus sp. and type C filaments (Fig. 6). While this uptake does not prove the use of acetate under natural conditions, in low-sulfate hot springs such as Mushroom Spring, Chloroflexaceae relatives are commonly considered to primarily feed photoheterotrophically, due to low concentrations of potential electron donors for autotrophy (43) and to the suppression of aerobic respiration in the light, at least in cultures of C. aurantiacus containing Bchls (11). It has been suggested that Chloroflexaceae relatives in these habitats may feed on low-molecular-weight organic compounds released by cyanobacteria (4, 5, 8, 11, 45, 51, 54). For such photoheterotrophs, an upper position within the mat may be advantageous due to strong light irradiation and close vicinity to the primary producers. In contrast, however, the natural carbon isotope composition of particular lipids in a microbial mat from Octopus Spring recently suggested that carbon fixation may occur through the 3-hydroxypropionate pathway, which is typical for C. aurantiacus and possibly also for related bacteria (52). At present, we cannot exclude the possibility that our microautoradiography experiments failed to detect the uptake of inorganic carbon by filamentous bacteria because of diffusion limitations or low specific activity of radiolabeled bicarbonate. Further investigations are needed to uncover the principal carbon sources of Chloroflexaceae-related bacteria in these habitats, taking into account that the availability of sulfide and hydrogen as possible electron donors for photoautotrophy may vary with light conditions and hence with daytime (43, 52).
While well-developed microbial mats are found at between approximately 40 and 70°C, most previous studies in Mushroom Spring and the very similar Octopus Spring have investigated sites that are from 48 to 70°C (6, 14-17, 41, 54, 56). Several different type C-like 16S rRNA gene sequences have been retrieved from mats in this temperature range (15-17, 41, 56), whereas sequences from Chloroflexus spp. had not previously been recovered. Through dot blot hybridizations, Ruff-Roberts et al. measured type C-like 16S rRNA in mats from 60 to 70°C and diminishing concentrations below 55°C (44). In the same study, another probe detected rRNA from C. aurantiacus only at the highest temperature investigated. Our present FISH results are in agreement with previous reports indicating predominance of type C and low abundance of Chloroflexus spp., the latter increasing with temperature. However, we did find small numbers of Chloroflexus spp. at 60°C; these were not previously detected through dot blot hybridizations, possibly due to the more general specificity of our probe CFX1238 than the C. aurantiacus probe used by Ruff-Roberts et al. The latter probe likely would not have detected rRNAs from, for example, C. aggregans (four mismatches) or the cluster around our Mushroom Spring enrichments (Fig. 1) (one mismatch), the nucleotide sequences of which had not been known at the time the probe was designed (44). However, small numbers of Chloroflexus spp. had previously been detected at low-temperature sites (50 to 55°C) through enrichment cultivation (46) and following artificial mat sample incubation at 67°C for 1 week (44).
Due to the use of oligonucleotide probes directed against rRNAs from entire clusters of the Roseiflexus assemblage and Chloroflexus spp. (Fig. 1), our present study did not investigate the distribution of within-cluster diversity. At present, it is therefore not clear if the observed distribution pattern is due to broad temperature tolerances of individual organisms or to the existence of numerous differentially adapted populations. The latter is suggested, however, by previous DGGE studies that detected different type C-like sequences at different temperatures (17). In addition, laboratory cultures of Chloroflexus spp. and R. castenholzii (the closest cultivated relative of type C) grow at 45 to 66°C and at 45 to 55°C, respectively (11, 21, 22), and hence do not cover the entire temperature range of occurrence of their relatives in nature. Since it is clear that these cultivated strains represent only a minor part of the natural diversity (Fig. 1), it seems likely that as-yet-uncultivated, genetically distinct populations of Chloroflexus spp. and type C organisms that are adapted to higher temperatures exist in the investigated hot spring.
Acknowledgments
We are indebted to D. Des Marais for the coordination of microbial mat research within the NASA Astrobiology Institute. We thank W. Schönhuber and D. A. Stahl for initial help with FISH, K. Slack and A. Scotti for field assistance with radiolabeling experiments, A. Glud for excellent technical assistance and microsensor construction, S. Hanada for cells of R. castenholzii and sharing unpublished data, E. Stackebrandt for inspiring discussions, and S. M. Holland for providing the freeware program aRarefactWin. The continued support provided by the U.S. National Park Service is greatly acknowledged.
Funding for this work was provided by the NASA Astrobiology Institute (CAN-97-01-OSS-004) through a cooperative agreement with the NASA Ames Research Center (NCC 2-1073), the NASA Exobiology Program (NAG5-8824), the NSF Ecology Program (BSR-9708136), the Danish Natural Science Research Council (to M.K.), and the Deutsche Sammlung von Mikroorganismen und Zellkulturen.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Amann, R. I., B. J. Binder, R. J. Olson, S. W. Chisholm, R. Devereux, and D. A. Stahl. 1990. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl. Environ. Microbiol. 56:1919-1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amann, R. I., B. Zarda, D. A. Stahl, and K. H. Schleifer. 1992. Identification of individual prokaryotic cells with enzyme-labeled, rRNA-targeted oligonucleotide probes. Appl. Environ. Microbiol. 58:3007-3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anderson, K. L., T. A. Tayne, and D. M. Ward. 1987. Formation and fate of fermentation products in hot spring cyanobacterial mats. Appl. Environ. Microbiol. 53:2343-2352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bateson, M. M., and D. M. Ward. 1988. Photoexcretion and fate of glycolate in a hot spring cyanobacterial mat. Appl. Environ. Microbiol. 54:1738-1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bauld, J., and T. D. Brock. 1973. Ecological studies of Chloroflexis, a gliding photosynthetic bacterium. Arch. Microbiol. 92:267-284. [Google Scholar]
- 7.Boomer, S. M., D. P. Lodge, B. E. Dutton, and B. K. Pierson. 2002. Molecular characterization of novel red-green nonsulfur bacteria from five distinct hot spring communities in Yellowstone National Park. Appl. Environ. Microbiol. 68:346-355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boomer, S. M., B. K. Pierson, R. Austinhirst, and R. W. Castenholz. 2000. Characterization of novel bacteriochlorophyll-a-containing red filaments from alkaline hot springs in Yellowstone National Park. Arch. Microbiol. 174:152-161. [DOI] [PubMed] [Google Scholar]
- 9.Castenholz, R. W. 1984. Composition of hot spring microbial mats: a summary, p. 101-119. In Y. Cohen, R. W. Castenholz, and H. O. Halvorson (ed.), Microbial mats: stromatolites. Alan R. Liss, Inc., New York, N.Y.
- 10.Castenholz, R. W. 1973. The possible photosynthetic use of sulfide by the filamentous phototrophic bacteria of hot springs. Limnol. Oceanogr. 18:863-876. [Google Scholar]
- 11.Castenholz, R. W., and B. K. Pierson. 1995. Ecology of thermophilic anoxygenic phototrophs, p. 87-103. In R. E. Blankenship, M. T. Madigan, and C. E. Bauer (ed.), Anoxygenic photosynthetic bacteria. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 12.Castenholz, R. W., and J. B. Waterbury. 1989. Oxygenic photosynthetic bacteria, group I. Cyanobacteria, p. 1710-1728. In M. P. Bryant, N. Pfennig, and J. G. Holt (ed.), Bergey's manual of systematic bacteriology. The Williams & Wilkins Co., Baltimore, Md.
- 13.Daims, H., A. Brühl, R. I. Amann, K.-H. Schleifer, and M. Wagner. 1999. The domain-specific probe EUB338 is insufficient for the detection of all Bacteria: development and evaluation of a more comprehensive probe set. Syst. Appl. Microbiol. 22:434-444. [DOI] [PubMed] [Google Scholar]
- 14.Doemel, W. N., and T. D. Brock. 1977. Structure, growth, and decomposition of laminated algal-bacterial mats in alkaline hot springs. Appl. Environ. Microbiol. 34:433-452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ferris, M. J., G. Muyzer, and D. M. Ward. 1996. Denaturing gradient gel electrophoresis profiles of 16S rRNA-defined populations inhabiting a hot spring microbial mat community. Appl. Environ. Microbiol. 62:340-346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ferris, M. J., S. C. Nold, N. P. Revsbech, and D. M. Ward. 1997. Population structure and physiological changes within a hot spring microbial mat community following disturbance. Appl. Environ. Microbiol. 63:1367-1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ferris, M. J., and D. M. Ward. 1997. Seasonal distributions of dominant 16S rRNA-defined populations in a hot spring microbial mat examined by denaturing gradient gel electrophoresis. Appl. Environ. Microbiol. 63:1375-1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garcia-Pichel, F., M. Mechling, and R. W. Castenholz. 1994. Diel migrations of microorganisms within a benthic, hypersaline mat community. Appl. Environ. Microbiol. 60:1500-1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gich, F., J. Garcia-Gil, and J. Overmann. 2001. Previously unknown and phylogenetially diverse members of the green nonsulfur bacteria are indigenous to freshwater lakes. Arch. Microbiol. 177:1-10. [DOI] [PubMed] [Google Scholar]
- 20.Giovannoni, S. J., N. P. Revsbech, D. M. Ward, and R. W. Castenholz. 1987. Obligately phototrophic Chloroflexus: primary production in anaerobic hot spring microbial mats. Arch. Microbiol. 147:80-87. [Google Scholar]
- 21.Hanada, S., A. Hiraishi, K. Shimada, and K. Matsuura. 1995. Chloroflexus aggregans sp. nov., a filamentous phototrophic bacterium which forms dense cell aggregates by active gliding movement. Int. J. Syst. Bacteriol. 45:676-681. [DOI] [PubMed] [Google Scholar]
- 22.Hanada, S., S. Takaichi, K. Matsuura, and K. Nakamura. 2002. Roseiflexus castenholzii gen. nov., sp. nov., a thermophilic, filamentous, photosynthetic bacterium that lacks chlorosomes. Int. J. Syst. Evol. Microbiol. 52:187-193. [DOI] [PubMed] [Google Scholar]
- 23.Hurlbert, S. H. 1971. The nonconcept of species diversity: a critique and alternative parameters. Ecology 52:577-586. [DOI] [PubMed] [Google Scholar]
- 24.Keppen, O. I., T. P. Tourova, B. B. Kuznetsov, R. N. Ivanovsky, and V. M. Gorlenko. 2000. Proposal of Oscillochloridaceae fam. nov. on the basis of a phylogenetic analysis of the filamentous anoxygenic phototrophic bacteria and emended description of Oscillochloris and Oscillochloris trichoides in comparison with further new isolates. Int. J. Syst. Evol. Microbiol. 50:1529-1537. [DOI] [PubMed] [Google Scholar]
- 25.Kopczynski, E. D., M. M. Bateson, and D. M. Ward. 1994. Recognition of chimeric small-subunit ribosomal DNAs composed of genes from uncultivated microorganisms. Appl. Environ. Microbiol. 60:746-748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kühl, M., and T. Fenchel. 2000. Bio-optical characteristics and the vertical distribution of photosynthetic pigments and photosynthesis in an artificial cyanobacterial mat. Microb. Ecol. 40:94-103. [DOI] [PubMed] [Google Scholar]
- 27.Lee, N., P. H. Nielsen, K. H. Andreasen, S. Juretschko, J. L. Nielsen, K. H. Schleifer, and M. Wagner. 1999. Combination of fluorescent in situ hybridization and microautoradiography: a new tool for structure-function analyses in microbial ecology. Appl. Environ. Microbiol. 65:1289-1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Madigan, M. T., S. R. Petersen, and T. D. Brock. 1974. Nutritional studies on Chloroflexus, a filamentous photosynthetic, gliding bacterium. Arch. Microbiol. 100:97-103. [Google Scholar]
- 29.Manz, W., R. I. Amann, W. Ludwig, M. Wagner, and K. H. Schleifer. 1992. Phylogenetic oligodeoxynucleotide probes for the major subclasses of proteobacteria: problems and solutions. Syst. Appl. Microbiol. 15:593-600. [Google Scholar]
- 30.Moré, M. I., J. B. Herrick, M. C. Silva, W. C. Ghiorse, and E. L. Madsen. 1994. Quantitative cell lysis of indigenous microorganisms and rapid extraction of microbial DNA from sediment. Appl. Environ. Microbiol. 60:1572-1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Muyzer, G., and K. Smalla. 1998. Application of denaturing gradient gel electrophoresis (DGGE) and temperature gradient gel electrophoresis (TGGE) in microbial ecology. Antonie Leeuwenhoek 73:127-141. [DOI] [PubMed] [Google Scholar]
- 32.Neef, A., A. Zaglauer, H. Meier, R. I. Amann, H. Lemmer, and K.-H. Schleifer. 1996. Population analysis in a denitrifying sand filter: conventional and in situ identification of Paracoccus spp. in methanol-fed biofilms. Appl. Environ. Microbiol. 62:4329-4339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nübel, U., M. M. Bateson, M. Kühl, M. T. Madigan, and D. M. Ward. 2001. Diversity and distribution in hypersaline microbial mats of bacteria related to Chloroflexus spp. Appl. Environ. Microbiol. 67:4365-4371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nübel, U., B. Engelen, A. Felske, J. Snaidr, A. Wieshuber, R. I. Amann, W. Ludwig, and H. Backhaus. 1996. Sequence heterogeneities of genes encoding 16S rRNAs in Paenibacillus polymyxa detected by temperature gradient gel electrophoresis. J. Bacteriol. 178:5636-5643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nübel, U., F. Garcia-Pichel, M. Kühl, and G. Muyzer. 1999. Spatial scale and the diversity of benthic cyanobacteria and diatoms in a salina. Hydrobiologia 401:199-206. [Google Scholar]
- 36.Ouverney, C. C., and J. A. Fuhrman. 1999. Combined microautoradiography-16S rRNA probe technique for determination of radioisotope uptake by specific microbial cell types in situ. Appl. Environ. Microbiol. 65:1746-1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pierson, B. K. 2001. Family I. “Chloroflexaceae”—filamentous anoxygenic phototrophic bacteria, p. 427-444. In D. R. Boon, R. W. Castenholz, and G. M. Garrity (ed.), Bergey's manual of systematic bacteriology, 2nd ed., vol. 1. Springer Verlag, Berlin, Germany.
- 38.Pierson, B. K., and R. W. Castenholz. 1974. A phototrophic gliding filamentous bacterium of hot springs. Chloroflexus aurantiacus, gen. and sp. nov. Arch. Microbiol. 100:5-24. [DOI] [PubMed] [Google Scholar]
- 39.Pierson, B. K., S. J. Giovannoni, D. A. Stahl, and R. W. Castenholz. 1985. Heliothrix oregonensis, gen. nov., sp. nov., a phototrophic filamentous gliding bacterium containing bacteriochlorophyll a. Arch. Microbiol. 142:164-167. [DOI] [PubMed] [Google Scholar]
- 40.Qiu, X., L. Wu, H. Huang, P. E. McDonel, A. V. Palumbo, J. M. Tiedje, and J. Zhou. 2001. Evaluation of PCR-generated chimeras, mutations, and heteroduplexes with 16S rRNA gene-based cloning. Appl. Environ. Microbiol. 67:880-887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ramsing, N. B., M. J. Ferris, and D. M. Ward. 2000. Highly ordered vertical structure of Synechococcus populations within the one-millimeter-thick photic zone of a hot spring cyanobacterial mat. Appl. Environ. Microbiol. 66:1038-1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Revsbech, N. P. 1989. An oxygen microelectrode with a guard cathode. Limnol. Oceanogr. 34:474-478. [Google Scholar]
- 43.Revsbech, N. P., and D. M. Ward. 1984. Microelectrode studies of interstitial water chemistry and photosynthetic activity in a hot spring microbial mat. Appl. Environ. Microbiol. 48:270-275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ruff-Roberts, A. L., J. G. Kuenen, and D. M. Ward. 1994. Distribution of cultivated and uncultivated cyanobacteria and Chloroflexus-like bacteria in hot spring microbial mats. Appl. Environ. Microbiol. 60:697-704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sandbeck, K. A., and D. M. Ward. 1981. Fate of immediate methane precursors in low-sulfate, hot-spring algal-bacterial mats. Appl. Environ. Microbiol. 41:775-782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Santegoeds, C. M., S. C. Nold, and D. M. Ward. 1996. Denaturing gradient gel electrophoresis used to monitor the enrichment culture of aerobic chemoorganotrophic bacteria from a hot spring cyanobacterial mat. Appl. Environ. Microbiol. 62:3922-3928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schönhuber, W., B. Fuchs, S. Juretschko, and R. I. Amann. 1997. Improved sensitivity of whole-cell hybridization by the combination of horseradish peroxidase-labeled oligonucleotides and tyramide signal amplification. Appl. Environ. Microbiol. 63:3268-3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schönhuber, W., B. Zarda, S. Eix, R. Rippka, M. Herdman, W. Ludwig, and R. I. Amann. 1999. In situ identification of cyanobacteria with horseradish peroxidase-labeled, rRNA-targeted oligonucleotide probes. Appl. Environ. Microbiol. 65:1259-1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Speksnijder, A. G. C. L., G. A. Kowalchuk, S. De Jong, E. Kline, J. R. Stephen, and H. J. Laanbroek. 2001. Microvariation artifacts introduced by PCR and cloning of closely related 16S rRNA gene sequences. Appl. Environ. Microbiol. 67:469-472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tayne, T. A., J. E. Cutler, and D. M. Ward. 1987. Use of Chloroflexus-specific antiserum to evaluate filamentous bacteria of a hot spring microbial mat. Appl. Environ. Microbiol. 53:1962-1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Teiser, M. L. O. 1993. Extracellular low molecular weight organic compounds produced by Synechococcus sp. and their roles in the food web of alkaline hot spring microbial mat communities. Ph.D. thesis. University of Oregon, Eugene.
- 52.van der Meer, M. T. J., S. Schouten, J. W. de Leeuw, and D. M. Ward. 2000. Autotrophy of green non-sulphur bacteria in hot spring microbial mats: biological explanations for isotopically heavy organic carbon in the geological record. Environ. Microbiol. 2:428-435. [DOI] [PubMed] [Google Scholar]
- 53.Wang, G. C.-Y., and Y. Wang. 1997. Frequency of formation of chimeric molecules as a consequence of PCR coamplification of 16S rRNA genes from mixed bacterial genomes. Appl. Environ. Microbiol. 63:4645-4650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ward, D. M., M. J. Ferris, S. J. Nold, and M. M. Bateson. 1998. A natural view of microbial biodiversity within hot spring cyanobacterial mat communities. Microbiol. Mol. Biol. Rev. 62:1353-1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ward, D. M., R. Weller, and M. M. Bateson. 1990. 16S ribosomal RNA sequences reveal numerous uncultured microorganisms in a natural community. Nature 345:63-65. [DOI] [PubMed] [Google Scholar]
- 56.Weller, R., M. M. Bateson, B. K. Heimbuch, E. D. Kopczynski, and D. M. Ward. 1992. Uncultivated cyanobacteria, Chloroflexus-like inhabitants, and Spirochete-like inhabitants of a hot spring microbial mat. Appl. Environ. Microbiol. 58:3964-3969. [DOI] [PMC free article] [PubMed] [Google Scholar]