Abstract
Sphingomonas paucimobilis SYK-6 degrades a lignin-related biphenyl compound, 5,5′-dehydrodivanillate (DDVA), to 5-carboxyvanillate (5CVA) by the enzyme reactions catalyzed by the DDVA O-demethylase (LigX), the ring cleavage oxygenase (LigZ), and the meta-cleavage compound hydrolase (LigY). In this study we examined the degradation step of 5CVA. 5CVA was transformed to vanillate, O-demethylated, and further degraded via the protocatechuate 4,5-cleavage pathway by this strain. A cosmid clone which conferred the 5CVA degradation activity to a host strain was isolated. In the 7.0-kb EcoRI fragment of the cosmid we found a 1,002-bp open reading frame responsible for the conversion of 5CVA to vanillate, and we designated it ligW. The gene product of ligW (LigW) catalyzed the decarboxylation of 5CVA to produce vanillate along with the specific incorporation of deuterium from deuterium oxide, indicating that LigW is a nonoxidative decarboxylase of 5CVA. LigW did not require any metal ions or cofactors for its activity. The decarboxylase activity was specific to 5CVA. Inhibition experiments with 5CVA analogs suggested that two carboxyl groups oriented meta to each other in 5CVA are important to the substrate recognition by LigW. Gene walking analysis indicated that the ligW gene was located on the 18-kb DNA region with other DDVA catabolic genes, including ligZ, ligY, and ligX.
Lignin is the most abundant natural aromatic compound, and the biodegradation of lignin is an important process in the carbon cycle. Biochemical and genetic analyses of the enzymes involved in the lignin biotransformation process would supply the strategies for the effective production of valuable products from lignin. It consists of phenylpropane units with a variety of linkages (8, 31). Biphenyl linkage is one of the key connections between phenylpropane units. The biphenyl structure is so stable that its decomposition seems to be the rate-limiting step in lignin degradation (26, 27).
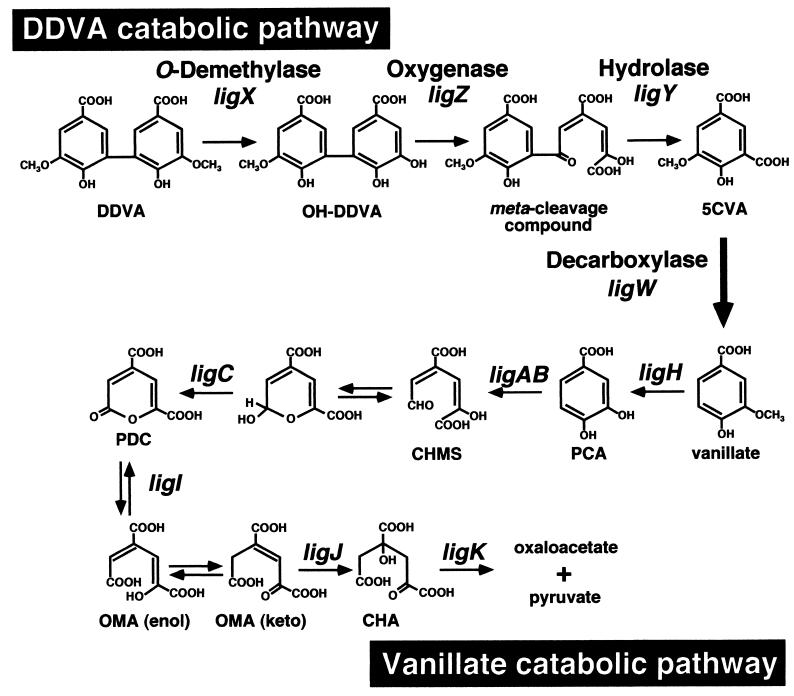
Sphingomonas paucimobilis SYK-6 has been isolated with a lignin-related biphenyl compound, 5,5′-dehydrodivanillate (DDVA), as the sole carbon and energy source (13, 19). This strain could also grow on vanillate, syringate, ferulate, and other dimeric lignin compounds, including β-aryl ether, diarylpropane, and phenylcoumarane. In SYK-6, the β-aryl ether having a guaiacyl moiety was shown to be degraded via vanillate as an intermediate (20). It was indicated that vanillate is initially converted to protocatechuate (PCA) by O-demethylation in which a gene, ligH, is involved (23) (Fig. 1). PCA is further decomposed to oxaloacetate and pyruvate through the PCA 4,5-cleavage pathway (10, 21, 22, 24, 34).
FIG. 1.
Proposed degradation pathway for DDVA in S. paucimobilis SYK-6. DDVA is transformed to 5CVA and is estimated to be decarboxylated to vanillate on the basis of results described in this study. Vanillate is converted to PCA by a specific O-demethylase and is further degraded through the PCA 4,5-cleavage pathway. Compound abbreviations: CHMS, 4-carboxy-2-hydroxymuconate-6-semialdehyde; PDC, 2-pyrone-4,6-dicarboxylate; OMA, 4-oxalomesaconate; CHA, 4-carboxy-4-hydroxy-2-oxoadipate. The following genes were used: ligX, encoding a component of DDVA O-demethylase; ligZ, encoding OH-DDVA meta-cleavage oxygenase; ligY, encoding OH-DDVA meta-cleavage compound hydrolase; ligW, encoding 5CVA decarboxylase; ligH, encoding a protein involved in the O-demethylation of vanillate and syringate; ligAB, encoding small and large subunits of PCA 4,5-dioxygenase; ligC, encoding CHMS dehydrogenase; ligI, encoding PDC hydrolase; ligJ, encoding OMA hydratase; ligK, encoding CHA aldolase.
In the case of DDVA degradation by SYK-6 (Fig. 1), DDVA is initially converted to a diol compound, 2,2′,3-trihydroxy-3′-methoxy-5,5′-dicarboxybiphenyl (OH-DDVA), by a multicomponent DDVA O-demethylase in which a gene, ligX, is involved (33). The ring fission of OH-DDVA to produce a meta-cleavage compound is catalyzed by OH-DDVA oxygenase, which is encoded by ligZ (26). The resulting meta-cleavage compound is hydrolyzed to form 5-carboxyvanillate (5CVA) by ligY-encoded hydrolase (27).
In this study we focused on the degradation pathway of 5CVA. In a previous study, 3-O-methylgallate (3MGA) was suggested to be an intermediate metabolite of 5CVA (12). Here we provide evidence that 5CVA is transformed not to 3MGA but to vanillate by SYK-6. We then characterized the gene for a specific decarboxylase involved in the 5CVA transformation.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The bacterial strains and plasmids used in this study are listed in Table 1. S. paucimobilis SYK-6, a bacterium that can grow on DDVA as the sole carbon and energy source, was isolated from a kraft pulp effluent (13). S. paucimobilis AKB is an SYK-6 mutant of which ligB encoding the β-subunit of PCA 4,5-dioxygenase was inactivated by the insertion of a kanamycin resistance gene (H. Aoshima, E. Masai, S. Nishikawa, Y. Katayama, and M. Fukuda, Abstr. 8th Int. Symp. Microb. Ecol. [ISME-8], abstr. 93, 1998). Pseudomonas putida PpY101 (9) was used as a host strain for the cloning of the 5CVA decarboxylase gene and for the preparation of 5CVA. A broad-host-range cosmid vector, pVK100, was used to construct the gene library of SYK-6 (6). Helper plasmid pRK2013 (7) was used to transfer the gene library from Escherichia coli HB101 to P. putida PpY101 by triparental mating.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s) | Reference or source |
|---|---|---|
| Strains | ||
| S. paucimobilis | ||
| SYK-6 | Wild type; Nalr Smr | 13 |
| AKB | Mutant derivative of SYK-6; ligB::Kmr; Nalr Smr Kmr | H. Aoshima et al., ISME-8, 1998 |
| P. putida PpY101 | Nalr Smr | 9 |
| E. coli JM109 | recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 Δ(lac-proAB) F′[traD36 proAB+lacIqlacZΔM15] | 36 |
| E. coli HB101 | supE44 hsdS20(rB− mB−) recA13 ara-14 proA2 lacY1 galK2 rpsL20 xyl-5 mtl-1 | 3 |
| Plasmids | ||
| pVK100 | Broad-host-range cosmid vector, Kmr Tetr | 6 |
| pRK2013 | Kmr Tra+ Mob+, ColE1 replicon | 7 |
| pKT230 | Broad-host-range vector; Kmr | 2 |
| pBluescript II KS(+) | Cloning vector; ColE1 replicon; Apr | 32 |
| pUC18 and pUC19 | Cloning vectors; ColE1 replicon; Apr | 36 |
| pCVA | pVK100 with an approx 20-kb DNA fragment carrying ligW | This study |
| pVK7E2 | pVK100 with a 7.0-kb EcoRI fragment of pCVA | This study |
| pKS7E2 | KS(+) with a 7.0-kb EcoRI fragment of pVK7E2 | This study |
| pKS7E1 | KS(+) carrying the same fragment as pKS7E2 but in the opposite direction | This study |
| pKV6, pKV9, pKV21 | Deletion derivatives of pKS7E2 | This study |
| p1N2 | KS(+) with a 3.3-kb NotI-EcoRI fragment of pKS7E2 | This study |
| pAH34, pAH47 | Deletion derivatives of p1N2 | This study |
| p1SC2 | KS(+) with a 2.4-kb SacI-EcoRI fragment of pKS7E2 | This study |
| pTE01 | pUC19 with a 15-kb EcoRI fragment carrying ligZ and ligY | 26 |
| pLE6 | pKT230 with a 6.0-kb EcoRI fragment carrying ligX | 33 |
| pHN139F | pUC18 with a 10.5-kb EcoRI fragment carrying ligJAB, a part of ligC, and ligI | 22 |
| pBE10 | pVK100 with an approx 24-kb DNA fragment carrying ligDFE | 17 |
Synthesis of model lignin compounds.
The method for the preparation of DDVA was described in a previous study (26). To synthesize 5CVA, vanillin (Tokyo Kasei Kogyo Co.) was used as an initial material to produce 5-formylvanillin by the method of Profft and Krause (28). P. putida PpY101, which has the ability to convert 5-formylvanillin to 5CVA, was used for the preparation of 5CVA. P. putida PpY101 was grown to the stationary phase at 30°C in Luria-Bertani (LB) medium. Cells were centrifuged at 4,500 × g for 10 min at 4°C, and a cell pellet was washed in fresh 1/10 LB medium (Bacto-tryptone, 1.0 g/liter; yeast extract, 0.5 g/liter; NaCl, 5 g/liter) and suspended in the same medium to an optical density at 600 nm (OD600) of 2.0. A 20% solution of 5-formylvanillin dissolved in dimethyl sulfoxide was added into the above medium to a final concentration of 0.02%. After incubation with shaking for 48 h at 30°C, the culture was centrifuged at 4,500 × g and the supernatant was collected. To obtain 5CVA, the supernatant was acidified with hydrochloric acid and was extracted with ethyl acetate. The organic phase was dried in vacuo and was derivatized with the trimethylsilyl (TMS) reagent. The resultant sample was confirmed by gas chromatography-mass spectrometry (GC-MS) analysis. The mass spectrum of the TMS derivative of the prepared 5CVA had a molecular ion at m/z 428 and major fragment ions at m/z 413 and 251. This molecular size was in good agreement with that described in a previous report (12).
Isophthalate, 4-hydroxyisophthalate, 3-methoxylsalicylate, and deuterium oxide (D2O; 99.75%) were purchased from Wako Pure Chemical Industries (Osaka, Japan).
Analysis of metabolites.
S. paucimobilis SYK-6 and the ligB mutant (AKB) were grown to an OD600 of 1.0 in LB medium at 30°C. Cells were washed twice with a mineral salts medium (W medium) (26) and were suspended to an OD600 of 0.2 in 10 ml of the same medium. After the addition of the substrates (DDVA or 5CVA) to a final concentration of 0.2%, the mixtures were shaken at 30°C. A 200-μl portion of the cultures was collected from 0 to 48 h at intervals of 4 h and was acidified with hydrochloric acid to pH 1. Finally, metabolites were extracted with 200 μl of ethyl acetate and were dried in vacuo. GC-MS analysis of the TMS derivatives of the metabolite was carried out.
GC-MS analysis.
GC-MS analysis was carried out by using a model 5971A (Hewlett-Packard Co., Palo Alto, Calif.) with an Ultra-2 capillary column (50 m by 0.2 mm; Hewlett-Packard Co.). The column temperature was increased initially from 100 to 150°C and then from 150 to 300°C at rates of 20 and 3°C per min, respectively. Temperatures of injection and detection were 220 and 300°C, respectively.
Cloning procedure.
A gene library of SYK-6 was constructed in E. coli HB101 by inserting the partial SalI digests of the total DNA into pVK100 and was introduced into P. putida PpY101 by triparental mating. P. putida PpY101 was not able to degrade 5CVA but could grow on vanillate. The transconjugants were grown in 3 ml of W medium containing nalidixic acid (25 mg/liter), kanamycin (50 mg/liter), methionine (40 mg/liter), 0.2% vanillate, and 0.002% 5CVA at 30°C for 10 days. Each 200 μl of culture was acidified with 10 μl of 6 N hydrochloric acid and was extracted with 60 μl of ethyl acetate. The resultant samples were analyzed on thin-layer chromatography (TLC) by using silica gel 60 F254 (E. Merck, Darmstadt, Germany). The developing solvent was chloroform-ethyl acetate-formic acid (10:8:2 [vol/vol/vol]). Compounds were visualized under UV light at 254 nm. 5CVA has an Rf value of 0.52 in this system.
DNA manipulations and nucleotide sequencing.
DNA manipulations were carried out essentially as described elsewhere (1, 29). A Kilosequence kit (Takara Shuzo Co., Ltd., Kyoto, Japan) was used to construct a series of deletion derivatives. The nucleotide sequences were determined by the dideoxy termination method with an ALFexpress DNA sequencer (Pharmacia Biotech, Milwaukee, Wis.). A Sanger reaction was carried out with the Thermosequenase fluorescent-labeled primer cycle sequencing kit with 7-deaza-dGTP (Amersham Pharmacia Biotech, Little Chalfont, United Kingdom) (30). Sequencing analysis and homology alignment were carried out with the GeneWorks programs (IntelliGenetics, Inc., Mountain View, Calif.). The DDBJ databases were used for searching homologous proteins. Southern hybridization analysis of SYK-6 was performed with the DIG System (Boehringer Mannheim Biochemicals, Indianapolis, Ind.) according to the procedure recommended by the manufacturer.
Enzyme purification and determination of N-terminal amino acid sequence.
Cells were grown in 200 ml of LB medium containing 100 mg of ampicillin/liter. Expression of the 5CVA decarboxylase gene was induced for 4 h by the addition of isopropyl-β-d-thiogalactopyranoside (IPTG) to a final concentration of 1 mM when the OD600 of the culture reached 0.5. Cells were harvested by centrifugation and were ruptured by a passage though a French pressure cell in 20 mM Tris-HCl (pH 7.5). The cell lysate was centrifuged at 15,000 × g, and the supernatant was collected. Streptomycin was added to a final concentration of 1% to the supernatant, and the resultant supernatant was incubated on ice for 10 min and centrifuged at 15,000 × g for 15 min to remove nucleic acids. The supernatant was then centrifuged at 100,000 × g for 60 min at 4°C, and the cell extract was obtained after concentration by ultrafiltration with a YM-10 membrane (Amicon, Beverly, Mass.). The protein concentration was measured by the method of Bradford (3a), with bovine serum albumin as the standard. Enzyme purification was performed according to the method described below by using a BioCAD700E apparatus (PerSeptive Biosystems, Framingham, Mass.). The cell extract was applied to a POROS PI (polyethyleneimine) column (4.6 by 100 mm) (PerSeptive Biosystems) previously equilibrated with 50 mM Tris-HCl (pH 8.0). The enzyme was eluted with 25 ml of a linear gradient of 0 to 0.5 M NaCl. The 5CVA decarboxylase was eluted at approximately 0.33 M.
To determine the N-terminal amino acid sequence, the partially purified enzyme was subjected to sodium dodecyl sulfate-12% polyacrylamide gel electrophoresis (SDS-PAGE) (14) and electroblotted onto a polyvinylidene difluoride membrane (Bio-Rad, Hercules, Calif.). The enzyme band was cut out and analyzed on a PPSQ-21 protein sequencer (Shimadzu Co., Kyoto, Japan).
Enzyme assays.
The decarboxylase activity with 5CVA was spectrophotometrically determined by measuring the decrease in the absorbance at 312 nm with a DU-7500 spectrophotometer (Beckman, Fullerton, Calif.). The reaction was initiated by the addition of 5CVA to a final concentration of 80 μM to 1 ml of 20 mM Tris-HCl buffer (pH 7.5) containing the cell extract (1 mg of protein) at 25°C. To determine the molar extinction coefficient of 5CVA and vanillate, each compound was added at various concentrations to 1 ml of 20 mM Tris-HCl buffer (pH 7.5). After measuring the absorption at 312 nm under 25°C, the molar extinction coefficients of 5CVA and vanillate were calculated to be 32,000 and 110 M−1 cm−1, respectively.
To examine the metal ion dependency, 100 μM EDTA or 100 μM metal salt solution containing either MgSO4 · 7H2O, Na2SO4 · 10H2O, FeSO4 · 7H2O, CuSO4 · 5H2O, CaSO4 · 2H2O, CoSO4 · 7H2O, ZnSO4 · 7H2O, or MnSO4 · 4H2O was added to 1 ml of 20 mM Tris-HCl (pH 7.5) containing the cell extract (1 mg of protein). After incubation for 30 min on ice, 5CVA was added to a final concentration of 80 μM to initiate the reaction, and the enzymatic activity was determined as described above. To examine the dependency of cofactors, either NAD+, NADH, NADP+, or NADPH was added to a final concentration of 120 μM to a 1-ml reaction mixture, and the enzyme activity was measured.
Inhibition of 5CVA decarboxylase activity by the substrate analogs was assessed by high-performance liquid chromatography (HPLC) analysis, because there is no significant difference between the absorption maxima of 5CVA and substrate analogs. Each substrate analog and 5CVA were added at a final concentration of 160 and 80 μM, respectively, to 1 ml of reaction mixture containing the cell extract (0.5 mg of protein) and were incubated for 30 s at 25°C. The reaction was stopped by thermal inactivation of the enzyme at 80°C for 3 min. The reaction mixture was centrifuged to remove the denatured protein, and the resultant supernatant was subjected to HPLC analysis.
HPLC analysis.
HPLC (LC Module I model; Waters, Randolph, Mass.) with a reverse-phase TSKgel ODS-80TM column (6 by 150 mm; Tosoh, Tokyo, Japan) was used in this study. The mobile phase used in this study consisted of 1% phosphoric acid and 20% acetonitrile in water. The flow rate was 1.0 ml/min. The absorbance of eluent was monitored at 312 nm.
Conversion of 5CVA to vanillate by LigW in D2O.
5CVA (0.4 mM) and LigW protein (10 μg) were incubated in the reaction mixture containing 0, 48, and 96% D2O, respectively. After incubation for 30 min at 25°C, the reaction product was extracted with ethyl acetate and analyzed by GC-MS as described above.
PFGE.
Total DNA prepared in an agarose block was digested with SpeI and was separated on a CHEF DRIII apparatus (Bio-Rad) with 1% low-melting-point agarose in Tris-borate-EDTA. Pulsed-field gel electrophoresis (PFGE) was run with a 50-s pulse for 23 h. The 1-Mb SpeI fragment was cut out from the gel and further digested with AseI or AflII. The digests were separated on the same apparatus with a 1% agarose gel. PFGE was run with a 20- to 30-s pulse for 24 h. A separation with a 1- to 6-s pulse for 10 h was carried out after PFGE with the conditions described above for AseI digests. Southern hybridization analysis was done with the digoxigenin-labeled plasmids pCVA, pTE01, pLE6, pHN139F, and pBE10 (Table 1), carrying ligW, ligZ and ligY, ligX, ligI and ligJABC, and ligDFE, respectively.
Nucleotide sequence accession number.
The nucleotide sequence of ligW has been deposited in the DDBJ, EMBL, and GenBank sequence databases under accession no. AB033664.
RESULTS
5CVA is converted into vanillate by S. paucimobilis SYK-6.
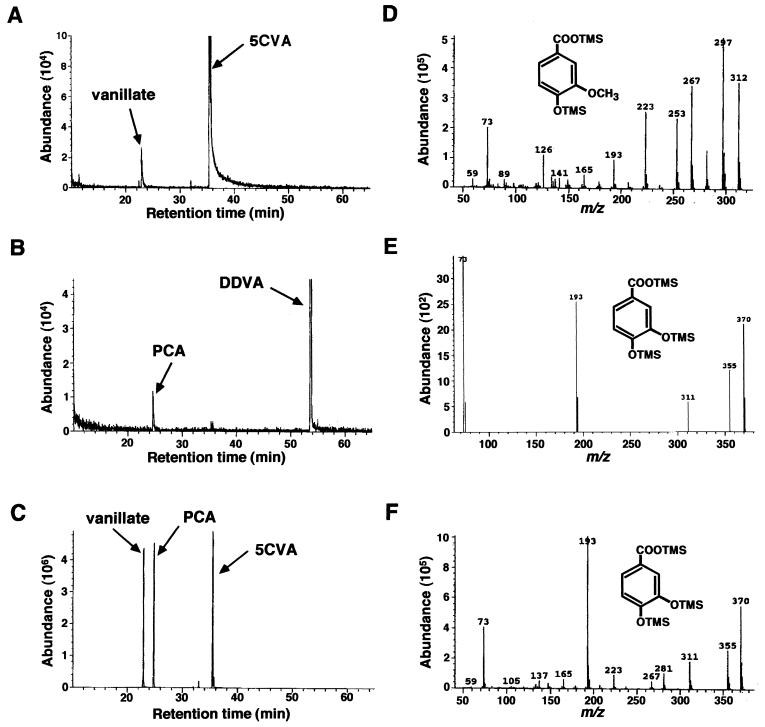
S. paucimobilis SYK-6 was cultivated in LB medium and suspended to an OD600 of 0.2 in W medium containing 0.2% 5CVA and was incubated with shaking at 30°C. After 32 h of incubation the culture was acidified and extracted with ethyl acetate. The TMS derivatives of the metabolites were analyzed by GC-MS. The production of a compound with a retention time at 22.9 min was observed (Fig. 2A). Its retention time and mass spectrum corresponded to those of authentic vanillate (Fig. 2D). Thus, 5CVA was thought to be degraded via vanillate as an intermediate. However, in a previous study resting cells of SYK-6 produced 3MGA from 5CVA (12). To address this discrepancy a ligB insertion mutant of S. paucimobilis SYK-6 was tested for growth with 5CVA. The ligB gene encodes the β subunit of PCA 4,5-dioxygenase (LigAB), which is required for catabolism of PCA (24), and it was previously shown that the 3MGA catabolic pathway in S. paucimobilis does not require LigAB (H. Aoshima et al., ISME-8, 1998). The ligB mutant did not grow with 5CVA, suggesting that 5CVA is catabolized to vanillate and then to PCA rather than being catabolized to 3MGA. After 32 h of incubation the TMS derivative of the metabolites from DDVA and 5CVA by the ligB mutant were analyzed by GC-MS. A metabolite with a retention time at 24.8 min was observed from both substrates (Fig. 2B and C), and it was determined to be PCA by comparison with the retention time and mass spectrum of authentic PCA (Fig. 2E and F). Vanillate was also detected in 5CVA degradation by the ligB mutant (Fig. 2C). On the basis of these results we concluded that 5CVA is metabolized to PCA via vanillate in SYK-6 and that a decarboxylase seems to be involved in the conversion of 5CVA to vanillate. 3MGA detected during the 5CVA degradation in our previous study is thought not to be a true metabolite of 5CVA; it might be generated in a nonphysiological condition, such as a resting-cell assay.
FIG. 2.
Identification of the degradation products of 5CVA by S. paucimobilis SYK-6 and the ligB mutant. Gas chromatograms of the TMS derivative of the metabolites produced from 5CVA degraded by SYK-6 (A), from DDVA degraded by the ligB mutant (B), and from 5CVA degraded by the ligB mutant (C). Mass spectra of the TMS derivative of metabolites appeared at the retention time of 22.9 min for panel A (D), 24.8 min for panel B (E), and 24.8 min for panel C (F).
Cloning and sequencing of the 5CVA decarboxylase gene.
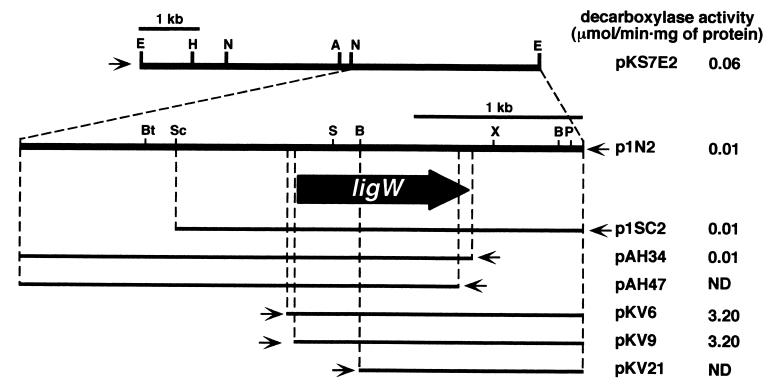
To isolate the gene responsible for the decarboxylation of 5CVA to vanillate, P. putida PpY101, which is not able to degrade 5CVA, was employed as a host strain. A gene library of SYK-6 was constructed in E. coli with a cosmid vector, pVK100, and the partially digested SalI fragments of total SYK-6 DNA were introduced into P. putida PpY101 by triparental mating. Approximately 3,300 transconjugants were tested by TLC for their ability to convert 5CVA into vanillate. A cosmid from a positive transconjugant that had an approximately 20-kb insert was isolated and designated pCVA. A subcloning experiment showed that the 7.0-kb EcoRI fragment of pCVA was required to confer the 5CVA conversion activity upon P. putida PpY101. This fragment was subcloned into pBluescript II KS(+), and the resultant plasmid was designated pKS7E2 (Fig. 3). A cell extract of E. coli JM109 containing pKS7E2 grown with IPTG had decarboxylase activity when 5CVA was added to the reaction mixture. This result suggested the existence of the 5CVA decarboxylase gene in this fragment. A series of subclones of the 7.0-kb EcoRI fragment was constructed by using restriction enzymes and E. coli exonuclease III, and these subclones were partially sequenced. To determine the localization of the 5CVA decarboxylase gene, the 5CVA decarboxylase activity of the cell extract of E. coli JM109 carrying each of these subclones was assayed by measuring the decrease in absorbance at 312 nm. The activity was not observed in E. coli carrying deletion plasmids pKV21 and pAH47. The activity in E. coli harboring pKV9 was approximately 300 times higher than that in the transformant containing pAH34, whose DNA insert was in the opposite orientation to that in pKV9. These results suggested that the decarboxylase gene was contained in the DNA shared by pKV9 and pAH34 and that the transcription of this gene was driven by the lac promoter in pKV9. The open reading frame that was predicted from these results to encode a 5CVA decarboxylase was designated ligW (Fig. 3). A putative ribosome binding site of AGGAGAGAG was located 6 bp upstream from the initiation codon of ligW. The G+C content of the ligW gene was 65% and was consistent with that of the other lignin degradation genes of SYK-6. The molecular mass of the ligW product (LigW) calculated from the deduced amino acid sequence was 36,920 Da. A BLAST search with the DDBJ database revealed that the deduced amino acid sequence of ligW has 31% identity over 276 amino acids that overlap with those of the 2-amino-3-carboxymuconate-6-semialdehyde decarboxylase gene with accession no. AB069781, which was recently isolated from Rattus norvegicus (35).
FIG. 3.
Deletion analysis of the 7.0-kb EcoRI fragment. The direction of transcription from the lac promoter in a vector is depicted by a thin arrow. The 5CVA decarboxylase activity of E. coli containing each plasmid is presented on the right. Abbreviations: A, ApaI; B, BamHI; Bt, BstXI; E, EcoRI; H, HindIII; N, NotI; P, PstI; S, SalI; Sc, SacI; X, XhoI. ND, not detected.
Expression of ligW in E. coli.
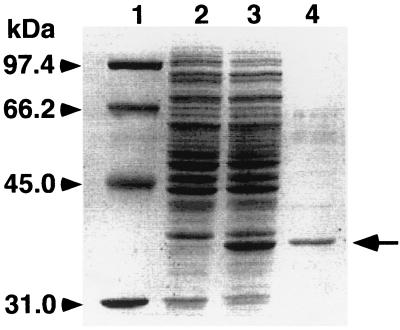
The ligW gene expression induced by IPTG in E. coli JM109 was examined with the plasmid pKV9. A 37-kDa protein, whose size was in good agreement with the value calculated from the deduced amino acid sequence of ligW (36,920 Da), was observed by SDS-PAGE (Fig. 4). After partial purification of this protein its N-terminal amino acid sequence was determined to be MRLIATEEAVTFQPVV, which corresponds to the deduced N-terminal amino acid sequence of ligW.
FIG. 4.
Expression of ligW in E. coli JM109 demonstrated on SDS-PAGE. Lane 1, molecular size markers; lane 2, cell extract of E. coli harboring pBluescript II KS(+); lane 3, cell extract of E. coli harboring pKV9; lane 4, an active fraction separated by a POROS PI column.
Conversion of 5CVA into vanillate in D2O by LigW.
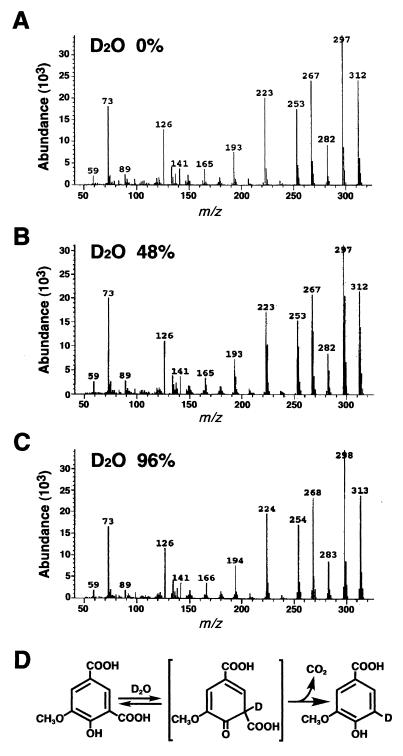
To confirm the reaction mechanism of LigW as a nonoxidative decarboxylase, the incorporation of a hydrogen atom into 5CVA was examined by using D2O, as demonstrated by Huang et al. (11). 5CVA was incubated for 30 min with the cell extract of E. coli JM109 harboring pKV9 in the presence of 0, 48, or 96% D2O. The reaction product, vanillate, was extracted and subjected to GC-MS analysis. Mass spectral profiles of unlabeled vanillate were compared to those of vanillate generated in the presence of 48 and 96% D2O (Fig. 5). Unlabeled vanillate showed an intense molecular ion at m/z 312 and major ion fragments at m/z 297 (M-CH3), 282 (M-2CH3), and 267 (M-3CH3). The relative abundance of ions at m/z 313, 298, 283, and 268 increased in proportion to the concentration of D2O in the reaction mixture. The mass spectrum for 96% D2O indicates that a deuterium derivative accounted for most of the vanillate produced. These results suggested that the 5-carboxyl group in 5CVA is replaced by a proton originating from H2O.
FIG. 5.
Conversion of 5CVA into vanillate by LigW in the presence of D2O. Mass spectra of vanillate produced in the presence of 0, 48, and 96% D2O are shown in panels A, B, and C, respectively. The deduced reaction pathway of LigW is illustrated in panel D on the basis of the result shown in this figure and that reported for vanillate decarboxylase of Rhodotorula rubra (11).
Substrate specificity of LigW.
The decarboxylation activity of LigW toward various 5CVA analogs, such as 3-methoxysalicylate (3MSA), 4-hydroxyisophthalate (4HIP), and isophthalate (IPA), was assessed by using a spectrophotometer. Enzymatic activity toward these compounds was not detected, indicating that the substrate specificity of LigW was restricted to 5CVA. On the other hand, the decarboxylation activity toward 5CVA was reduced to 51 and 79% in the presence of 4HIP and IPA, respectively, in the reaction mixture containing 80 μM 5CVA and 160 μM analog. 3MSA did not inhibit the activity. These results indicate that the substrate recognition of LigW seems to depend strictly on two carboxyl groups oriented meta to each other in 5CVA. The lack of enzymatic activity toward 4HIP seems to suggest that the 3-methoxyl group is also important for the catalytic activity.
Addition of EDTA and various metal ions, such as Na+, Mg2+, Fe2+, Cu2+, Ca2+, Co2+, Zn2+, and Mn2+, to the assay mixture had no significant effect on the decarboxylase activity toward 5CVA. Furthermore, the decarboxylation activity was not stimulated by the addition of the cofactors NADH, NADPH, NAD+, and NADP+. LigW seems to be a nonoxidative decarboxylase that requires no cofactors.
Mapping of the DDVA catabolic genes.
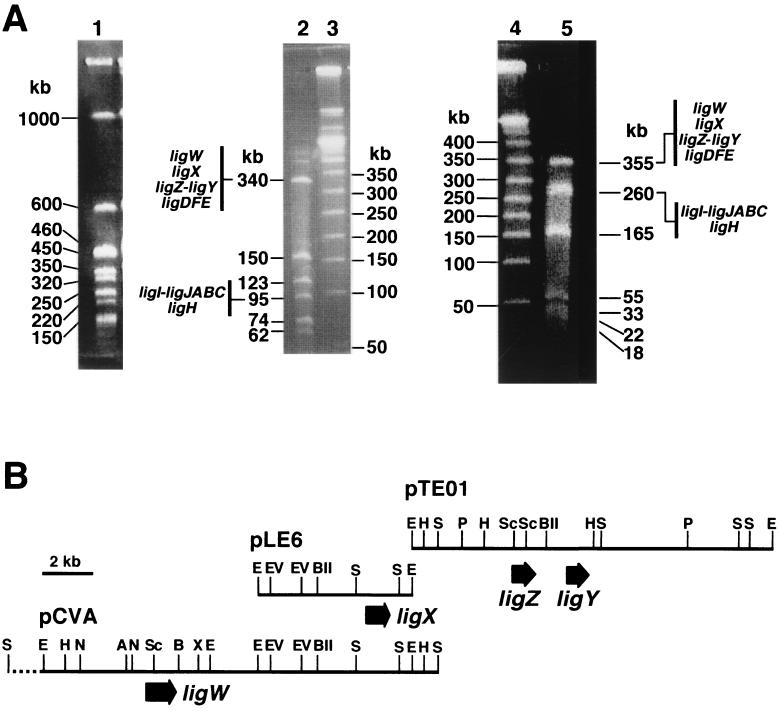
From SYK-6 we have isolated ligX (33) and ligZ-ligY (26, 27), which are involved in the DDVA catabolic pathway; ligH (23) and ligI-ligJABC (10, 21, 22, 24), which are involved in the vanillate catabolic pathway; and ligDFE (16-18), which is involved in the β-aryl ether catabolic pathway. We were interested in knowing how these genes are organized in the SYK-6 chromosome in relation to ligW. Thus, hybridization experiments with large restriction fragments separated on PFGE were performed to address the distribution of the lig genes on the SYK-6 genome. Because the SYK-6 genome has a high G+C content, SpeI was used to digest the total DNA. Total DNA was prepared in an agarose block and was digested with SpeI. It was separated into nine fragments, and the genome size of SYK-6 was estimated to be approximately 3.8 Mb (Fig. 6A). Southern hybridization analysis indicated that all the genes described above lie in the same 1-Mb SpeI fragment. This fragment was then isolated as an agarose block and was further digested with AseI or AflII (Fig. 6A). Southern hybridization analyses indicated that the ligX, ligW, ligZ-ligY, and ligDFE genes lie in the 340-kb AseI-digested fragment and also in the 355-kb AflII-digested fragment. On the other hand, the ligH and ligI-ligJABC genes lie in the 95-kb AseI-digested fragment and also in the 260-kb AflII-digested fragment (data not shown). The catabolic genes for the dimeric compounds DDVA and β-aryl ether seem to be separated from those for the monomeric compound vanillate.
FIG. 6.
Localization of the lignin degradation genes in the genome of S. paucimobilis SYK-6. PFGE profiles and a summary of the lig gene localizations are indicated in panel A. Lane 1, SpeI digests of the SYK-6 total DNA; lane 2, AseI digests of the 1-Mb SpeI fragment shown in lane 1; lanes 3 and 4, lambda ladder markers; lane 5, AflII digests of the 1-Mb SpeI digest. The restriction maps of pCVA, pLE6, and pTE01 containing ligW, ligX, and ligZ-ligY, respectively, are presented in panel B. The large arrows indicate the coding region of each gene. The restriction map of the region shown by a dashed line in pCVA was not determined. Abbreviations: A, ApaI; B, BamHI; BII, BglII; E, EcoRI; EV, EcoRV; H, HindIII; N, NotI; P, PstI; S, SalI; Sc, SacI; X, XhoI.
To determine the detailed localization of the DDVA catabolic genes, Southern hybridization analysis of pLE6, pCVA, and pTE01 containing ligX, ligW, and ligZ-ligY, respectively, was carried out with each plasmid as a probe. The pCVA probe hybridized with the 6.0-kb EcoRI fragment of pLE6 and the 15-kb EcoRI fragment of pTE01. On the other hand, the pTE01 probe hybridized with the 1.7-kb SalI fragment of pCVA (data not shown). The restriction map of pCVA was constructed in detail and was compared with those of pLE6 and pTE01. This analysis indicated that the four genes for conversion of DDVA to vanillate were all within an 18-kb region (Fig. 6).
DISCUSSION
In this study we concluded that 5CVA was converted into vanillate and was further degraded through the PCA 4,5-cleavage pathway after O-demethylation on the basis of the following reasons. (i) Vanillate was detected as an intermediate in growing cells of SYK-6 with 5CVA. (ii) The ligB insertion mutant of SYK-6 could not grow on 5CVA. (iii) The accumulation of PCA from DDVA and 5CVA degraded by the ligB mutant was observed. (iv) The 37-kDa protein produced in E. coli carrying the ligW gene from SYK-6 catalyzed the decarboxylation of 5CVA to generate vanillate. This is the first report on the genetic analysis of 5CVA decarboxylase, which plays a key role as an interface between DDVA and vanillate catabolic pathways in SYK-6.
The mapping of lig genes indicated that ligW as well as the other DDVA catabolic genes, ligX, ligZ, and ligY, were localized within an 18-kb region, suggesting that these genes constitute a DDVA catabolic gene cluster. These results also indicated that the DDVA catabolic gene cluster was separated from the vanillate catabolic gene cluster; however, the relative location of these gene clusters and β-aryl ether catabolic genes was not determined.
It is known that the aromatic acid decarboxylases are classified into two types. One is the nonoxidative (reductive) decarboxylase, which is involved in elimination of the carboxyl group from the aromatic nucleus (11). This kind of reaction has been reported for the transformation of vanillate to guaiacol. The nonoxidative decarboxylase reported previously did not require the external addition of any cofactor for its activity. The other is the oxidative decarboxylase, which catalyzes hydroxylation and requires NAD(P)H for its enzymatic activity. For example, vanillate hydroxylase, which is a flavoprotein, catalyzes decarboxylation by concomitant hydroxylation at the relevant carbon atom (4, 5). LigW does not require any cofactor for its activity, and MS analysis of the product from the reaction with D2O clearly indicated that one deuterium atom was incorporated into vanillate from D2O. We concluded that LigW is a nonoxidative decarboxylase. On the basis of the mechanism for nonoxidative decarboxylation proposed by Huang et al. (11), we illustrate the detailed reaction where a quinoid tautomer is generated as an intermediate through proton replacement during the decarboxylation of 5CVA (Fig. 5D).
Several nonoxidative decarboxylases have been purified and characterized previously, and two genes from Comamonas testosteroni (15) and P. putida (25), phtC and pht5, respectively, encoding the same enzyme (4,5-dihydroxyphthalate decarboxylase) have been isolated and characterized. The deduced amino acid sequence of phtC has 78% identity with that of pht5. However, that of ligW has no significant identity with those of phtC and pht5. Recently the 2-amino-3-carboxymuconate-6-semialdehyde decarboxylase gene has been isolated from R. norvegicus, which encodes a nonoxidative decarboxylase involved in tryptophan metabolism (35). This protein had 31% identity over 276 amino acid residues with the deduced amino acid sequence of LigW. In addition, a comparison of LigW with the LigJ 4-oxalomesaconate hydratase and the LigY meta-cleavage compound hydrolase revealed 25% identity over 191 amino acids and 24% identity over 249 amino acids, respectively. An activity in common between these enzymes is incorporation of a water-derived hydrogen atom into the substrate, a fact that may account for their shared amino acid sequence.
Acknowledgments
This work was supported in part by a Grant-in-Aid for the Encouragement of Young Scientists (No. 09760077) from the Ministry of Education, Science, Sports and Culture, Japan, to E.M.
REFERENCES
- 1.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1990. Current protocols in molecular biology. John Wiley & Sons, Inc., New York, N.Y.
- 2.Bagdasarian, M., R. Lurz, B. Rückert, F. C. H. Franklin, M. M. Bagdasarian, J. Frey, and K. N. Timmis. 1981. Specific-purpose plasmid cloning vectors. II. Broad-host-range, high-copy-number, RSF1010-derived vectors, and a host-vector system for gene cloning in Pseudomonas. Gene 16:237-247. [DOI] [PubMed] [Google Scholar]
- 3.Bolivar, F., and K. Backman. 1979. Plasmids of Escherichia coli as cloning vectors. Methods Enzymol. 68:245-267. [DOI] [PubMed] [Google Scholar]
- 3a.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 4.Buswell, J. A., K.-E. E. Eriksson, and B. Pettersson. 1981. Purification and partial characterization of vanillate hydroxylase (decarboxylase) from Sporotrichum pulverulentum. J. Chromatogr. 215:99-108. [Google Scholar]
- 5.Buswell, J. A., P. Pettersson, and K.-E. E. Eriksson. 1979. Oxidative decarboxylation of vanillic acid by Sporotrichum pulverulentum. FEBS Lett. 103:98-101. [DOI] [PubMed] [Google Scholar]
- 6.Ditta, G., S. Stanfield, D. Corbin, and D. R. Helinski. 1980. Broad host range DNA cloning system for gram-negative bacteria: construction of gene bank of Rhizobium meliloti. Proc. Natl. Acad. Sci. USA 77:7347-7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Figurshi, D. H., and D. R. Helinski. 1979. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc. Natl. Acad. Sci. USA 76:1648-1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Freudenberg, K. 1968. The constitution and biosynthesis of lignin, p. 47-122. In A. C. Neish and K. Freudenberg (ed.), Constitution and biosynthesis of lignin. Springer-Verlag, New York, N.Y.
- 9.Fukuda, M., and K. Yano. 1985. Construction of broad host range cloning vectors for Gram-negative bacteria. Agric. Biol. Chem. 49:2719-2724. [Google Scholar]
- 10.Hara, H., E. Masai, Y. Katayama, and M. Fukuda. 2000. The 4-oxalomesaconate hydratase gene, involved in the protocatechuate 4,5-cleavage pathway, is essential to vanillate and syringate degradation in Sphingomonas paucimobilis SYK-6. J. Bacteriol. 182:6950-6957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang, Z., L. Dostal, and J. P. N. Rosazza. 1993. Metabolism of ferulic acid conversions to vanillic acid and guaiacol by Rhodotorula rubra. J. Biol. Chem. 268:23954-23958. [PubMed] [Google Scholar]
- 12.Katayama, Y., S. Nishikawa, A. Murayama, M. Yamasaki, N. Morohoshi, and T. Haraguchi. 1988. The metabolism of biphenyl structures in lignin by the soil bacterium (Pseudomonas paucimobilis SYK-6). FEBS Lett. 233:129-133. [DOI] [PubMed] [Google Scholar]
- 13.Katayama, Y., S. Nishikawa, M. Nakamura, K. Yano, M. Yamasaki, N. Morohoshi, and T. Haraguchi. 1987. Cloning and expression of Pseudomonas paucimobilis SYK-6 genes involved in the degradation of vanillate and protocatechuate in P. putida. Mokuzai Gakkaishi 33:77-79. [Google Scholar]
- 14.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 227:680-685. [DOI] [PubMed] [Google Scholar]
- 15.Lee, J.-H., T. Omori, and T. Kodama. 1994. Identification of the metabolic intermediates of phthalate by Tn5 mutants of Pseudomonas testosteroni and analysis of the 4,5-dihydroxyphthalate decarboxylase gene. J. Ferment. Bioeng. 77:591-597. [Google Scholar]
- 16.Masai, E., S. Kubota, Y. Katayama, S. Kawai, M. Yamasaki, and N. Morohoshi. 1993. Characterization of the Cα-dehydrogenase gene involved in the cleavage of β-aryl ether by Pseudomonas paucimobilis. Biosci. Biotechnol. Biochem. 57:1655-1659. [DOI] [PubMed] [Google Scholar]
- 17.Masai, E., Y. Katayama, S. Kawai, S. Nishikawa, M. Yamasaki, and N. Morohoshi. 1991. Cloning and sequencing of the gene for a Pseudomonas paucimobilis enzyme that cleaves β-aryl ether. J. Bacteriol. 173:7950-7955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Masai, E., Y. Katayama, S. Kubota, S. Kawai, M. Yamasaki, and N. Morohoshi. 1993. A bacterial enzyme degrading the model lignin compound β-aryl etherase is a member of the glutathione-S-transferase superfamily. FEBS Lett. 323:135-140. [DOI] [PubMed] [Google Scholar]
- 19.Masai, E., Y. Katayama, S. Nishikawa, and M. Fukuda. 1999. Characterization of Sphingomonas paucimobilis SYK-6 genes involved in degradation of lignin-related compounds. J. Ind. Microbiol. Biotechnol. 23:364-373. [DOI] [PubMed] [Google Scholar]
- 20.Masai, E., Y. Katayama, S. Nishikawa, M. Yamasaki, N. Morohoshi, and T. Haraguchi. 1989. Detection and localization of a new enzyme catalyzing the β-aryl ether cleavage in the soil bacterium (Pseudomonas paucimobilis SYK-6). FEBS Lett. 249:348-352. [DOI] [PubMed] [Google Scholar]
- 21.Masai, E., K. Momose, H. Hara, S. Nishikawa, Y. Katayama, and M. Fukuda. 2000. Genetic and biochemical characterization of 4-carboxy-2-hydroxymuconate-6-semialdehyde dehydrogenase and its role in the protocatechuate 4,5-cleavage pathway in Sphingomonas paucimobilis SYK-6. J. Bacteriol. 182:6651-6658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Masai, E., S. Shinohara, H. Hara, S. Nishikawa, Y. Katayama, and M. Fukuda. 1999. Genetic and biochemical characterization of a 2-pyrone-4,6-dicarboxylic acid hydrolase involved in the protocatechuate 4,5-cleavage pathway of Sphingomonas paucimobilis SYK-6. J. Bacteriol. 181:55-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nishikawa, S., T. Sonoki, T. Kasahara, T. Obi, S. Kubota, S. Kawai, N. Morohoshi, and Y. Katayama. 1998. Cloning and sequencing of the Sphingomonas (Pseudomonas) paucimobilis gene essential for the O demethylation of vanillate and syringate. Appl. Environ. Microbiol. 64:836-842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Noda, Y., S. Nishikawa, K. Shiozuka, H. Kadokura, H. Nakajima, K. Yoda, Y. Katayama, N. Morohoshi, T. Haraguchi, and M. Yamasaki. 1990. Molecular cloning of the protocatechuate 4,5-dioxygenase genes of Pseudomonas paucimobilis. J. Bacteriol. 172:2704-2709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nomura, Y., M. Nakagawa, N. Ogawa, S. Harashima, and Y. Oshima. 1992. Genes in PHT plasmid encoding the initial degradation pathway of phthalate in Pseudomonas putida. J. Ferment. Bioeng. 74:333-344. [Google Scholar]
- 26.Peng, X., T. Egashira, K. Hanashiro, E. Masai, S. Nishikawa, Y. Katayama, K. Kimbara, and M. Fukuda. 1998. Cloning of a Sphingomonas paucimobilis SYK-6 gene encoding a novel oxygenase that cleaves lignin-related biphenyl and characterization of the enzyme. Appl. Environ. Microbiol. 64:2520-2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peng, X., E. Masai, Y. Katayama, and M. Fukuda. 1999. Characterization of the meta-cleavage compound hydrolase gene involved in the degradation of the lignin-related biphenyl structure by Sphingomonas paucimobilis SYK-6. Appl. Environ. Microbiol. 65:2789-2793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Profft, E., and W. Krause. 1964. Über die chlormethylierung des o- und novo-vanillins und die gewinnung von 4-hydroxy-5-alkoxyisophthalaldehyden. Arch. Pharm. 298:148-162. [DOI] [PubMed] [Google Scholar]
- 29.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 30.Sanger, F., S. Nicklen, and A. R. Coulson. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 74:5463-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sarkanen, K. V., and C. H. Ludwig. 1971. Lignins. Occurrence, formation, structure and reactions. Wiley-Interscience, New York, N.Y.
- 32.Short, J. M., J. M. Férnandez, J. A. Sorge, and W. Huse. 1988. λZAP: a bacteriophage λ expression vector with in vivo excision properties. Nucleic Acids Res. 16:7583-7600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sonoki, T., T. Obi, S. Kubota, M. Higashi, E. Masai, and Y. Katayama. 2000. Coexistence of two different O-demethylase systems in lignin metabolism by Sphingomonas paucimobilis SYK-6: cloning and sequencing of the lignin biphenyl-specific O-demethylase (LigX) gene. Appl. Environ. Microbiol. 66:2125-2132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sugimoto, K., T. Senda, H. Aoshima, E. Masai, M. Fukuda, and Y. Mitsui. 1999. Crystal structure of an aromatic-ring-opening dioxygenase LigAB which is a protocatechuate 4,5-dioxygenase. Struct. Fold. Des. 15:953-965. [DOI] [PubMed] [Google Scholar]
- 35.Tanabe, A., Y. Egashira, S. Fukuoka, K. Shibata, and H. Sanada. 2002. Purification and molecular cloning of rat 2-amino-3-carboxymuconate-6-semialdehyde decarboxylase. Biochem. J. 361:567-575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]