Abstract
Biofilms were grown from wild-type (WT) Pseudomonas aeruginosa PAO1 and the cell signaling lasI mutant PAO1-JP1 under laminar and turbulent flows to investigate the relative contributions of hydrodynamics and cell signaling for biofilm formation. Various biofilm morphological parameters were quantified using Image Structure Analyzer software. Multivariate analysis demonstrated that both cell signaling and hydrodynamics significantly (P < 0.000) influenced biofilm structure. In turbulent flow, both biofilms formed streamlined patches, which in some cases developed ripple-like wave structures which flowed downstream along the surface of the flow cell. In laminar flow, both biofilms formed monolayers interspersed with small circular microcolonies. Ripple-like structures also formed in four out of six WT biofilms, although their velocity was approximately 10 times less than that of those that formed in the turbulent flow cells. The movement of biofilm cell clusters over solid surfaces may have important clinical implications for the dissemination of biofilm subject to fluid shear, such as that found in catheters. The ability of the cell signaling mutant to form biofilms in high shear flow demonstrates that signaling mechanisms are not required for the formation of strongly adhered biofilms. Similarity between biofilm morphologies in WT and mutant biofilms suggests that the dilution of signal molecules by mass transfer effects in faster flowing systems mollifies the dramatic influence of signal molecules on biofilm structure reported in previous studies.
Cells in bacterial biofilms are often less susceptible to host immune responses and antibiotics than cells grown in suspension (18). Biofilms may also provide a protective environment for pathogens, which, when released from the biofilm, may result in contamination of drinking water and medical fluids in delivery devices such as dialysis machines, venous catheters, dental water lines, and airway ventilators. Life-threatening infection caused by Pseudomonas aeruginosa in cystic fibrosis patients is a well-known example (8). Since biofilm formation in itself can be considered a virulence factor, it is important to understand the mechanisms which influence biofilm accumulation, structure, and behavior. Both hydrodynamics and cell signaling have been found to influence the structure of P. aeruginosa PAO1 biofilms. Stoodley et al. (27) reported that, under conditions of low-shear laminar flow, the biofilm consisted of a monolayer of cells with mound-shaped circular microcolonies but under high-shear, turbulent flow conditions, the biofilm formed filamentous streamers. Davies et al. (3) found that N-3-oxo-dodecanoyl homoserine lactone (OdDHL), a cell signal molecule involved in quorum sensing (QS) (reports regarding putative regulatory QS pathways and the role of QS in pathogenicity can be found elsewhere [5, 16, 20]), was required for the differentiation of biofilms into complex mushroom- and tower-like structures, which they described as characteristic of normal biofilms. However, Heydorn et al. reported in a recent study (11) that in their system, the wild-type (WT) PAO1 biofilm was relatively flat and there was no statistically significant difference in structure between the WT and the lasI JP1 mutant. Additionally, Heydorn et al. reported that the structure of laboratory-grown biofilms is often highly variable (10). A complicating factor in the role of QS in biofilm formation is the possible effect that an overlying flowing fluid will have on the concentration of signal molecules within a biofilm. QS is not solely a function of high cell density but is more directly related to high signal concentrations. In closed-system batch cultures, these tend to coincide in the stationary phase. However, in open, flowing systems in which the bulk water is continually refreshed, it is possible for diffusible signal molecules to be washed out of the biofilm. Even though mass transport studies demonstrate that diffusion is the principal transport mechanism within cell clusters (4), the removal of signal molecules from the flowing bulk fluid surrounding the cell clusters increases the concentration gradient across the biofilm-bulk liquid interface, driving the diffusive flux of signal molecules out of the biofilm. The signal concentration in the cell clusters in an open system would, therefore, be expected to depend on the production rate, the rate of diffusion through the biofilm, and the hydrodynamic conditions of the bulk liquid. An understanding of the effects of QS on biofilm structure and behavior under different flow conditions has important applications in industry and medicine.
The goal of the study was to investigate the role of QS in biofilm structure and behavior under different flow regimens. We grew biofilms from the WT P. aeruginosa PAO1 strain and the OdDHL-deficient JP1 mutant strain under conditions of either turbulent (high shear) or laminar (low shear) flow. Digital time-lapse microscopy (DTLM) was used to quantify biofilm accumulation, structural morphology, and dynamic behavior. The influence of QS and flow on biofilm structure was assessed by univariate and multivariate analysis of spatial and textural image parameters, which were quantified using Image Structure Analyzer (ISA) software developed for biofilm analysis.
(This paper is to be used as partial fulfillment for the M.S. degree in Microbiology of B.P.)
MATERIALS AND METHODS
Bacterial strains and nutrients.
Biofilms were grown from WT P. aeruginosa PAO1 (12) and JP1, a ΔlasI::tet, lasI null mutant derived from PAO1 which does not produce the QS signal OdDHL (21). Luria-Bertani (LB) broth (1/50 strength, 4 g/liter) was used as the growth medium for biofilms. Full-strength LB broth was used to grow the flask cultures (grown with 24 h of shaking) used for initial inoculation.
Biofilm reactor system.
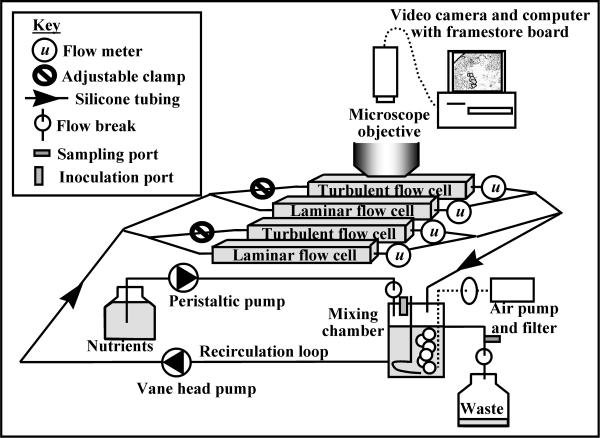
Biofilms were grown in two glass tube flow cells, 20 cm long and 3 mm in width and height (Friedrick & Dimmock, Millville, N.J.), which were incorporated in parallel positions into a recirculating continuous-culture system (Fig. 1). The flow velocity (u) was maintained at u = 0.033 m/s in one flow cell for laminar flow and at u = 1.0 m/s in the other flow cell for turbulent flow. At these flow velocities, the Reynolds (Re) numbers were 100 and 3,000, respectively. The Re number is a comparative indicator of hydrodynamic conditions in different flowing systems. Flow velocity was measured with in-line flow sensors (McMillan model 101T Flo-Sensor, serial no. 3724 and 3835; Cole-Parmer, Niles, Ill.). The flow cells were positioned in a polycarbonate holder, which was mounted on the stage of an Olympus BH2 upright microscope so that the biofilm could be imaged in situ without interrupting flow. A septum-sealed sampling port was positioned between two flow breaks in the effluent line. The system, including a mixing chamber designed for aseptic aeration and nutrient addition, is described in detail elsewhere (29). Independent triplicate experiments (runs 1, 2, and 3) were run for 6 days, each system consisting of side-by-side laminar and turbulent flow cells in duplicate. Under operating conditions, the water temperature in the reactor system was 23°C, and all experiments were performed at this temperature.
FIG. 1.
Schematic representing the main components of the flow system and orientation of flow cells with respect to the microscopic objective.
Reactor sterilization.
The reactor system, except the thermally sensitive flow sensors, was autoclaved at 121°C for 15 min. The sensors were sterilized with 70% ethanol for 15 min, NaOCl solution for 15 min, and 70% ethanol for 30 min (29). The sterility of the reactor system was confirmed by plating 0.1-ml aliquots of effluent onto LB agar (LA).
Inoculum and media.
The reaction mixture, containing 1/50-strength LB broth, was inoculated with 1 ml of an overnight LB broth (20 g/liter; 37°C) shake flask culture of PAO1 and JP1. The reactor was initially run as a recirculating batch culture for 24 h, to allow attachment, before being switched to continuous culture mode. The system was switched to continuous culture mode by delivering 1/50-strength LB broth to the mixing chamber via peristaltic pump (serial no. 7553-80; Cole-Parmer). The influent flow rate was maintained at 4.3 ml/min, giving a dilution rate of 0.025 h−1 (hydraulic residence time = 40 min). This rate was above that of washout (the growth rates of PAO1 and JP1 on LB broth were 0.15 ± 0.01 h−1 (n = 3) and 0.10 ± 0.02 h−1 (n = 3), respectively) to minimize suspended growth and encourage biofilm growth. Effluent samples were taken periodically to monitor the detached population and to confirm culture purity. The JP1 effluent was plated on both LA and LA with tetracycline (50 μg/ml) to confirm the culture purity and integrity of the mutant. Daily comparisons showed no significant difference (P > 0.05).
Biofilm cell concentration.
At the end of each experiment, the flow cells were aseptically separated from the system and 2-cm-thick sections were cut (using a diamond knife) from the inlet, middle, and outlet of the flow cells. The sections of glass tubes were sonicated for 5 min and vortexed in test tubes with 5 ml of Ringer's solution to remove biofilm cells (29). This procedure was repeated three times. A serial dilution was prepared, and six 10-μl aliquots were plated onto LA and LA plates with tetracycline (50 μg/ml). The plates were incubated at 37°C for 24 h.
Microscopy.
The developing biofilm was visualized in situ by using transmitted light and 5×, 10×, and 50× objective lenses with an Olympus BH2 microscope. Images were collected using a COHU 4612-5000 charge-coupled device camera (Cohu, Inc., San Diego, Calif.) and captured with a VG-5 PCI framestone board (Scion Inc., Frederick, Md.). The Scion image software was used to collect time-lapse sequences and for image enhancement and analysis. A 1-mm-long graticule with 10-μm divisions was used to calibrate length measurements. For the ripple dimensions, the length was defined as the longer dimension, running perpendicularly to the flow, and the width was defined as the shorter dimension, running parallel to the flow. On days 5 and 6, the distance traveled by individual ripples over a 16-h monitoring period was also measured, at hourly intervals. Linear regression was used to calculate the average ripple travel velocity over this time period. The biofilm thickness and surface area coverage were measured on each day at five random locations in the biofilm area for each flow cell (29).
ISA.
The ISA software package, which was developed by the Biofilm Structure and Function Research Group at the Center for Biofilm Engineering (www.erc.montana.edu/CBEssentials-SW/research/ImageStructureAnalyzer/default.htm), was used to quantify nine spatial and textural parameters from individual biofilm images for statistical comparison (32). The ISA software was operated in a MATLAB 6.1 program (The MathWorks). Low-power images, taken with a 10× lens objective, were used to quantify the larger-scale biofilm structures and patterns, which could not be seen at higher power magnifications. The calculated biofilm cell cluster dimensions were horizontal run length (average length of the cell clusters, which in our setup were parallel with flow direction), vertical run length (average length of the cell clusters perpendicular to flow), average diffusion distance (average distance from the cells in the cluster to the nearest interstitial space, similar to an equivalent radius), and maximum diffusion distance (maximum distance from the interior of the cluster to the edge). The fractal dimension (a measure of the roughness of the biofilm cell clusters) and porosity (the proportion of void areas) were also quantified. These parameters were calculated from automatically thresholded binary images to remove subjectivity from the analysis (33). ISA also calculates three textural parameters from the gray scale images which describe microscale heterogeneities in the image: textural entropy (a measure of randomness between pixels in the biofilm image), angular second moment (a measure of directional repeating patterns of pixels), and inverse difference moment (a measure of spatially repeating patterns of pixels).
Statistical analysis.
Statistical comparisons of thickness and manually measured parameters from Scion Image were analyzed by analysis of variance (ANOVA) using Minitab software (version 13.3; Minitab, Inc., State College, Pa.). Data were reported as means ± 1 standard error. Univariate ANOVA and multivariate ANOVA (MANOVA) were used to statistically compare quantified parameters from ISA. Data from biofilm images taken on days 4 and 5 were pooled to increase statistical rigor. A two-factor (P. aeruginosa strain and flow rate) additive ANOVA and MANOVA calculation was performed on the nine-variable matrix. Differences were considered significant for P values of <0.05.
RESULTS
Biofilm development, morphology, and behavior. (i) Biofilms grown under laminar flow.
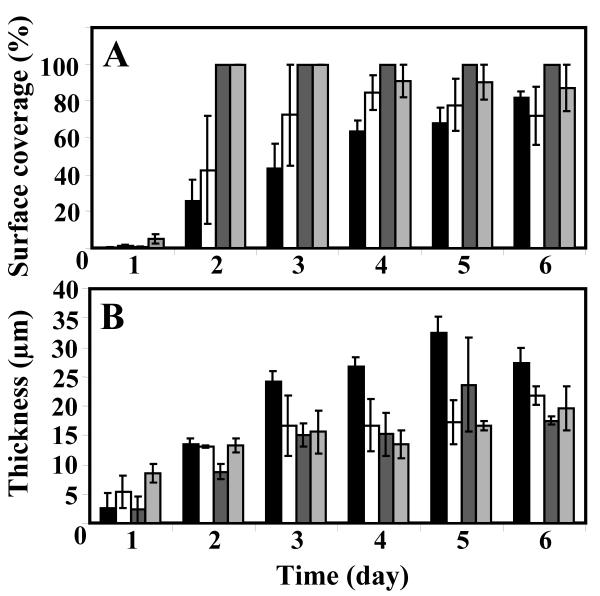
Within 24 h of the inoculation period, the biofilms for both the PAO1 and JP1 strains consisted of a sparse layer of cells (Fig. 2). By day 3, circular microcolonies approximately 15 μm thick had developed in both biofilms. By this time, there was a monolayer of cells between the colonies such that the surface area coverage had reached 100% in both biofilms (Fig. 3). The thickness and surface area coverage did not change significantly (P > 0.05), and at the end of the run (day 6), the PAO1 and JP1 biofilms were 17.5 ± 0.7 μm and 19.6 ± 3.7 μm thick, respectively. The average surface coverage of the PAO1 biofilm had decreased slightly to 87.3 ± 12.7%, whereas the JP1 biofilm remained at 100 ± 0%. There was no significant difference between the measurements of daily thickness (all P values of >0.06) and surface area coverage (all P values of >0.19) of the PAO1 and JP1 biofilms on any of the 6 days. In runs 1 and 2, the PAO1 biofilms formed highly organized ripple-like structures (regularly spaced ridges running perpendicularly to the flow direction) (Fig. 4, panels A to C). The ripples were only evident when the biofilm was viewed under low-power magnification (10×) and were not apparent when viewed with higher (50×) magnification objectives (Fig. 4D and F). Although ripple motion was not apparent in real time, DTLM showed that the ripples were traveling at a constant downstream velocity of 0.51 ± 0.06 μm/h (n = 17) (Fig. 5). It appeared that the individual ripples moved over an underlying layer of bacterial cells that were more firmly attached to the surface. Ripple structures were not seen in any of the JP1 biofilms.
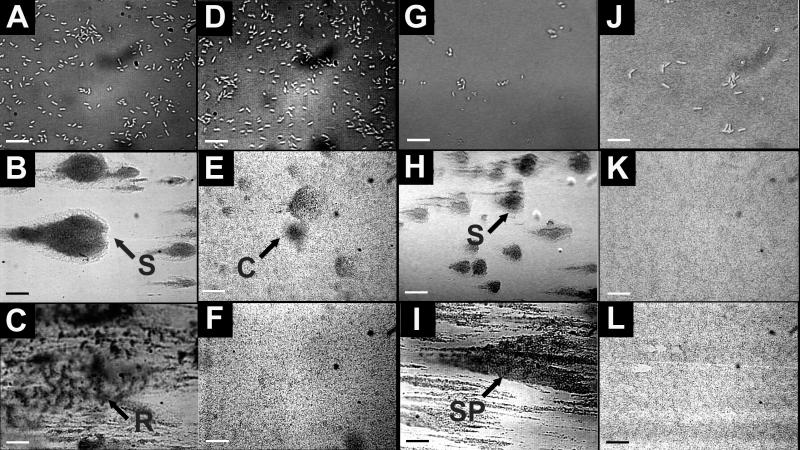
FIG. 2.
P. aeruginosa PAO1 and JP1 biofilms grown in turbulent and laminar flow cells. PAO1 biofilm in turbulent (A to C) and laminar (G to I) flow cells and JP1 biofilm in turbulent (D to F) and laminar (J to L) flow cells on days 1 (top row), 3 (middle row), and 6 (bottom row) (scale bars = 20 μm, 100 μm, and 100 μm, respectively) are shown. By day 3 in turbulent flow, both the PAO1 and JP1 strains had formed similar streamlined patchy biofilms. In laminar flow, both PAO1 and JP1 formed a flat monolayer of cells with occasional circular colonies. C, circular colony; R, ripple structure; S, streamers; SP, streamlined patches. The flow direction is from right to left on each panel.
FIG. 3.
Biofilm development and accumulation measured by surface area coverage (A) and thickness (B) over the course of the experiments. JP1 in turbulent conditions (solid bars), PAO1 in turbulent conditions (open bars), JP1 in laminar conditions (dark gray bars), and PAO1 in laminar conditions (light gray bars) are shown. Error bars represent 1 standard error; n = 15.
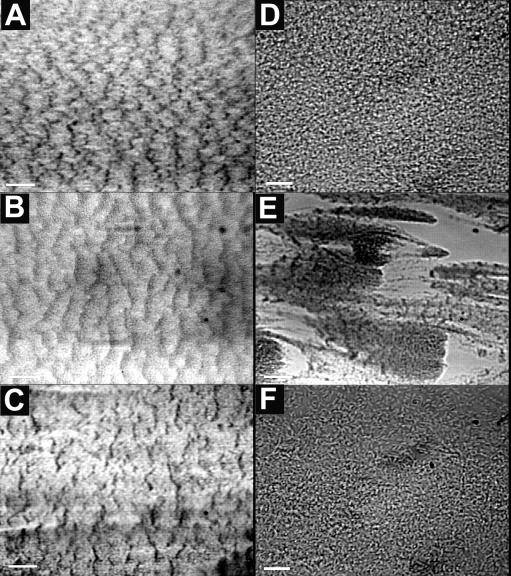
FIG. 4.
Ripple structures formed in PAO1 and JP1 biofilms. Images of PAO1 biofilm ripple structures in the biofilms growing in the turbulent flow cell (A) and the laminar flow cell (B), taken at days 4 and 5, respectively, are shown. The ripples were aligned perpendicularly to the flow direction (right to left). Scale bar, 200 μm. (C) JP1 biofilm ripple structures in the turbulent flow cell taken on day 6. Scale bar, 200 μm. The ripple structures were much less evident under higher magnification (the images in panels D and F are of the same fields as those in panels A and C, respectively). Scale bar, 20 μm. (E) Patchy PAO1 biofilm structures in the turbulent flow cell, run 3, day 5. Scale bar, 100 μm.
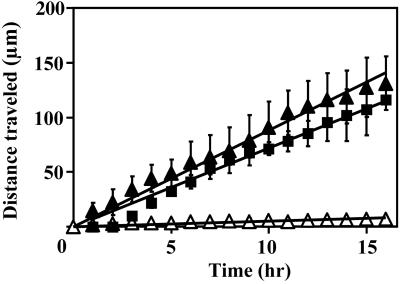
FIG. 5.
Downstream transport of PAO1 ripples that formed in laminar (open triangles) and turbulent (solid triangles) flow and in the turbulent JP1 biofilm (solid squares) over a 16-h monitoring period. Measurements were made on day 5 of run 1 for turbulent biofilms and day 6 of run 1 for the laminar biofilm. The positions of five different ripples were used to calculate the mean distance traveled during each hour. Error bars represent 1 standard deviation. The solid linear regression curves were used to determine the average ripple travel velocity.
(ii) Biofilms grown under turbulent flow.
The mean surface coverage of both PAO1 and JP1 biofilms steadily increased over the growth period and by day 6 reached 82.1 ± 3.4% and 72.2 ± 1.4%, respectively. The JP1 biofilm was generally thicker than the PAO1 biofilm and after 6 days was 27.2 ± 2.6 μm thick compared to 21.7 ± 15.8 μm. However, in similarity to the biofilms grown in laminar flow, there were no statistical differences between the daily thickness (all P values of >0.05) and surface coverage (all P values of >0.05) measurements over the course of the experiment. Morphologically, both biofilms formed large streamlined patches and filamentous streamers (Fig. 2). Between the streamers, the substratum was covered with single cells, whose arrangement ranged from a sparse covering to a confluent monolayer. In some areas, the thicker patches of biofilms appeared to have joined to form a continuous layer. All three of the PAO1 biofilms and two of the JP1 biofilms also formed ripple structures similar to those seen in the PAO1 biofilms grown in laminar flow. Between days 3 and 5 of growth, the ripples formed both in the streamlined patches and in the more extensively covered areas. These ripples had higher contrast, making them easier to visualize than those that developed in laminar flow. However, the ripple dimensions in the PAO1 biofilms were not significantly different from those that formed in laminar flow (length, P = 0.45; width, P = 0.79; and spacing, P = 0.35). The ripples that formed in the JP1 turbulent biofilm were smaller than those in the PAO1 biofilm (all length, width, and spacing P values, <0.001). DTLM from run 1 on day 5 showed that the ripples traveled downstream at an average velocity of 8.7 ± 1.6 μm/h (n = 17; r2= 0.97) in the PAO1 biofilm and 7.2 μm/h (n = 17; r2 = 0.98) in the JP1 biofilm (Fig. 5). In run 3, the PAO1 biofilm ripples were traveling at an average velocity of 14 μm/h (n = 15; day 6). These velocities were more than 10 times those measured in the PAO1 biofilms grown in laminar flow conditions (0.51 μm/h, r2= 0.61, n = 17).
Video time lapse movies showing the biofilm ripple structures flowing downstream are available at the following website: www.erc.montana.edu/Res-Lib99-SW/Movies/default.htm.
ISA and statistical analysis.
ISA results and univariate ANOVA results from ISA are shown in Table 1. The five dimensional parameters showed that the cell clusters in the biofilms grown under turbulent flow were larger than those grown under laminar flow. These differences were highly significant between the different flows for all of the parameters (P < 0.002), while between the WT and mutant biofilms, three of the five dimensional parameters were significantly different. There was a significant difference between bacterial strains in two of the three textural parameters and between the different flow rates in one of the three textural parameters. MANOVA showed that both strain and flow had a significant influence on the ISA parameters (both P values, <0.001), but there was not a significant univariate interaction between flow and strain (P = 0.091). Recalculation using an additive model with 1 degree of freedom also yielded P values of <0.001 for both flow and strain.
TABLE 1.
Quantified parameters from ISA analysis of low-power images taken of P. aeruginosa WT (PAO1) and mutant (JP1) biofilms grown under laminar (L) and turbulent (T) flow conditions on days 4 and 5
| Parametera | Strainc
|
ANOVA (P value)b
|
||||
|---|---|---|---|---|---|---|
| PAO1L (n = 20) | PAO1T (n = 28) | JP1L (n = 12) | JP1T (n = 41) | Strain | Flow | |
| Porosity | 0.62 ± 0.12 | 0.60 ± 0.09 | 0.66 ± 0.07 | 0.62 ± 0.08 | 0.219 | 0.152 |
| Average diffusion distance (μm) | 6.49 ± 6.53 | 18.71 ± 18.90 | 2.38 ± 0.39 | 9.99 ± 5.30 | 0.003d | 0.000d |
| Maximum diffusion distance (μm) | 64.94 ± 65.71 | 80.00 ± 55.35 | 58.46 ± 36.60 | 77.57 ± 39.43 | 0.016d | 0.002d |
| Horizontal run length (μm) | 10.75 ± 8.06 | 17.29 ± 10.16 | 15.48 ± 9.12 | 20.93 ± 8.33 | 0.178 | 0.000d |
| Vertical run length (μm) | 8.47 ± 6.05 | 11.19 ± 7.28 | 10.32 ± 7.33 | 12.27 ± 4.60 | 0.029d | 0.000d |
| Fractal dimension | 1.53 ± 0.12 | 1.37 ± 0.11 | 1.56 ± 0.08 | 1.34 ± 0.12 | 0.536 | 0.000d |
| Angular second moment (× 10−3) | 4.26 ± 1.37 | 3.93 ± 1.13 | 2.83 ± 1.19 | 0.99 ± 1.23 | 0.032d | 0.105 |
| Textural entropy | 8.31 ± 0.35 | 8.40 ± 0.27 | 8.96 ± 0.51 | 8.47 ± 0.61 | 0.015d | 0.128 |
| Inverse difference moment | 0.12 ± 0.03 | 0.16 ± 0.03 | 0.10 ± 0.03 | 0.17 ± 0.06 | 0.796 | 0.000d |
See Materials and Methods for explanations of parameters.
The ANOVA columns show P values from univariate analysis for comparisons between P. aeruginosa strain and flow regimen.
n, number of samples.
Statistically significant difference (P < 0.05).
Biofilm cell concentration.
The average biofilm cell concentration in the three PAO1 runs was similar (P = 0.28) in both turbulent and laminar flow cells (1.2 ± 0.1 × 107 CFU/cm2 [n = 54] and 8.1 ± 0.7 × 106 CFU/cm2 [n = 61], respectively). In the JP1 biofilms, there was also no significant difference (P = 0.79) between cell concentration in the turbulent and laminar flow cells (2.1 ± 0.9 × 106 and 1.7 ± 0.6 × 106 CFU/cm2, respectively). Differences were not significant between the PAO1 and JP1 biofilms grown in turbulent flow (P = 0.09) but were significant between the PAO1 and JP1 biofilms grown in laminar flow (P = 0.01).
Effluent cell concentration.
After 3 days, the average effluent concentration in the three runs in the PAO1 and JP1 biofilms increased to 8.9 × 107 ± 3.4 × 107 CFU/ml and 1.6 × 108 ± 2.4 × 108 CFU/ml, respectively. Over the remaining 3 days of the experiment, these concentrations varied no more than 1 order of magnitude.
DISCUSSION
Biofilm structure.
There was little difference in the accumulation rates of the WT and QS mutant biofilms grown under either laminar or turbulent conditions, as estimated by thickness, surface area coverage, or viable biofilm cell concentration values. After 6 days, the thickness of all the biofilms had stabilized at approximately 20 μm and the surface area coverage was over 80%. We noted few visual differences between the WT and mutant biofilms, in contrast to more obvious differences between biofilms grown under the two flow regimens. Biofilms grown under laminar flow consisted predominantly of a thin layer of cells interspersed with distinct circular microcolonies. In some cases in the WT biofilm, ripple-like ridges running perpendicularly to the flow appeared in the biofilm. The ripples were only apparent at lower magnification. Similar structures have been reported in mixed-species laboratory biofilms grown in turbulent flow (28), and in river water biofilms (17). The ripples consisted of densely packed bacteria. In turbulent flow, both WT and mutant biofilms developed streamlined patches, which were tapered in the downstream direction. Ripple structures formed in each of six flow cells with WT biofilms but in only four out of six of the mutant biofilms, demonstrating the inherent variability commonly encountered in flow cell studies. Manual measurement of ripple dimensions revealed that the WT ripples were significantly larger that the ripples formed in the QS mutant (P < 0.05). Ripples also formed in four out of six of the WT biofilms grown under laminar flow but in none of the mutant biofilms. It is possible that this variability may be explained by subtle differences in the composition and cohesiveness of the extrapolysaccharide slime matrix formed by the different strains under the two different growth conditions.
Structural analysis of biofilm images and statistical comparisons.
The ISA image analysis package was used to nonsubjectively quantify various spatial and textural parameters of the biofilm images, allowing us to statistically assess the influence of cell signaling and flow on biofilm structure. As noted by Heydorn et al. (10, 11), the inherent heterogeneity common in biofilm structure makes qualitative comparisons difficult. To overcome this difficulty, the Danish group developed the software package COMSTAT, which is based on single-cell resolution in a three-dimensional volume collected by confocal microscopy. ISA quantifies two-dimensional grayscale images, and although ISA does not incorporate three-dimensional information, it is useful for quantifying the shapes and dimensions of biofilm microcolonies and patterns which are visible at the larger scales (i.e., at the millimeter level). It can be used with images collected by conventional bright-field or fluorescence-based microscopy. Although visually it was difficult to distinguish structural differences between the WT and QS mutant biofilms in comparison to the more obvious differences due to flow regimens, MANOVA of the ISA data revealed significant structural differences due to both strain and flow. While the parameters quantifying size dimensions were easily interpretable, textural information was also useful for the statistical comparison of data sets, even though physical interpretation of the data was less obvious.
OdDHL is not required for the formation of strongly attached, cohesive biofilms.
Although none of our biofilms developed the distinct mushroom structures which formed in the PAO1 biofilms in the study of Davies et al. (3), both the WT and mutant strains did form significant biofilms in both laminar and turbulent flow. Unlike the results of the Davies study, which suggested that, in the absence of OdDHL, there was no true biofilm formation but only a loose accretion of cells, in our flow system, which was operated at much higher shear stresses, the biofilms must have been strongly adhered and cohesive to remain attached. Our data suggest that cell signaling is not required for biofilm formation but possibly plays a role in the structural heterogeneity of the biofilm. What is becoming apparent is that biofilm structure is highly sensitive to growth conditions. In the present study, both the WT and mutant biofilms grown in laminar flow were more similar to the flat WT biofilms described by Hentzer et al. (9) and Heydorn et al. (10, 11), and interestingly, to the cell signaling JP1 mutant biofilm in the study of Davies et al. than were those grown in turbulent flow conditions. Henzter et al. (9) attributed the structural differences between the PAO1 WT biofilm in their study and the WT biofilm in the Davies study to differences in the growth medium used. In the present study, we used the same nutrients (2% LB broth) as Hentzer et al. (9), which may explain why our biofilms were also relatively flat. These data challenge the ever-growing acceptance of a generalized, normal biofilm structure as consisting of mushrooms and towers.
Hydrodynamic effects on mass transfer, cell signaling, and biofilm structure.
Hydrodynamic conditions can also strongly influence biofilm structure. In a previous study of the influence of cell signaling and hydrodynamics on the structure of P. aeruginosa biofilms, Stoodley et al. found that hydrodynamic conditions had a greater influence than null mutations on structure in the cell signaling regulators lasR and rhlIR (27). In the study of Davies et al. (3), the biofilms were grown at a Re of 0.17. The calculated wall shear stress value, however, was comparable to that calculated for our laminar flow cell (approximately 0.1 Nm−2), suggesting that differences in observed WT biofilm structure were more likely related to rates of mass transfer than to shear-related detachment (which may be expected to result in flatter, rather than mushroom-shaped, biofilms). It is possible that in our flow system, the higher rates of mass transfer, even in our laminar flow cell, had a dilution effect on signaling molecules. In the absence of QS-inducing concentrations, it may be expected that there would be little difference between the WT and QS mutant biofilms. In addition to influencing structure, hydrodynamic conditions also influence biofilm density (31) and strength (1, 26), which in turn may be expected to influence the diffusion of nutrients and signals through the biofilm.
Movement of biofilm ripples over solid surfaces.
DTLM revealed that the ripples traveled downstream along the channel walls of the glass. This movement was not noticeable in real time. Although there were no significant differences in the morphologies of the ripples grown in the two flow regimens, the velocity of the ripples in the turbulent flow cell was 8.7 μm/h, approximately 10 times faster than that of those that formed under laminar flow. Although these velocities appear low, they may represent a large downstream flux of microorganisms. For example, if half of the approximately 107 CFU of biofilm cells/cm2 were traveling in ripples at 8.7 μm/h, the downstream flux (which is dependent on the tube geometry) would be approximately 5 × 103 CFU cm−1 h−1. The flux in the laminar flow cell would be 10-fold less. With the exception of detachment events, biofilms are often depicted as immobilized layers in which the cell clusters remain at the same location on the substratum (2, 19). The present study demonstrates that biofilms can move along solid surfaces while remaining attached to those surfaces. The only previous reports of traveling ripples in bacterial communities have been for the myxobacteria (23, 24). However, we believe that there are some fundamental differences in the mechanisms of migration described in those reports from those reported here. For Myxococcus xanthus, a species of gliding bacteria, ripples are formed because of the coordinated motility of individual cells, which is controlled by signaling molecules (23). These ripples are not flow dependent and can occur on solid surfaces in quiescent air or water. In our biofilms, the migration velocity and ripple structure varied as a function of the fluid shear stress (28), suggesting that this behavior was a physical phenomenon. The apparent flow of biofilms may be related to the fluid-like properties reported for mixed- and pure-culture PAO1 biofilms (29) and to the hydrogel nature of the polymer matrix (7, 30). Nevertheless, shear-mediated migration of biofilms represents a previously unrecognized mechanism for dissemination in flowing systems and may have important consequences for contamination and infection in industrial or clinical environments.
Flowing biofilms have been implicated in ventilator-associated cases of pneumonia (13). In that clinical study, 23 of 50 tracheal tubes were found to contain biofilms with what were termed “wave-like” patterns. This led Inglis et al. (13, 14) to infer that the biofilm had been flowing along the tube and that this flow may be related to dissemination into the lungs. Our studies directly demonstrated that this phenomenon is possible. Unlike dissemination via detached planktonic cells, surface-associated transport allows the spread of entire biofilm structures, presumably with preserved resistance to various antibiotics and chemical disinfectants (25). The flow of biofilms along pipe walls may also be an important consideration in infection from venous catheters (6), dental unit water lines (22), or dialysis machines (15). Further work is required to determine the significance of surface-associated biofilm transport in the dissemination of microbial pathogens in both clinical and industrial settings.
Concluding remarks.
In agreement with previous studies (11, 27), our results suggest that QS alone is not necessarily required for biofilm formation and that other factors of the growth environment, such as nutrients and hydrodynamic conditions, can play a role of equal if not greater significance in determining the biofilm structure. The relative contribution of each of these interlinked factors under different growth conditions has yet to be established. However, until a connection is made that relates the formation of specific biofilm structures to biofilm virulence in both clinical and industrial contexts, interference in cell signaling pathways may not be the magic bullet for biofilm control it was initially thought to be.
Acknowledgments
This work was funded by the National Institutes of Health RO1 grant GM60052-02 and in part by cooperative agreement EEC-8907039 between the National Science Foundation and Montana State University—Bozeman and the W. M. Keck Foundation.
From Montana State University, we thank Michael Franklin for experimental discussion, Marty Hamilton and Margo Schurman for statistical analysis, Haluk Beyenal and Zbigniew Lewandowski for providing ISA, and Suzanne Wilson for experimental assistance.
REFERENCES
- 1.Beyenal, H., and Z. Lewandowski. 2002. Internal and external mass transfer in biofilms grown at various flow velocities. Biotechnol. Prog. 18:55-61. [DOI] [PubMed] [Google Scholar]
- 2.Costerton, J. W., P. S. Stewart, and E. P. Greenberg. 1999. Bacterial biofilms: a common cause of persistent infections. Science 284:1318-1322. [DOI] [PubMed] [Google Scholar]
- 3.Davies, D. G., M. R. Parsek, J. P. Pearson, B. H. Iglewski, J. W. Costerton, and E. P. Greenberg. 1998. The involvement of cell-to-cell signals in the development of a bacterial biofilm. Science 280:295-298. [DOI] [PubMed] [Google Scholar]
- 4.deBeer, D., and P. Stoodley. 1995. Relation between the structure of an aerobic biofilm and mass transport phenomena. Water Sci. Technol. 32:11-18. [Google Scholar]
- 5.deKievit, T. R., and B. H. Iglewski. 2000. Bacterial quorum sensing in pathogenic relationships. Infect. Immun. 68:4839-4849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Donlan, R. M., R. Murga, M. Bell, C. M. Toscano, J. H. Carr, T. J. Novicki, C. Zuckerman, L. C. Corey, and J. M. Miller. 2001. Protocol for detection of biofilms on needleless connectors attached to central venous catheters. J. Clin. Microbiol. 39:750-753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Flemming, H.-C., J. Wingender, C. Mayer, V. Körstgens, and W. Borchard. 2000. Cohesiveness in biofilm matrix polymers, p. 87-105. In D. Allison, P. Gilbert, H. M. Lappin-Scott, and M. Wilson (ed.), Community structure and cooperation in biofilms. SGM Symposium Series 59. Cambridge University Press, Cambridge, United Kingdom.
- 8.Govan, J. R., and V. Deretic. 1996. Microbial pathogenesis in cystic fibrosis: mucoid Pseudomonas aeruginosa and Burkholderia cepacia. Microbiol. Rev. 60:539-574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hentzer, M., G. M. Tietzel, G. J. Balzer, A. Heydorn, S. Molin, M. Givskov, and M. R. Parsek. 2001. Alginate overproduction affects Pseudomonas aeruginosa biofilm structure and function. J. Bacteriol. 183:5395-5401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Heydorn, A., A. T. Nielsen, M. Hentzer, C. Sternberg, M. Givskov, B. K. Ersbøll, and S. Molin. 2000. Quantification of biofilm structures by the novel computer program COMSTAT. Microbiology 146:2395-2407. [DOI] [PubMed] [Google Scholar]
- 11.Heydorn, A., B. K. Ersbøll, J. Kato, M. Hentzer, M. R. Parsek, A. T. Nielsen, M. Givskov, and S. Molin. 2002. Statistical analysis of Pseudomonas aeruginosa biofilm development: impact of mutations in genes involved in twitching motility, cell-to-cell signaling, and stationary-phase sigma factor expression. Appl. Environ. Microbiol. 68:2008-2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Holloway, B. W., V. Krishnapillai, and A. F. Morgan. 1979. Chromosomal genetics of Pseudomonas. Microbiol. Rev. 43:73-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Inglis, T. J. J. 1993. Evidence for dynamic phenomena in residual tracheal tube biofilm. Br. J. Anaesth. 70:22-24. [DOI] [PubMed] [Google Scholar]
- 14.Inglis, T. J. J., M. R. Millar, J. G. Jones, and D. A. Robinson. 1989. Tracheal tube biofilm as a source of bacterial colonization of the lung. J. Clin. Microbiol. 27:2014-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Man, N. K., A. Degremont, J. C. Darbord, M. Collet, and P. Vaillant. 1998. Evidence of bacterial biofilm in tubing from hydraulic pathway of hemodialysis system. Artif. Organs 22:596-600. [DOI] [PubMed] [Google Scholar]
- 16.Miller, M. B., and B. L. Bassler. 2001. Quorum sensing in bacteria. Annu. Rev. Microbiol. 55:165-199. [DOI] [PubMed] [Google Scholar]
- 17.Neu, R. T., and J. R. Lawrence. 1997. Development and structure of microbial biofilms in river water studied by confocal laser scanning microscopy. FEMS Microbiol. Ecol. 24:11-25. [Google Scholar]
- 18.Nickel, J. C., I. Ruseska, J. B. Wright, and J. W. Costerton. 1985. Tobramycin resistance of Pseudomonas aeruginosa cells growing as a biofilm on urinary catheter material. Antimicrob. Agents Chemother. 27:619-624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Palmer, R. J., and D. C. White. 1997. Developmental biology of biofilms: implications for treatment and control. Trends Microbiol. 435:435-440. [DOI] [PubMed] [Google Scholar]
- 20.Passador, L., J. M. Cook, M. J. Gambello, L. Rust, and B. H. Iglewski. 1993. Expression of Pseudomonas aeruginosa virulence genes requires cell-to-cell communication. Science 260:1127-1130. [DOI] [PubMed] [Google Scholar]
- 21.Pearson, J. P., E. C. Pesci, and B. H. Iglewski. 1997. Roles of Pseudomonas aeruginosa las and rhl quorum-sensing systems in control of elastase and rhamnolipid biosynthesis genes. J. Bacteriol. 179:5756-5767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Putnins, E. E., D. Di Giovanni, and A. S. Bhullar. 2001. Dental unit waterline contamination and its possible implications during periodontal surgery. J. Periodontol. 72:393-400. [DOI] [PubMed] [Google Scholar]
- 23.Sager, B., and D. Kaiser. 1994. Intercellular C-signaling and the traveling waves of Myxococcus. Genes Dev. 8:2793-2804. [DOI] [PubMed] [Google Scholar]
- 24.Shimkets, L. J., and D. Kaiser. 1982. Induction of coordinated movement of Myxococcus xanthus cells. J. Bacteriol. 152:451-461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stewart, P. S., and J. W. Costerton. 2001. Antibiotic resistance of bacteria in biofilms. Lancet 358:135-138. [DOI] [PubMed] [Google Scholar]
- 26.Stoodley, P., A. Jacobsen, B. C. Dunsmore, B. Purevdorj, S. Wilson, H. M. Lappin-Scott, and J. W. Costerton. 2001. The influence of fluid shear and AlCl3 on the material properties of Pseudomonas aeruginosa PAO1 and Desulfovibrio sp. EX265 biofilms. Water Sci. Technol. 43:113-120. [PubMed] [Google Scholar]
- 27.Stoodley, P., F. Jorgensen, P. Williams, and H. M. Lappin-Scott. 1999. The role of hydrodynamics and ahl signaling molecules as determinants of structure of Pseudomonas aeruginosa biofilms, p. 323-330. In R. Bayston et al. (ed.), Biofilms: the good, the bad, and the ugly. BioLine Press, Cardiff, United Kingdom.
- 28.Stoodley, P., Z. Lewandowski, J. D. Boyle, and H. M. Lappin-Scott. 1999. The formation of migratory ripples in a mixed species bacterial biofilm growing in turbulent flow. Environ. Microbiol. 1:447-455. [DOI] [PubMed] [Google Scholar]
- 29.Stoodley, P., Z. Lewandowski, J. D. Boyle, and H. M. Lappin-Scott. 1999. Structural deformation of bacterial biofilms caused by short-term fluctuations in fluid-shear: an in situ investigation of biofilm rheology. Biotechnol. Bioeng. 65:83-93. [PubMed] [Google Scholar]
- 30.Sutherland, I. 2001. Biofilm exopolysaccharides: a strong and sticky framework. Microbiology 147:3-9. [DOI] [PubMed] [Google Scholar]
- 31.Vieira, M. J., L. F. Melo, and M. M. Pinheiro. 1993. Biofilm formation: hydrodynamic effects on internal diffusion and structure. Biofouling 7:67. [Google Scholar]
- 32.Yang, X., H. Beyenal, G. Harkin, and Z. Lewandowski. 2000. Quantifying biofilm structure using image analysis. J. Microbiol. Methods 39:109-119. [DOI] [PubMed] [Google Scholar]
- 33.Yang, X., H. Beyenal, G. Harkin, and Z. Lewandowski. 2001. Evaluation of biofilm image thresholding methods. Water Res. 35:1149-1158. [DOI] [PubMed] [Google Scholar]