Abstract
Pseudomonas sp. strain DSS73 isolated from the sugar beet rhizosphere produces the cyclic lipopeptide amphisin, which inhibits the growth of plant-pathogenic fungi. By Tn5::luxAB mutagenesis, we obtained two nonproducing mutant strains, DSS73-15C2 and DSS73-12H8. The gene interrupted by the transposon in strain DSS73-15C2 (amsY) encoded a protein with homology to peptide synthetases that was designated amphisin synthetase. DSS73-12H8 carried the transposon in a regulatory gene encoding a protein with homology to the sensor kinase GacS. Growth of strain DSS73-15C2 (amsY) was impaired during the transition to stationary phase in a minimal medium amended with an exudate of sugar beet seeds. This growth phenotype could be complemented by purified amphisin. Seed exudate further induced expression of bioluminescence from the amsY::luxAB reporter during the transition to stationary phase. This agreed with an increase in amphisin production by the DSS73 wild-type strain during early stationary phase. Amphisin synthesis in DSS73 was strictly dependent on GacS, and even induction by seed exudate depended on a functional gacS locus. Hence, a signal triggering the GacS/GacA two-component system appeared to be present in the seed exudate.
Several Pseudomonas strains isolated from soil produce cyclic lipopeptides (38, 39). The purified lipopeptides show in vitro antagonistic activity against several fungi, including root pathogens such as Rhizoctonia solani (38, 39) and Pythium ultimum (29, 38, 39). These studies demonstrate a potential role for lipopeptide-producing bacteria in the biocontrol of fungal diseases, as also documented by laboratory and field experiments (55, 56). An antagonistic activity toward fungi has also been reported for tolaasiin produced by Pseudomonas tolaasii, which elicits the symptoms of the brown blotch disease of the mushroom Agaricus bisporus (5). Furthermore, syringomycin contributes to the pathogenicity of the plant pathogen Pseudomonas syringae (3, 48). The primary antagonistic mechanism of these lipopeptides is the formation of transmembrane ion channels that disrupt the electrical potential across the plant or fungal cell membrane (5, 23).
Lipopeptides may also function as biosurfactants (11), which can facilitate bacterial growth on water-insoluble carbon sources (26, 46) or their interaction with hydrophobic surfaces (37), e.g., surface motility (30). However, the significance of cyclic lipopeptides for growth and survival of the producing bacteria in soil or at soil-plant interphases has yet to be demonstrated.
The peptide moieties of lipopeptides are biosynthesized nonribosomally on large multienzyme complexes called peptide synthetases (3, 35, 51). The peptide synthetases are composed of modules, each of which incorporates a specific amino acid into the lipopeptide (31). It has been reported that the carbon source is important for lipopeptide production, but there is limited knowledge about the regulation of lipopeptide synthesis in soil pseudomonads (38, 39). Pseudomonas spp. possess a number of two-component regulatory systems that modulate their cellular activity in response to various environmental signals (45, 53). The GacA/GacS system controls the production of several extracellular products, including proteases (9, 14, 17, 22), chitinases (15), hydrogen cyanide (9, 43), various antibiotics (9, 14), and the lipopeptides syringomycin and tolaasiin (17, 22). Syringomycin synthesis is induced by plant signal molecules (33, 34). However, it is unknown whether these signals are channeled through the GacA/GacS system (44) and the signal(s) to which the GacA/GacS system responds remains unknown (9, 57).
Pseudomonas sp. strain DSS73 isolated from the sugar beet rhizosphere produces the cyclic lipopeptide amphisin, which has antifungal activities (50). In the present work, we identify a peptide synthetase gene involved in amphisin production. Interaction between biocontrol strains such as DSS73 and pathogens such as P. ultimum takes place on sugar beet seeds as early as 3 to 4 h after sowing in infested soil (40). As seeds take up water, significant amounts of solutes and various antimicrobial compounds are released into the surrounding medium (6, 7, 10, 20, 54). We demonstrate here that components of sugar beet seed exudate influence production of amphisin, expression of the peptide synthetase gene, and growth of a peptide synthetase mutant. Furthermore, we present evidence indicating that these effects are channeled through the GacA/GacS system.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. Pseudomonas strains were cultured at 20°C in Davis minimal medium (DMM; Difco Laboratories, Detroit, Mich.) or Luria-Bertani broth (LB) (47). Escherichia coli strains were cultured in LB at 37°C. Growth of the cultures was measured as optical density at 600 nm (OD600) with a Shimadzu UV-160A spectrophotometer. Tests for growth on glycyl-l-glutamic acid were performed in DMM with 0.4% glycyl-l-glutamic acid (Sigma Chemical Co., St. Louis, Mo.) as the only carbon source. In experiments employing amphisin-amended growth media, amphisin was dissolved in methanol at 5 mg ml−1 and added to the growth media to obtain a final concentration of 0.1 mg ml−1. Methanol (2% final concentration) was added to the corresponding controls without amphisin.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant genotype and/or characteristicsa | Source or reference |
|---|---|---|
| E. coli strains | ||
| XL1-Blue | recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 lac[F′ proAB lacIqZΔM15 Tn10 (Tetr)] | Stratagene |
| DH5α | F−endA1 hsdR17 (rK− mK+) supE44 thi-1 recA1 gyrA96 relA1 φ80dlacZΔM15 λ− | 47 |
| Pseudomonas strains | ||
| DSS73 | Wild type | 50 |
| DSS73-15C2 | KmramsY::Tn5 mutant of DSS73 | This study |
| DSS73-12H8 | KmrgacS::Tn5 mutant of DSS73 | This study |
| DSS73-MM | Kmr spontaneous gacS mutant of DSS73-15C2 | This study |
| Plasmids | ||
| p15C2 | Kmr; contains Tn5 and flanking chromosomal DNA from DSS73-15C2 | This study |
| p12H8 | Kmr; contains Tn5 and flanking chromosomal DNA from DSS73-12H8 | This study |
| pRK415 | Tcr; IncP1 replicon; polylinker of pUC19 | 24 |
| pJEL5771 | Tcr; contains functional gacS gene from P. fluorescens Pf5 | 9 |
| pEMH97 | Tcr; contains functional gacS gene from P. syringae pv. syringae B728a | 22 |
| pRL1063 | Kmr; delivery plasmid for Tn5 and luxAB | 58 |
| pRK2013 | Kmr; mobilizing plasmid | 13 |
Kmr and Tcr, resistance to kanamycin and tetracycline, respectively.
The phenolic compounds arbutin, salicin, and phenyl-β-d-glucopyranoside (Sigma) were added to DMM to final concentrations of 100 μM to 1 mM. Antibiotics were added to liquid or solid media at the following concentrations when appropriate: nystatin, 50 μg ml−1; ampicillin, 100 μg ml−1; kanamycin, 25 μg ml−1; tetracycline, 10 μg ml−1.
CFU were determined on LB agar plates containing 1.5% Bacto Agar (Becton Dickinson, Sparks, Md.) after incubation for approximately 24 h at 30°C.
Seeds and seed exudate amendment procedure.
Sugar beet (cv. Madison) seeds were obtained from Danisco Seed A/S, Holeby, Denmark. Seed exudate-amended DMM was made by incubating 40 g of seeds in 1 liter of DMM (without glucose). The medium contained kanamycin or ampicillin, as appropriate, and nystatin. Incubation was for 16 to 20 h at 28°C with shaking. After incubation, the exudate was filtered through a 0.2-μm-pore-size bottle top filter (Nalgene; Nunc International, Rochester, N.Y.). Glucose (0.4% final concentration) was added prior to inoculation with bacteria. Growth media were inoculated with exponentially growing cells to initial OD600s of 0.002 for growth experiments with DSS73-15C2 (amsY) and 0.1 to 0.3 for all other studies.
Recombinant DNA techniques.
Restriction digestions, ligations, agarose gel electrophoresis, and plasmid DNA isolation were performed by standard methods (47). Plasmid DNA used for sequencing and electroporation was purified with the Qiagen plasmid kit (Qiagen, Hilden, Germany). PCRs were carried out with DyNAzyme II DNA polymerase (Finnzymes Oy, Espoo, Finland) or with Deep Vent DNA polymerase (New England Biolabs, Beverly, Mass.). PCR products used for sequencing or as a probe for Southern hybridization were purified with the QIAquick PCR purification kit (Qiagen). Pseudomonas chromosomal DNA was purified with the Wizard genomic DNA purification kit (Promega, Madison, Wis.) as recommended by the manufacturer.
Plasmids pJEL5771 (9), pEMH97 (22), and pRK415 (24) were introduced into relevant Pseudomonas strains by electroporation as previously described (21). Electroporation of E. coli was carried out on cells harvested in exponential phase and washed with sterile water (2).
For transposon mutagenesis, Tn5 delivery plasmid pRL1063 (58) and helper plasmid pRK2013 (13) were transferred from E. coli DH5α into Pseudomonas sp. strain DSS73 by triparental mating as previously described (28). Transposon mutants were selected on LB agar plates containing kanamycin and ampicillin. To clone the Tn5-tagged genes in selected mutants, chromosomal DNA was cut with EcoRI, ligated, and electroporated into E. coli XL-1 Blue. The plasmids containing Tn5 and flanking Pseudomonas DNA are able to replicate in E. coli because of oriV in Tn5 (58).
Southern blots were prepared with nylon membranes (Hybond-N; Amersham, Piscataway, N.J.) in accordance with the supplier's directions. The DNA probe was a 1,634-bp PCR product amplified from pRL1063 with the primers Tn5-7451 (5′ ACC ACC TCT TTG AGT TAT CGC C 3′) and Tn5-5817 (5′ TGA AAT CGC ACC TGC CCA TC 3′) obtained from TAG Copenhagen A/S (Copenhagen, Denmark). The probe was labeled with digoxigenin-11-dUTP (Boehringer Mannheim, Mannheim, Germany). Labeling and detection were performed as recommended by the manufacturer.
DNA sequencing and sequence analysis.
Sequencing of plasmids containing Tn5 and flanking Pseudomonas chromosomal DNA was done by GATC GmbH (Constance, Germany) by primer walking. The first sequence reactions were carried out with primers recognizing the right and left ends of the transposon, respectively, hence providing information on the orientation of the insert. The sequence data were analyzed by using the University of Wisconsin Genetics Computer Group package, version 10.2 (12). BLAST searches (1) of the nonredundant database comprising GenBank coding sequence translations plus Protein Data Base plus SwissProt plus Protein Identification Resources were made courtesy of the National Center for Biotechnology Information, Bethesda, Md.
Measurements of bioluminescence.
Bioluminescence was measured with a luminometer (Bio-Orbit 1253; Struers KEBO Laboratory, Albertslund, Denmark). To eliminate effects of the metabolic state of cultured cells on bioluminescence, an assay for potential bioluminescence was used (32). In brief, culture samples were amended with 523 medium (including sodium citrate at 10 mg ml−1) (32). The maximal level of potential bioluminescence was reached immediately after exposure of the cells to this medium (data not shown). Subsequently, the substrate for luciferase (2.5 μl of a 10% [vol/vol] n-decanal solution in 96% ethanol) was added to the sample and mixed by vortexing for 15 s. Bioluminescence was measured for 3 × 10 s starting 90 s after vortexing.
Measurement of chitinase and protease activities and detection of HCN.
Chitinolytic activity was detected as clearing zones on potato dextrose agar (Becton Dickinson) plates containing dialyzed carboxymethyl chitin Remazol brilliant violet (Loewe Biochemica GmbH, Otterfing, Germany) at 1 mg ml−1 or measured in the supernatant from liquid cultures essentially as previously described (36). Protease activity was detected as clearing zones on skim milk agar plates (49). Production of cyanide was detected by inoculating bacteria on King's B agar (25) supplemented with glycine (4 g liter−1) and following the color shift from yellow to orange red of a filter paper, wetted with 0.5% picric acid (8), that was attached to the lid of the petri dish.
Isolation of a double mutant.
Spontaneous GacA/GacS mutants often accumulate during growth in rich media (14). To isolate spontaneous GacS or GacA mutants of DSS73-15C2, the strain was grown in LB at 28°C and reinoculated several times from stationary-phase cultures. Screening for proteinase-deficient mutants was performed on skim milk agar plates.
Measurements of amphisin production and purification of amphisin.
The presence of amphisin in growth media was detected by a drop collapse test as described by Bodour and Miller-Maier (4). Quantitative analyses for amphisin were carried out by high-performance liquid chromatography (HPLC) with a Hypersil base-deactivated silica C18 HPLC column described in detail elsewhere (38a). To purify amphisin, DSS73 was cultivated in DMM for 3 days at 15°C. The broth was then exhaustively extracted with ethyl acetate, which was evaporated under reduced pressure to yield a crude extract. The crude extract was subjected to solid-phase extraction (Waters Sep-Pak Vac 35 cc [10 g] C18 cartridge; 100-ml step gradient of 10:90, 90:10, and 100:0 acetonitrile/water plus 0.1% trifluoroacetic acid). To obtain pure amphisin, the second fraction was subjected to preparative HPLC (Hypersil Hyperprep C18 column [250 by 10 mm; 8-μm particle diameter]; isocratic 65:35 acetonitrile/water plus 0.1% trifluoroacetic acid; eluent flow, 6 ml per min).
Nucleic acid sequence accession numbers.
The EMBL accession numbers for the partial DNA sequence of the amsY and gacS genes of Pseudomonas sp. strain DSS73 are AJ416154 and AJ416155, respectively.
RESULTS AND DISCUSSION
Sequence analysis of amphisin-deficient mutants generated by Tn5 mutagenesis.
To identify genes required for amphisin synthesis, we generated a panel of approximately 1,300 Tn5 mutants of amphisin-producing Pseudomonas sp. strain DSS73. Five mutant strains were scored as amphisin deficient by the drop collapse test (4) and by HPLC analysis for the lipopeptide. Southern hybridization analysis revealed that each of these strains carried a single Tn5 insertion in the genome (data not shown).
For three mutants, sequence analysis of the chromosomal DNA flanking the transposon showed homologies to lipopeptide synthetases. For one selected strain, DSS73-15C2, a region of 1,770 nucleotides flanking the transposon was sequenced. The partial open reading frame encoded by this region showed the greatest homology (54.3% identity) to SyrE, which is involved in syringomycin synthesis in P. syringae (19). SyrE contains peptide synthetase modules carrying adenylation, thiolation, and condensation domains. The sequenced region corresponds to the major part of a module and contains conserved motifs characteristic of peptide synthetase adenylation and thiolation domains (Fig. 1) including the motifs involved in ATP binding and hydrolysis (16). Stachelhaus et al. (52) defined consensus sequences for substrate binding pockets of adenylation domains. The sequence obtained from DSS73-15C2 had the best match to domains binding leucine. Leucine accounts for 5 of the 11 amino acids in amphisin (Fig. 1).
FIG. 1.
(A) Comparison of conserved adenylation motifs (A3 to A10) and the conserved thiolation motif (T), as defined by Konz and Marahiel (27), for peptide synthetases (upper line) with motifs found in the syringomycin synthetase encoded by syrE (19) (middle line) and with the motifs identified in the sequenced part of the amphisin synthetase encoded by amsY (lower line). Residues in amphisin synthetase corresponding to one or both of the sequences used for comparison are in bold. (B) Comparison of one of the three leucine-specific consensus sequences found in the leucine binding domains of BacA, LicA, LchAA, LicB, LchAB, SrfAA, and SrfAB (52) with the corresponding residues in the sequenced part of amphisin synthetase.
In conclusion, the tagged gene encodes an enzyme belonging to the class of peptide synthetases that catalyze the nonribosomal synthesis of a diverse group of peptides found in both bacteria and fungi (31). We consequently refer to the tagged gene as amphisin synthetase, which is encoded by amsY.
For strain DSS73-12H8, a region of 1,128 nucleotides flanking the transposon was sequenced. The deduced amino acid sequence of 376 amino acids showed homology to several GacS proteins from Pseudomonas spp. The highest score was 90% identity to residues 11 to 386 of GacS from Pseudomonas chlororaphis (accession no. AF192795). The last mutant, DSS73-12D7, showed no homology to known sequences in the databases and was not analyzed further.
Phenotypic characterization of amphisin-deficient DSS73 mutant strains.
Strain DSS73-15C2 (amsY) did not synthesize amphisin, but its production of the extracellular products hydrogen cyanide, protease, and chitinase was unaffected compared to that in the wild-type strain (Table 2). In minimal medium, the mutant grew with the same doubling time and survived during stationary phase to the same extent as the wild type, as judged by OD600 determinations (Fig. 2A) and CFU counts (Fig. 2B).
TABLE 2.
Phenotypes of Pseudomonas sp. strain DSS73 and DSS73 mutant strains
| Strain | Amphisin production | Chitinase activity | Protease activity | Cyanide production | Growth on glycyl-l- glutamic acid |
|---|---|---|---|---|---|
| DSS73 | + | + | + | + | + |
| DSS73-15C2 (amsY) | − | + | + | + | + |
| DSS73-12H8 (gacS) | − | − | − | − | − |
| DSS73-12H8 (gacS)(pJEL5771) | + | + | +a | − | + |
| DSS73-12H8 (gacS)(pEMH97) | + | + | + | + | + |
| DSS73-MM (amsY gacS) | − | − | − | − | − |
| DSS73-MM (amsY gacS)(pJEL5771) | − | + | +a | − | + |
| DSS73-MM (amsY gacS)(pEMH97) | − | + | + | + | + |
Weak reaction.
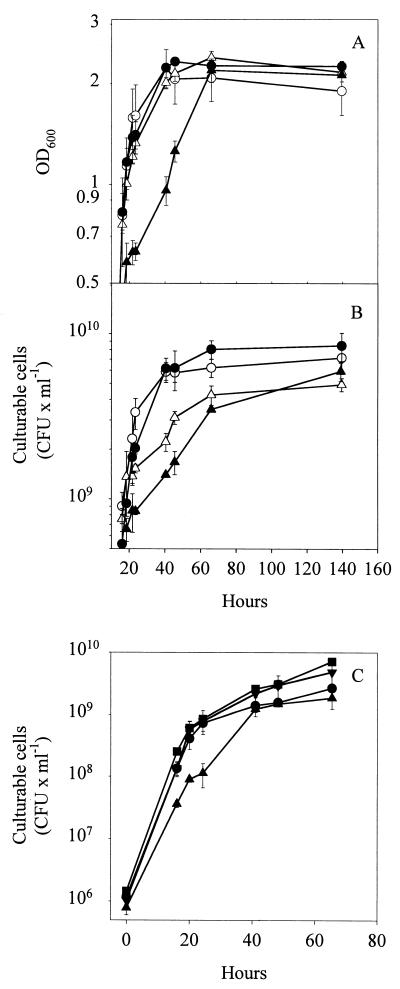
FIG. 2.
Growth of Pseudomonas sp. strain DSS73 (open symbols) and Pseudomonas sp. strain DSS73-15C2 (amsY) (closed symbols) in DMM (○, •) and in DMM amended with seed exudate (▴, ▵), measured as OD600 (A) and CFU (B). (C) Growth of Pseudomonas sp. strain DSS73-15C2 (amsY) in DMM (•), DMM amended with seed exudate (▴), DMM plus amphisin (100 μg ml−1) (▾), and DMM amended with seed exudate plus amphisin (100 μg ml−1) (▪). The data shown are mean values from a representative experiment performed in triplicate. Standard deviations are shown as bars. The experiment was independently repeated twice.
Next, we determined the growth phenotypes of DSS73 and DSS73-15C2 (amsY) in the presence of seed exudates. In minimal medium (DMM) amended with an exudate of sugar beet seeds, OD600 and CFU measurements showed that DSS73-15C2 (amsY) grew slower than the wild-type strain during the transition from the exponential to the stationary growth phase (Fig. 2A and B), although the difference was most pronounced in OD600 measurements. Furthermore, the growth effect could be complemented by addition of purified amphisin at 100 μg ml−1 to cultures of DSS73-15C2 (amsY) growing in seed exudate-amended DMM (Fig. 2C). This concentration of amphisin is comparable to that produced by DSS73 in this medium (see below).
Strain DSS73-12H8 (gacS) did not produce detectable amounts of amphisin, protease, chitinase, or hydrogen cyanide and was unable to grow on the dipeptide glycyl-l-glutamic acid as the sole carbon source (Table 2). These phenotypes have previously been reported for Pseudomonas gacS or gacA mutants (9, 14, 17, 22, 43). Next, we verified that these phenotypes could be complemented by the introduction of plasmid pEMH97, which carries the heterologous gacS (lemA) gene from P. syringae (22) (Table 2).
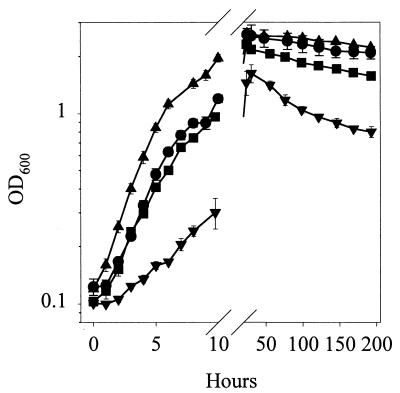
DSS73-12H8 (gacS) grew slightly faster than the wild type in the exponential phase, but their final population sizes in the early stationary phase (24 h) did not differ significantly (Fig. 3). In accordance, a previous study of the growth characteristics of gacA/gacS mutants of P. tolaasii showed a significantly higher growth rate for the gacS (pheN) mutant than for the wild type (17). Introduction of pEMH97 normalized the growth rate and caused a small reduction in the final population size in the stationary phase (Fig. 3). In contrast, introduction of pJEL5771, carrying the heterologous gacS (adpA) gene from Pseudomonas fluorescens Pf5 (9), led to dramatic decreases in both parameters. Introduction of the vector pRK415 (24), used for construction of pJEL5771, into DSS73-12H8 (gac) caused a smaller growth reduction (data not shown). Hence, the inhibition of growth is possibly due to overexpression of gacS, as a similar gene dosage effect was observed by Reimmann et al. (43) for gacA in P. aeruginosa. Survival after 1 week in the stationary phase did not differ significantly between DSS73 and DSS73-12H8 (gacS), while complementation by adpA led to faster dying off on the basis of both OD600 (Fig. 3) and CFU measurements (data not shown).
FIG. 3.
Growth of Pseudomonas sp. strains DSS73 (•), DSS73-12H8 (gacS) (▴), DSS73-12H8 (gacS)(pEMH97) (▪), and DSS73-12H8 (gacS)(pJEL5771) (▾) in DMM. The data shown are mean values from a representative experiment performed in triplicate. Standard deviations are shown as bars. The experiment was independently repeated twice.
Spontaneous mutations in the gacA/gacS system occur in Pseudomonas laboratory cultures (14, 15, 17, 44), e.g., after repeated cycling of batch cultures through the stationary phase (44). It has been speculated that this phenomenon could be due to an advantage for the mutants in the stationary phase (14, 44). Our results indicate that the selection of GacA/GacS mutants in laboratory media may be linked to rapid growth rather than to differences in stationary-phase survival.
Regulation of amsY expression and effect of seed exudate.
DSS73-15C2 (amsY) carries a transcriptional fusion between amsY and the promoterless luxAB genes, and we exploited this reporter facility to study how different environmental conditions influence expression of the tagged locus. In selected experiments, we combined measurements of bioluminescence from DSS73-15C2 (amsY) with HPLC analysis of amphisin production by DSS73.
Expression of amsY in minimal medium was highest during the transition from the exponential to the stationary phase of growth. For other Pseudomonas strains producing the lipopeptides tensin and viscosinamide, lipopeptide production has been found to be tightly coupled to cell proliferation in several growth media (38, 39). In contrast, syringomycin is primarily produced by P. syringae in the stationary phase (18). These differences in growth phase dependency of lipopeptide expression, as well as the diversity of their structures and the differences in membrane association (38, 39, 42), might well reflect a large functional diversity.
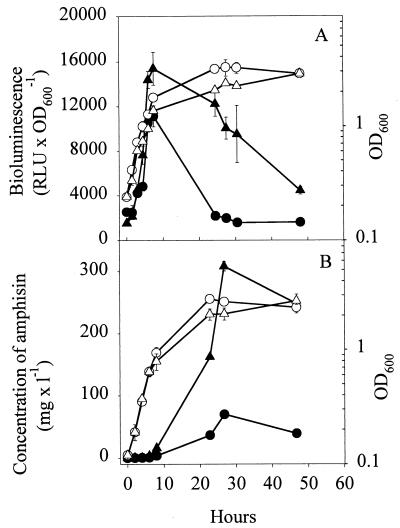
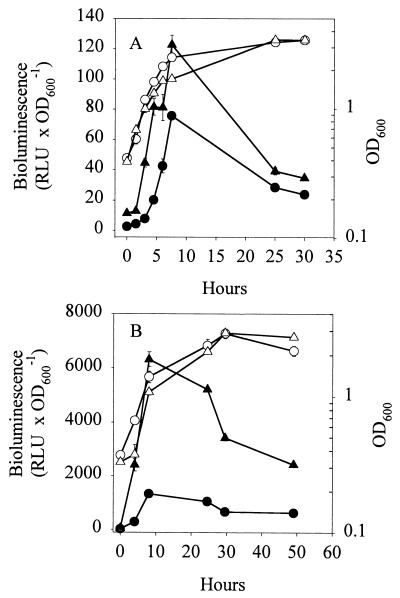
When strain DSS73-15C2 (amsY) was grown in DMM amended with seed exudate, the expression of amsY was greater than in DMM (Fig. 4A). This was in agreement with the greater production of amphisin by the wild-type strain (Fig. 4B). The largest difference in expression occurred in the early stationary phase (approximately sixfold), leading to a comparable sixfold increase in the amount of amphisin in the stationary phase.
FIG. 4.
(A) Expression of the PamsY-luxAB transcriptional fusion in Pseudomonas sp. strain DSS73-15C2 (amsY) (closed symbols) and OD600 (open symbols) of Pseudomonas sp. strain DSS73-15C2 (amsY) during growth in DMM (•, ○) and in DMM amended with sugar beet exudate (▴, ▵). RLU × OD600−1, relative light units per unit of OD600. (B) Production of amphisin by Pseudomonas sp. strain DSS73 (closed symbols) and OD (open symbols) of Pseudomonas sp. strain DSS73 during growth in DMM (•, ○) and in DMM amended with sugar beet exudate (▴, ▵). The data shown are mean values from a representative experiment performed in triplicate. Standard deviations are shown as bars. The experiment was independently repeated twice. mg × l−1, milligrams per liter.
Hence, sugar beet seed exudate had a negative influence on growth and a positive effect on amphisin production during the transition between exponential growth and the stationary phase. Compounds in the exudate might constitute a challenge for Pseudomonas by being toxic or by decreasing the biological availability of nutrients in the medium. We speculate that amphisin could play a previously unknown role in protecting the producing organism against stress.
The active compound(s) of the seed exudate could be extracted with both aqueous solutions and methanol. The compound(s) was resistant to boiling and could be removed by dialysis, indicating that small organic molecules are involved (data not shown). In most strains of P. syringae, syringomycin synthesis is influenced by the presence of the plant phenolics arbutin, salicin, and phenyl-β-d-glucoside (34, 41). However, these compounds (or Casamino Acids) were not able to induce the expression of bioluminescence in DSS73-15C2 (amsY) when added to DMM (data not shown). Consequently, lipopeptide production in pseudomonads colonizing seeds and developing roots might respond to plant compounds other than those to which the leaf-colonizing bacterium P. syringae responds.
GacS mediates induction of amphisin synthetase by seed exudate.
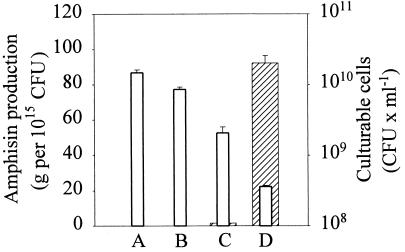
In DSS73-12H8 (gacS), amphisin production was detected neither in the absence nor in the presence of seed exudate but introduction of pEMH97 (data not shown) or pJEL5771 (Fig. 5) restored amphisin production. Furthermore, the seed exudate-inducible production of amphisin was also restored, indicating that the putative signal transmission required a functional GacS homologue (Fig. 5). To study amsY expression in a gacS mutant of strain DSS73-15C2 (amsY), we selected a spontaneous gacS mutant of this strain. The selected mutant, DSS73-MM (gacS amsY), showed phenotypes similar to those of strain DSS73-12H8 (gacS), and the relevant deficiencies were complemented by introduction of pEMH97 or pJEL5771 (Table 2). The strict dependence of amphisin production on GacS is consistent with previous data on pathogenic strains (17, 22) and point to the GacA/GacS system as a central regulator of lipopeptide synthesis in Pseudomonas.
FIG. 5.
Amphisin production, in grams per 1015 CFU (hatched bars), and CFU counts (open bars) of Pseudomonas sp. strains DSS73-12H8 (gacS) (column A) and DSS73-12H8 (gacS)(pJEL5771) (column C) grown to stationary phase in DMM and Pseudomonas sp. strains DSS73-12H8 (gacS) (column B) and DSS73-12H8 (gacS)(pJEL5771) (column D) grown to stationary phase in DMM amended with sugar beet exudate. The data shown are mean values from a representative experiment performed in triplicate. Standard deviations are shown as bars. The experiment was independently repeated twice.
The expression of bioluminescence in DSS73-MM (gacS amsY) was approximately 1,000-fold lower than in DSS73-15C2 (amsY) but could still be measured (Fig. 6A). Seed exudate doubled the bioluminescence output in early stationary-phase cultures of DSS73-MM (gacS amsY) (Fig. 6A). However, a comparable effect on three randomly selected DSS73 Tn5::luxAB mutants was observed, indicating that the effect of seed exudate on bioluminescence was not specific to the amsY locus but rather due to increased metabolic activity of the cells, which would then, in turn, stimulate the bioluminescence reaction (data not shown). Introduction of pJEL5771 or pEMH97 increased luxAB expression considerably and restored the response to seed exudate, as shown for pJEL5771 in Fig. 6B. These results strongly indicate that the observed induction of amphisin production in the presence of sugar beet seed exudate is channeled through the GacA/GacS two-component regulatory system. For P. syringae, it was found that the addition of arbutin (and fructose) restored syringomycin production to several spontaneous gacA-like mutants but not to gacA or gacS insertion mutants (44). Hence, the question of whether arbutin and related phenolic compounds (33, 34) can serve as signals for the GacA/GacS system in P. syringae has not been unambiguously answered.
FIG. 6.
(A) Expression of the PamsY-luxAB transcriptional fusion (closed symbols) and culture turbidity (OD600) (open symbols) of Pseudomonas sp. strain DSS73-MM (gacS amsY) during growth in DMM (•, ○) and in DMM amended with sugar beet exudate (▴, ▵). (B) Expression of the PamsY-luxAB transcriptional fusion (closed symbols) and culture turbidity (OD600) (open symbols) of Pseudomonas sp. strain DSS73-MM (gacS amsY)(pJEL5771) during growth in DMM (•, ○) and in DMM amended with sugar beet exudate (▴, ▵). The data shown are mean values from a representative experiment performed in triplicate. Standard deviations are shown as bars. The experiment was independently repeated twice. RLU × OD600−1, relative light units per unit of OD600.
Besides the inducibility of amphisin synthetase by compounds in the seed exudate, we found a rather high expression level during growth in minimal medium. As stated by Corbell and Loper (9), GacA/GacS-dependent phenotypes are expressed in both culture media of different composition and rhizosphere environments. This might indicate that GacS responds to multiple signals, some of which could be growth phase dependent and others of which could originate directly from the extracellular environment. Interestingly, GacS belongs to the group of unorthodox histidine kinases that possess a histidine phospho-transfer (Hpt) module and a phosphate receiver domain (45). Although the functional role of the Hpt domain remains unsolved (22), it might represent an additional regulatory checkpoint besides the phosphate transmitter domain involved in classical signal transduction (45).
More research is needed to address the significance of lipopeptide production for pseudomonads at plant-soil interfaces. One approach, as outlined in this study, is to provide more information on genetic regulatory mechanisms and environmental signals. Another is to address the importance of these compounds for the environmental fitness of pseudomonads. By combining these approaches, we hope to improve our understanding of the roles that selected lipopeptides play in the producing organism and in biocontrol.
Acknowledgments
This work was supported by the Danish Agricultural and Veterinary Research Council (grant 9702796).
We thank J. Loper for providing plasmid pJEL5771, K. Willis for providing plasmid pEMH97, and N. T. Keen for providing plasmid pRK415. May-Britt Prahm is thanked for excellent technical assistance.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1990. Current protocols in molecular biology. Greene Publishing Associates, New York, N.Y.
- 3.Bender, C. L., F. Alarcon-Chaidez, and D. C. Gross. 1999. Pseudomonas syringae phytotoxins: mode of action, regulation, and biosynthesis by peptide and polyketide synthetases. Microbiol. Mol. Biol. Rev. 63:266-292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bodour, A. A., and R. M. Miller-Maier. 1998. Application of a modified drop-collapse technique for surfactant quantitation and screening of biosurfactant-producing microorganisms. J. Microbiol. Methods 32:273-280. [Google Scholar]
- 5.Brodney, C. L., P. B. Rainey, M. Tester, and K. Johnstone. 1991. Bacterial blotch disease of the cultivated mushroom is caused by an ion channel forming lipodepsipepside toxin. Mol. Plant-Microbe Interact. 4:407-411. [Google Scholar]
- 6.Broekaert, W. F., B. P. A. Cammue, M. F. C. DeBolle, K. Thevissen, G. W. DeSamblanx, and R. W. Osborn. 1997. Antimicrobial peptides from plants. Crit. Rev. Plant Sci. 16:297-323. [Google Scholar]
- 7.Broekaert, W. F., F. R. G. Terras, B. P. A. Cammue, and R. W. Osborn. 1995. Plant defensins—novel antimicrobial peptides as components of the host-defense system. Plant Physiol. 108:1353-1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Castric, P. A. 1975. Hydrogen cyanide, a secondary metabolite of Pseudomonas aeruginosa. Can. J. Microbiol. 21:613-618. [DOI] [PubMed] [Google Scholar]
- 9.Corbell, N., and J. E. Loper. 1995. A global regulator of secondary metabolite production in Pseudomonas fluorescens Pf-5. J. Bacteriol. 177:6230-6236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Curl, E. A., and B. Truelove. 1986. The rhizosphere. Springer-Verlag, Berlin, Germany.
- 11.Desai, J. D., and I. M. Banat. 1997. Microbial production of surfactants and their commercial potential. Microbiol. Mol. Biol. Rev. 61:47-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Devereux, J., P. Haeberli, and O. Smithies. 1984. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 12:387-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ditta, G., S. Stanfield, D. Corbin, and D. R. Helinski. 1980. Broad host range DNA cloning system for gram-negative bacteria: construction of a gene bank of Rhizobium meliloti. Proc. Natl. Acad. Sci. USA 77:7347-7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Duffy, B. K., and G. Defago. 2000. Controlling instability in gacS-gacA regulatory genes during inoculant production of Pseudomonas fluorescens biocontrol strains. Appl. Environ. Microbiol. 66:3142-3150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gaffney, T. D., S. T. Lam, J. Ligon, K. Gates, A. Frazelle, J. Di Maio, S. Hill, S. Goodwin, N. Torkewitz, and A. M. Allshouse. 1994. Global regulation of expression of antifungal factors by a Pseudomonas fluorescens biological control strain. Mol. Plant-Microbe Interact. 7:455-463. [DOI] [PubMed] [Google Scholar]
- 16.Gocht, M., and M. A. Marahiel. 1994. Analysis of core sequences in the d-Phe activating domain of the multifunctional peptide synthetase TycA by site-directed mutagenesis. J. Bacteriol. 176:2654-2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grewal, S. I., B. Han, and K. Johnstone. 1995. Identification and characterization of a locus which regulates multiple functions in Pseudomonas tolaasii, the cause of brown blotch disease of Agaricus bisporus. J. Bacteriol. 177:4658-4668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gross, D. C. 1985. Regulation of syringomycin synthesis in Pseudomonas syringae pv. syringae and defined conditions for its production. J. Appl. Microbiol. 58:174.. [DOI] [PubMed] [Google Scholar]
- 19.Guenzi, E., G. Galli, I. Grgurina, D. C. Gross, and G. Grandi. 1998. Characterization of the syringomycin synthetase gene cluster. A link between prokaryotic and eukaryotic peptide synthetases. J. Biol. Chem. 273:32857-32863. [DOI] [PubMed] [Google Scholar]
- 20.Hatano, T., H. Uebayashi, H. Ito, S. Shiota, T. Tsuchiya, and T. Yoshida. 1999. Phenolic constituents of cassia seeds and antibacterial effect of some naphthalenes and anthraquinones on methicillin-resistant Staphylococcus aureus. Chem. Pharm. Bull. 47:1121-1127. [DOI] [PubMed] [Google Scholar]
- 21.Højberg, O., U. Schnider, H. V. Winteler, J. Sørensen, and D. Haas. 1999. Oxygen-sensing reporter strain of Pseudomonas fluorescens for monitoring the distribution of low-oxygen habitats in soil. Appl. Environ. Microbiol. 65:4085-4093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hrabak, E. M., and D. K. Willis. 1992. The lemA gene required for pathogenicity of Pseudomonas syringae pv. syringae on bean is a member of a family of two-component regulators. J. Bacteriol. 174:3011-3020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hutchison, M. L., M. A. Tester, and D. C. Gross. 1995. Role of biosurfactants and ion channel-forming activities of syringomycin in transmembrane ion flux—a model for the mechanism of action in the plant-pathogen interaction. Mol. Plant-Microbe Interact. 8:610-620. [DOI] [PubMed] [Google Scholar]
- 24.Keen, N. T., S. Tamaki, D. Kobayashi, and D. Trollinger. 1988. Improved broad-host-range plasmids for DNA cloning in gram-negative bacteria. Gene 70:191-197. [DOI] [PubMed] [Google Scholar]
- 25.King, E. O., M. K. Ward, and D. E. Raney. 1954. Two simple media for the demonstration of pyocyanin and fluorescein. J. Lab. Clin. Med. 44:301-307. [PubMed] [Google Scholar]
- 26.Koch, A. K., O. Kappeli, A. Fiechter, and J. Reiser. 1991. Hydrocarbon assimilation and biosurfactant production in Pseudomonas aeruginosa mutants. J. Bacteriol. 173:4212-4219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Konz, D., and M. A. Marahiel. 1999. How do peptide synthetases generate structural diversity? Chem. Biol. 6:R39-R48. [DOI] [PubMed] [Google Scholar]
- 28.Kragelund, L., B. Christoffersen, O. Nybroe, and F. J. de Bruijn. 1995. Isolation of lux reporter gene fusions in Pseudomonas fluorescens DF57 inducible by nitrogen or phosphorus starvation. FEMS Microbiol. Ecol. 17:95-106. [Google Scholar]
- 29.Lee, C. H., H. J. Kempf, Y. Lim, and Y. H. Cho. 2000. Biocontrol activity of Pseudomonas cepacia AF2001 and anthelmintic activity of its novel metabolite, cepacidine A. J. Microbiol. Biotechnol. 10:568-571. [Google Scholar]
- 30.Lindum, P. W., U. Anthoni, C. Christophersen, L. Eberl, S. Molin, and M. Givskov. 1998. N-Acyl-l-homoserine lactone autoinducers control production of an extracellular lipopeptide biosurfactant required for swarming motility of Serratia liquefaciens MG1. J. Bacteriol. 180:6384-6388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marahiel, M. A. 1992. Multidomain enzymes involved in peptide-synthesis. FEBS Lett. 307:40-43. [DOI] [PubMed] [Google Scholar]
- 32.Meikle, A., A. Glover, K. Killham, and J. I. Prosser. 1994. Potential luminescence as an indicator of activation of genetically-modified Pseudomonas fluorescens in liquid culture and in soil. Soil Biol. Biochem. 26:747-755. [Google Scholar]
- 33.Mo, Y. Y., M. Geibel, R. F. Bonsall, and D. C. Gross. 1995. Analysis of sweet cherry (Prunus avium L.) leaves for plant signal molecules that activate the syrB gene required for synthesis of the phytotoxin, syringomycin, by Pseudomonas syringae pv. syringae. Plant Physiol. 107:603-612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mo, Y.-Y., and D. C. Gross. 1991. Plant signal molecules activate the syrB gene, which is required for syringomycin production by Pseudomonas syringae pv. syringae. J. Bacteriol. 173:5784-5792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moffitt, M. C., and B. A. Neilan. 2000. The expansion of mechanistic and organismic diversity associated with non-ribosomal peptides. FEMS Microbiol. Lett. 191:159-167. [DOI] [PubMed] [Google Scholar]
- 36.Neiendam Nielsen, M., and J. Sørensen. 1999. Chitinolytic activity of Pseudomonas fluorescens isolates from barley and sugarbeet rhizosphere. FEMS Microbiol. Ecol. 30:217-227. [DOI] [PubMed] [Google Scholar]
- 37.Neu, T. R. 1996. Significance of bacterial surface-active compounds in interaction of bacteria with interfaces. Microbiol. Rev. 60:151-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nielsen, T. H., C. Christophersen, U. Anthoni, and J. Sørensen. 1999. Viscosinamide, a new cyclic depsipeptide with surfactant and antifungal properties produced by Pseudomonas fluorescens DR54. J. Appl. Microbiol. 87:80-90. [DOI] [PubMed] [Google Scholar]
- 38a.Nielsen, T. H., D. Sørensen, C. Tobiasen, J. B. Andersen, C. Christophersen, M. Givskov, and J. Sørensen. 2002. Antibiotic and biosurfactant properties of cyclic lipopeptides produced by fluorescent Pseudomonas spp. from the sugar beet rhizosphere. Appl. Environ. Microbiol. 68:3416-3423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nielsen, T. H., C. Thrane, C. Christophersen, U. Anthoni, and J. Sørensen. 2000. Structure, production characteristics and fungal antagonism of tensin—a new antifungal cyclic lipopeptide from Pseudomonas fluorescens strain 96.578. J. Appl. Microbiol. 89:992-1001. [DOI] [PubMed] [Google Scholar]
- 40.Osburn, R. M., M. N. Schroth, J. G. Hancock, and M. Hendson. 1989. Dynamics of sugar beet seed colonization by Pythium ultimum and Pseudomonas species: effects on seed rot and damping off. Phytopathology 79:709-716. [Google Scholar]
- 41.Quigley, N. B., and D. C. Gross. 1994. Syringomycin production among strains of Pseudomonas syringae pv. syringae: conservation of the syrB and syrD genes and activation of phytotoxin production by plant signal molecules. Mol. Plant-Microbe Interact. 7:78-90. [DOI] [PubMed] [Google Scholar]
- 42.Quigley, N. B., Y. Y. Mo, and D. C. Gross. 1993. SyrD is required for syringomycin production by Pseudomonas syringae pathovar syringae and is related to a family of ATP-binding secretion proteins. Mol. Microbiol. 9:787-801. [DOI] [PubMed] [Google Scholar]
- 43.Reimmann, C., M. Beyeler, A. Latifi, H. Winteler, M. Foglino, A. Lazdunski, and D. Haas. 1997. The global activator GacA of Pseudomonas aeruginosa PAO positively controls the production of the autoinducer N-butyryl-homoserine lactone and the formation of the virulence factors pyocyanin, cyanide, and lipase. Mol. Microbiol. 24:309-319. [DOI] [PubMed] [Google Scholar]
- 44.Rich, J. J., T. G. Kinscherf, T. Kitten, and D. K. Willis. 1994. Genetic evidence that the gacA gene encodes the cognate response regulator for the lemA sensor in Pseudomonas syringae. J. Bacteriol. 176:7468-7475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rodrigue, A., Y. Quentin, A. Lazdunski, V. Mejean, and M. Foglino. 2000. Two-component systems in Pseudomonas aeruginosa: why so many? Trends Microbiol. 8:498-504. [DOI] [PubMed] [Google Scholar]
- 46.Ron, E. Z., and E. Rosenberg. 2001. Natural roles of biosurfactants. Environ. Microbiol. 3:229-236. [DOI] [PubMed] [Google Scholar]
- 47.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor Laboratory, N.Y.
- 48.Scholz-Schroeder, B. K., M. L. Hutchison, I. Grgurina, and D. C. Gross. 2001. The contribution of syringopeptin and syringomycin to virulence of Pseudomonas syringae pv. syringae strain B301D on the basis of sypA and syrB1 biosynthesis mutant analysis. Mol. Plant-Microbe Interact. 14:336-348. [DOI] [PubMed] [Google Scholar]
- 49.Smibert, R. M., and N. R. Krieg. 1994. Phenotypic characterization, p. 607-654. In P. Gerhardt, R. G. E. Murray, W. A. Wood, and N. R. Krieg (ed.), Methods for general and molecular bacteriology. American Society for Microbiology, Washington, D.C.
- 50.Sørensen, D., T. H. Nielsen, C. Christophersen, J. Sørensen, and M. Gajhede. 2001. Cyclic lipoundecapeptide amphisin from Pseudomonas sp. strain DSS73. Acta Crystallogr. Sect. C Cryst. Struct. Commun. 57:1123-1124. [DOI] [PubMed] [Google Scholar]
- 51.Stachelhaus, T., and M. A. Marahiel. 1995. Modular structure of genes encoding multifunctional peptide synthetases required for non-ribosomal peptide synthesis. FEMS Microbiol. Lett. 125:3-14. [DOI] [PubMed] [Google Scholar]
- 52.Stachelhaus, T., H. D. Mootz, and M. A. Marahiel. 1999. The specificity-conferring code of adenylation domains in nonribosomal peptide synthetases. Chem. Biol. 6:493-505. [DOI] [PubMed] [Google Scholar]
- 53.Stock, J. B., A. J. Ninfa, and A. M. Stock. 1989. Protein phosphorylation and regulation of adaptive responses in bacteria. Microbiol. Rev. 53:450-490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tesaki, S., S. Tanabe, H. Ono, E. Fukushi, J. Kawabata, and M. Watanabe. 1998. 4-Hydroxy-3-nitrophenylacetic and sinapic acids as antibacterial compounds from mustard seeds. Biosci. Biotechnol. Biochem. 62:998-1000. [DOI] [PubMed] [Google Scholar]
- 55.Thrane, C., M. N. Nielsen, J. Sørensen, and S. Olsson. 2001. Pseudomonas fluorescens DR54 reduces sclerotia formation, biomass development, and disease incidence of Rhizoctonia solani causing damping-off in sugar beet. Microb. Ecol. 42:438-445. [DOI] [PubMed] [Google Scholar]
- 56.Thrane, C., T. H. Nielsen, M. N. Nielsen, J. Sørensen, and S. Olsson. 2000. Viscosinamide-producing Pseudomonas fluorescens DR54 exerts a biocontrol effect on Pythium ultimum in sugar beet rhizosphere. FEMS Microbiol. Ecol. 33:139-146. [DOI] [PubMed] [Google Scholar]
- 57.Willis, D. K., J. J. Holmstadt, and T. G. Kinscherf. 2001. Genetic evidence that loss of virulence associated with gacS or gacA mutations in Pseudomonas syringae B728a does not result from effects on alginate production. Appl. Environ. Microbiol. 67:1400-1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wolk, C. P., Y. Cai, and J.-M. Panoff. 1991. Use of a transposon with luciferase as a reporter to identify environmentally responsive genes in a cyanobacterium. Proc. Natl. Acad. Sci. USA 88:5355-5359. [DOI] [PMC free article] [PubMed] [Google Scholar]