Abstract
This paper describes the characterization of an intracellular β-glucosidase enzyme BGLII (Cel1a) and its gene (bgl2) from the cellulolytic fungus Trichoderma reesei (Hypocrea jecorina). The expression pattern of bgl2 is similar to that of other cellulase genes known from this fungus, and the gene would appear to be under the control of carbon catabolite repression mediated by the cre1 gene. The BGLII protein was produced in Escherichia coli, and its enzymatic properties were analyzed. It was shown to be a specific β-glucosidase, having no β-galactosidase side activity. It hydrolyzed both cellotriose and cellotetraose. BGLII exhibited transglycosylation activity, producing mainly cellotriose from cellobiose and sophorose and cellobiose from glucose. Antibodies raised against BGLII showed the presence of the enzyme in T. reesei cell lysates but not in the culture supernatant. Activity measurements and Western blot analysis of T. reesei strains expressing bgl2 from a constitutive promoter further confirmed the intracellular localization of this β-glucosidase.
Three categories of cellulases, endoglucanases (EC 3.2.1.4), cellobiohydrolases (EC 3.2.1.91), and β-glucosidases (EC 3.2.1.21), are produced by saprophytic filamentous fungi for the degradation of insoluble cellulose into glucose. The endoglucanases mainly hydrolyze internal bonds in the cellulose polymer, producing new chain ends. The cellobiohydrolases, also known as exoglucanases, act processively from the chain ends, mainly producing cellobiose. This disaccharide and other short cello-oligomers are broken down to glucose by β-glucosidases. Trichoderma reesei possesses one of the best-known cellulase enzyme systems, and it has served as a model for fungal cellulose degradation (see chapters in the work cited in reference 10). The cellobiohydrolases and the endoglucanases from this fungus have been especially extensively characterized. The β-glucosidases of T. reesei, however, are less well characterized.
T. reesei has been reported to produce extracellular (5), cell-wall-bound (38), and intracellular (15) β-glucosidases. However, some data suggest that the extracellular and a major part of the cell-wall-bound activities could be due to the same enzyme (12, 26). The gene bgl1 (3), encoding an extracellular β-glucosidase, has been isolated from T. reesei, and most probably it encodes the extracellular protein isolated previously (5). The corresponding BGLI enzyme has sequence similarity with other β-glucosidases belonging to family 3 of the glycosyl hydrolases (3). It has been shown that by increasing the copy number of bgl1 and thus the amount of the BGLI enzyme in the cellulase mixture produced by T. reesei, the rate of reducing sugar formation from cellulose could be enhanced (3). This suggests that β-glucosidase activity is limiting for total hydrolysis of cellulose in the cellulase mixture. The bgl1 gene has been deleted from T. reesei (8, 21), and a minor amount of extracellular β-glucosidase activity was still produced. This would indicate that there are other genes encoding extracellular β-glucosidases in the T. reesei genome. The disruption of the bgl1 gene resulted in a delay in induction of the other cellulase genes by cellulose but not by sophorose, a β-1-2-linked disaccharide known as an efficient cellulase inducer (8). This suggests that BGLI is involved in formation of the soluble inducer of the cellulolytic enzymes. Results with similar implications were obtained with bgl1 disruptants and multicopy strains by Mach et al. (21). The amount of BGLI present affected cellulase induction by cellulose or nonsaturating sophorose concentrations.
Information about a T. reesei gene (bgl2) encoding a second β-glucosidase belonging to glycosyl hydrolase family 1 was published recently (35). Some enzymatic properties of the corresponding BGLII (Cel1A) enzyme were reported. The same gene was cloned in our laboratory, and in this article we describe the analysis of the bgl2 gene expression pattern and characterization of the BGLII protein. In contrast to the assumption of Takashima et al. regarding BGLII (35), data are presented in this paper indicating that BGLII is an intracellular enzyme.
MATERIALS AND METHODS
Strains.
The T. reesei bgl2 cDNA was cloned in the Saccharomyces cerevisiae strain H1152 (a, sso2-1, leu2-3, trp1-1, ura3-1, sso1::HIS3 [Markku Aalto and Sirkka Keränen, unpublished data]). The Escherichia coli strains TOP 10F′ (Invitrogen) and DH5α [F−, endA1, hsdR17(rk−, mk+), supE44, thi-1, λ−, recA1, gyrA96, relA1, Δ(argFlacZYA)U169, Φ80lacZΔM15] were used as plasmid hosts. BGLII was produced in the E. coli strain RV308 (su−, ΔlacX74, galISH, OP308, strA). The T. reesei strains QM9414 (23), Rut-C30 (27), and RutC-30, transformed with the cre1 gene (13), were used in studies of bgl2 regulation. The ace2 disruptant strains and their parental strain ALKO2221 (2) were used when the effect of the ace2 activator gene on bgl2 induction was examined. The T. reesei strain RL-P37 (34) was used in Western analysis of BGLII localization. T. reesei QM9414 (23) was the host for constitutive bgl2 expression.
Bgl2 expression studies.
To examine the effect of the carbon source on the expression of bgl2, the strain QM9414 was grown in shake flasks (200 rpm, 28°C) in minimal medium (29) with 3% glucose, 3% lactose, 2% cellobiose, or 3% cellulose (Solka Floc) for 2 (glucose) or 3 days. In experiments in which induction by sophorose was tested, QM9414 was grown in a minimal medium with 2% sorbitol or glycerol or 5% glucose for 72 or 57 h (glucose) and 1 mM sophorose was added at that time and 10 h later. The growth was continued for another 15 h, and the cells were harvested. Cultures, carried out with no sophorose additions for the total length of the experiment, were grown as controls of the induction. The glucose-depleted sample was derived from a glucose batch fermentation reaction mixture at 125 h, 50 h after the glucose in the fermentation had run out (14). The cre1-1 mutant strain RutC-30 and the same strain transformed with the wild-type cre1 gene (13) were cultivated in minimal medium with 2% glucose for 3 days. The ace2 disruptant and its parental strain were grown in minimal medium with glycerol as previously described (2), and the mycelium was transferred into a medium with Solka Floc cellulose. The induction of bgl2 was studied by Northern hybridization from samples harvested 9, 12, 15, 18, and 32 h after the medium change. The mycelia were collected by filtering, washed by 0.9% NaCl, and stored frozen. Total RNA was isolated as previously described (4) or by the Trizol reagent (Life Technologies). Northern hybridization was performed with the 32P-labeled bgl2 cDNA on Hybond N nylon filters as instructed by the manufacturer. The digital images were created by scanning films or Western filters with a Hewlett Packard Scanjet 4c scanner and processed with the Microsoft Powerpoint program.
Production of BGLII in E. coli.
A DNA fragment with the bgl2 protein-coding region of the cDNA fused with a C-terminal addition of six histidine codons was made by PCR with the primers TAGGATGAATTCATGTTGCCCAAGGACTTTCAG (forward primer with an EcoRI site) and ATACGAAAGCTTTCAGTGGTGGTGGTGGTGGTGCGCCGCCGCAATCAGCTCGT (reverse primer with six histidine codons, a stop codon, and a HindIII site). The PCR product was cloned between the EcoRI and HindIII sites of the pKKtac vector (36). Strain RV308 with the bgl2 expression construct was grown at 37°C to an optical density at 600 nm of 0.9, 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) was added as an inducer, and growth was continued for 4 h at 30°C. The BGLII protein was detected by Western blotting with anti-His6 antibody (Roche) and purified with Ni-nitrilotriacetic acid-agarose (Qiagen) according to the instructions of the manufacturer of the QIAexpressionist kit (Qiagen).
Activity measurements.
Both substrate analogs (p-nitrophenyl [PNP]-α-d-galactopyranoside, PNP-β-d-mannopyranoside, PNP-β-d-xylopyranoside, PNP-α-l-arabinofuranoside, PNP-β-l-arabinopyranoside, PNP-β-d-glucopyranoside [Sigma], 4-methylumbelliferyl-β-d-glucoside [MUG, Sigma], 5-bromo-4-chloro-3-indolyl-β-d-glucoside [X-Glu; Sigma], and 5-bromo-4-chloro-3-indolyl-β-d-galactoside [X-Gal; Sigma]) and native oligosaccharides (cellobiose, cellotriose, cellotetraose, lactose [Merck], and sophorose [Serva]) were used to test the substrate specificity of BGLII. The activity towards the substrate analogs was determined by incubating 100 μl of the substrate (the PNP substrates [10 mM], 0. 75 mg of X-Glu and X-Gal/ml, 1 mM of MUG) and 10 nkat of enzyme (as measured with PNP-β-d-glucopyranoside) at 45°C for 24 h in 50 mM Na-phosphate buffer, pH 7.0. The activity was measured spectrophotometrically at 405 nm (PNP substrates) or detected visually in UV light for MUG and in daylight for X-Glu and X-Gal. The activity towards native substrates was determined by incubating 1 ml (0.5% wt/vol) of the substrate and 2.5 nkat of purified BGLII in 50 mM sodium phosphate buffer, pH 7.0, at 45°C for 24 h. The samples were boiled for 5 min and analyzed by using high-pressure liquid chromatography (HPLC) as previously described (37).
The transglycosylation activity of BGLII was measured by incubating 1 ml of 20% or 40% glucose or 1 ml of 10% or 20% cellobiose with 100 nkat of purified BGLII/g of substrate in 50 mM Na-phosphate buffer (pH 7.0) at 45°C for 24 h. The samples were boiled for 5 min and analyzed using HPLC.
Western detection of BGLII.
Polyclonal antibodies were raised against BGLII produced in E. coli. Two rabbits were immunized with 400 μg of the protein, and two booster injections of 200 μg were given at 3-week intervals. The antiserum was used at a 1:10,000 dilution in Western blotting. To detect the BGLII protein in T. reesei and to study its localization, the fungus was grown for 3 days in minimal medium with 0.2% peptone and 2% glucose, 2% lactose, or 2% Solka Floc cellulose as the carbon source. The mycelia were washed with 0.9% NaCl, and lysates were prepared by grinding them into fine powder under liquid nitrogen and resuspending into 50 mM Na-phosphate buffer (pH 7.0) with protease inhibitor cocktail (Roche). Cell debris was removed by centrifugation at 14,000 × g for 10 min. The total protein concentrations of the lysates were measured as previously described (20). Sodium dodecyl sulfate (SDS) gels from the cell lysate samples containing 20 μg of total protein and culture medium samples of 30 μl were processed as previously described (18), and Western detection was carried out with the alkaline phosphatase-secondary antibody conjugate (Bio-Rad).
Constitutive bgl2 expression in T. reesei.
The bgl2 cDNA was released from the yeast expression vector pAJ401 (31) by an EcoRI-SspI digestion and cloned into the pAN52-1NotI vector (30), digested with NcoI, and treated with mung bean nuclease. A hygromycin resistance marker cassette was cloned into the NotI site of the resulting plasmid. The final expression vector, pJKP4, was transformed into T. reesei QM9414 as previously described (29). The transformants were selected on medium with 100 μg of hygromycin B/ml and purified through single-spore cultures before further analysis. Two transformants and the parental strain were grown in minimal medium (29) with 0.5% peptone and either 2% glucose or 2% cellobiose as the carbon source, and samples from the mycelia and supernatants were collected after 2 and 3 days of growth. Genomic DNA was isolated from the parental strain and the two transformants, and Southern hybridization with the bgl2 cDNA from the DNA as a probe was performed as previously described (32). Cell-wall-associated β-glucosidase activity was measured from the culture grown for 3 days with glucose as the carbon source. After the growth, the mycelia were harvested immediately from 1 ml of the cultures by centrifugation at 6,000 × g for 3 min and washed twice with 0.9% NaCl. The cells were then resuspended into 50 mM Na-phosphate buffer (pH 7.0) with 10 mM PNP-β-d-glucoside substrate, and β-glucosidase activity was measured. Total protein lysates were prepared from the mycelia for Western detection of BGLII as described above. The protein content of each lysate was measured as previously described (20), and β-glucosidase activity was measured with PNP-β-d-glucoside from samples containing 5 μg of total protein. Using the same substrate, activity was also measured for 100 μl of samples taken from the culture supernatants. All the activity measurements were carried out from triplicate samples. Western detection of BGLII was carried out as described above from 40 μl of culture supernatant samples and cell lysate samples containing 20 μg of total protein.
RESULTS
Isolation of the bgl2 gene.
An attempt was made to clone components of the T. reesei secretory pathway by complementation in yeast of the sso2 gene involved in exocytosis (1). The temperature-sensitive sso2-1 yeast strain was transformed with a T. reesei expression library in a yeast vector (24), and several clones supporting growth of the strain in the restrictive temperature were found. One of these contained a cDNA encoding a novel β-glucosidase belonging to family 1 of the glycosyl hydrolases. Both the cDNA and the chromosomal copy were sequenced. The isolated gene was identical to the T. reesei β-glucosidase gene bgl2, whose sequence was published by Takashima et al. (35).
Expression pattern of the bgl2 gene.
The cellulase and hemicellulase genes of T. reesei are regulated according to the carbon source and are under carbon catabolite repression mediated by the cre1 gene (13, 14, 25). Therefore, bgl2 expression was studied by Northern hybridization using total RNA samples derived from cultures performed with different carbon sources and using strains with mutant and wild-type cre1 genes. An activator gene, ace2, affecting the induction of the T. reesei cellulase gene, has been cloned recently (2), and the effect of ace2 disruption on bgl2 activation was also studied in this work by Northern hybridization. In addition to the bgl2 experimentation, the Northern blots were probed with the actin-encoding act1 gene as a loading control. Because act1 mRNA levels are relatively variable on different carbon sources, a staining of the RNA gel with acridine orange is shown in Fig. 1 as an additional loading control.
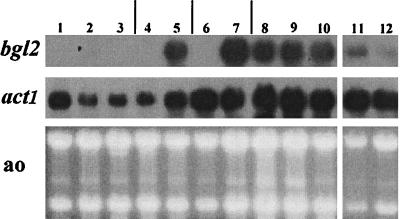
FIG. 1.
Northern analysis of the bgl2 gene expression. T. reesei was grown on minimal medium with glucose (lanes 1 to 3 and 11 and 12), sorbitol (lanes 4 and 5), glycerol (lanes 6 and 7), lactose (lane 8), cellobiose (lane 9), or cellulose (lane 10) as the carbon source. The effect of sophorose on the bgl2 mRNA level is shown in lanes 3, 5, and 7. Lane 2 contains a sample from a carbon-source-depleted stage of a glucose culture. The involvement of the cre1 gene in bgl2 regulation was studied in the cre1-1 mutant strain RutC-30 (lane 11) and in RutC-30 transformed with the wild type cre1 (lane 12). A control probing with the actin gene (act1) and a staining of the gel with acridine orange (ao) are shown.
No bgl2 mRNA could be detected in samples derived from cultures carried out with glucose, sorbitol, or glycerol (Fig. 1, lanes 1, 4, and 6). The other T. reesei cellulases have been found to be induced in glucose-grown cultures in late stages when the carbon source has run out (14), but bgl2 was not induced at this stage (lane 2). The bgl2 gene was strongly expressed on lactose, cellobiose, and cellulose (lanes 8, 9, and 10). When sophorose, a disaccharide capable of inducing the other cellulase genes, was added to cultures grown on sorbitol or glycerol, the bgl2 gene was strongly induced. This is in agreement with the findings presented in a previous paper (19), in which it was shown that intracellular β-glucosidase activity is induced by sophorose. In our experiment, sophorose did not induce bgl2 when it was added to a glucose-grown culture. One explanation for this could be inhibition of sophorose uptake by a glucose-inhibited β-diglucoside permease (17).
In the strain RutC-30, which is a cre1 mutant (13), the bgl2 gene was expressed in a culture performed on glucose (Fig. 1, lane 11). In the derivative of RutC-30 that was transformed with the wild-type cre1, the expression level of bgl2 was lower than that in RutC-30 in glucose cultures (Fig. 1, lane 12). The apparent involvement of cre1 in bgl2 regulation is in agreement with the fact that two putative binding sites of the CREI protein can be found in the bgl2 promoter region (data not shown). In an ace2 disruption strain, bgl2 induction by cellulose was delayed and was not as strong as that in the parental strain (data not shown), in similarity to findings for the cellulase genes cbh1, cbh2, and egl1 and the xylanase gene xyn2 (2). In conclusion, the bgl2 gene is regulated in a manner largely similar to that of other T. reesei cellulase genes.
The hydrolytic properties of BGLII.
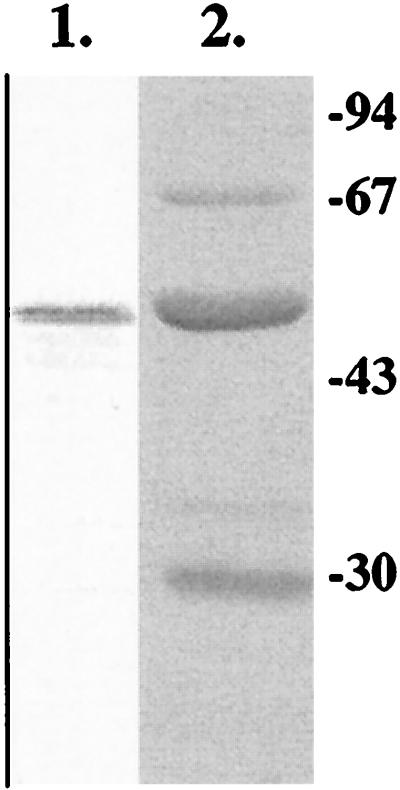
In order to be able to purify the T. reesei BGLII protein, the protein-coding region of bgl2 was expressed in E. coli from the tac promoter as a C-terminal histidine tag fusion. Clear activity towards β-glucosidase substrates was detected in lysates of E. coli cells carrying the bgl2 expression plasmids, whereas control cells with the vector alone did not produce any activity (data not shown). The BGLII-HisTag protein was purified from E. coli lysate by Ni-affinity chromatography, with a yield of about 5 mg per liter of culture. After purification, the protein was 50 to 60% pure (Fig. 2). Because no β-glucosidase activity was detected in the host strain, not even in prolonged incubations, the decision was made to use the partially purified BGLII protein in activity assays.
FIG. 2.
SDS gel analysis of BGLII produced in E. coli. Using Western blotting, the protein was detected by anti-HisTag antibodies (lane 1). The purified BGLII protein is shown in a Coomassie-stained gel (lane 2).
The pH profile of BGLII was measured with PNP-β-d-glucoside as the substrate in the pH range of 3 to 8. The enzyme had a broad pH optimum in the range from 5.5 to 7 (data not shown).
The substrate specificity of BGLII was analyzed by hydrolysis tests with chromogenic substrates in which glucose, galactose, mannose, arabinose, or xylose was linked to the chromogenic group by a β linkage. These were used to test whether BGLII would have activity towards lactose or disaccharides derived from the degradation of different hemicelluloses. PNP-α-d-galactopyranoside and PNP-α-l-arabinofuranoside were used to test whether BGLII would have α-galactosidase or α-arabinofuranosidase side activities, respectively. The only substrates hydrolyzed by BGLII were those with glucose linked to the chromogenic group via a β-1-4 linkage, i.e., methylumbelliferyl-β-d-glucoside, PNP-β-d-glucopyranoside, and X-Glu (Table 1). Thus, according to this experiment, BGLII is a specific β-glucosidase. That BGLII does not have β-xylosidase activity was a somewhat unexpected finding, since PNP-β-d-xylopyranoside closely resembles PNP-β-d-glucopyranoside. d-Xylopyranoside has the same hydroxyl configuration as d-glucopyranoside but without the primary hydroxyl group at C-5. This suggests that the primary hydroxyl is necessary for substrate recognition.
TABLE 1.
Hydrolysis of different chromogenic substrates by BGLII
| Substrate | Hydrolysis |
|---|---|
| Methylumbelliferyl-β-d-glucoside (MUG) | +++ |
| PNP-α-d-galactopyranoside | − |
| PNP-α-l-arabinofuranoside | − |
| PNP-β-d-glucopyranoside | +++ |
| PNP-β-d-mannopyranoside | − |
| PNP-β-d-xylopyranoside | − |
| PNP-β-l-arabinopyranoside | − |
| X-Gal | − |
| X-Glu | +++ |
In the next experiment, we tested whether BGLII is active on longer cello-oligosaccharides, cellotriose, and cellotetraose or the disaccharide sophorose that has two glucose units linked by a β-1-2 linkage. As mentioned above, sophorose is a potent inducer of the cellulase genes. A further hydrolysis test was made with lactose, to confirm the negative hydrolysis result obtained with X-Gal in the previous experiment. The di- or oligosaccharides were incubated in 0.5% concentrations in the presence of BGLII, and the hydrolysis products were analyzed by HPLC. To reveal possible impurities or instability in the substrates, the incubations were also done without BGLII and the reaction mixtures were analyzed similarly. The results (Table 2) show that BGLII has activity towards both cellotriose and cellotetraose. The formation of the expected products, glucose and cellobiose, could be detected from cleavage of cellotriose. Interestingly, a small amount of cellotetraose was also produced by BGLII from cellotriose, showing that BGLII has transglycosylation activity. Glucose and cellotriose were formed from cellotetraose, suggesting that BGLII can hydrolyze only a terminal linkage in the substrate and not the central one. Analysis of the reaction products by HPLC does not reveal whether the enzyme cleaves the glucose from the reducing or the nonreducing end. Lactose was not hydrolyzed at all by BGLII in this experiment. Sophorose appeared to be a substrate for BGLII, since a small amount of glucose was formed from it during incubation.
TABLE 2.
Hydrolysis experiments of different oligosaccharides with BGLIIa
| Hydrolyzer | Concn (mg/liter) of:
|
|||||
|---|---|---|---|---|---|---|
| Glucose | Cellobiose | Cellotriose | Cellotetraose | Lactose | Sophorose | |
| Control cellotriose | 0 | 0 | 3,800 | 32.5 | 0 | 0 |
| BGLII cellotriose | 57 | 132.5 | 3,650 | 70.5 | 0 | 0 |
| Control cellotetraose | 0 | 0 | 305 | 4,300 | 0 | 0 |
| BGLII cellotetraose | 45 | 0 | 380 | 4,050 | 0 | 0 |
| Control lactose | 0 | 0 | 0 | 0 | 4,300 | 0 |
| BGLII lactose | 0 | 0 | 0 | 0 | 4,250 | 0 |
| Control sophorose | 14 | 0 | 0 | 0 | 0 | 5,050 |
| BGLII sophorose | 180 | 0 | 0 | 0 | 0 | 4,750 |
The controls were incubated without enzyme addition in a manner similar to that for the samples containing BGLII. The concentrations of the indicated sugars in the reaction mixtures after the incubations are shown.
Transglycosylation studies.
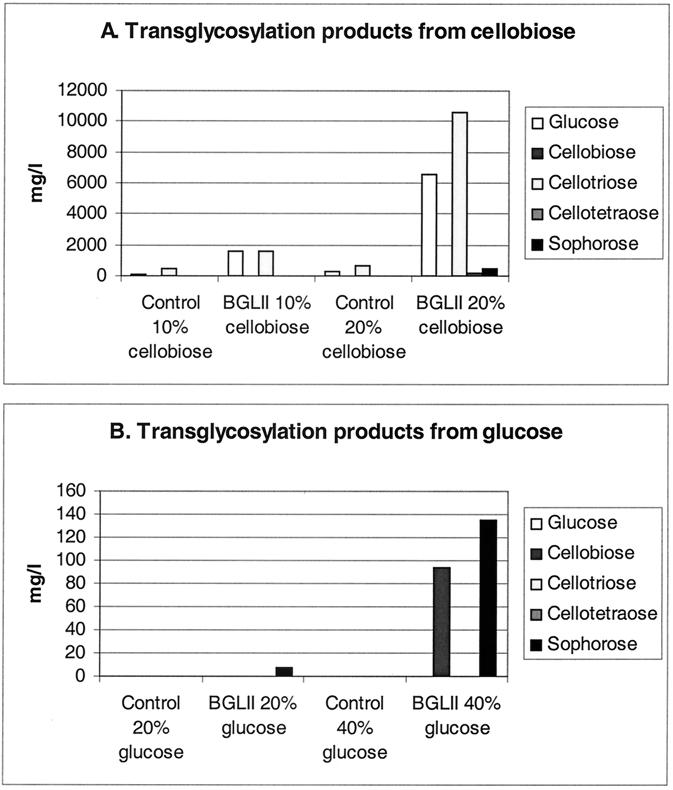
To examine more thoroughly the transglycosylation activity of BGLII, the enzyme was incubated with 10% or 20% cellobiose and 20% and 40% glucose. After the incubations, the reaction mixtures were analyzed by HPLC. Samples treated similarly but having no BGLII were prepared as controls. With 10% cellobiose, about equal amounts of glucose and cellotriose were produced (Fig. 3). With 20% cellobiose, the final concentration of cellotriose was higher than that of glucose, indicating that transglycosylation had a higher rate than hydrolysis. Small amounts of cellotetraose and sophorose were also detected as end products of the incubation of 20% cellobiose with BGLII. From 20% glucose, a small amount of sophorose was formed by BGLII. When incubated with 40% glucose, BGLII produced substantial amounts of sophorose and cellobiose by transglycosylation (Fig. 3).
FIG. 3.
The transglycosylation activity of BGLII was studied with 10% and 20% cellobiose (A) and 20% and 40% glucose (B). The incubations were done with or without (control) the BGLII enzyme.
Localization of BGLII in T. reesei.
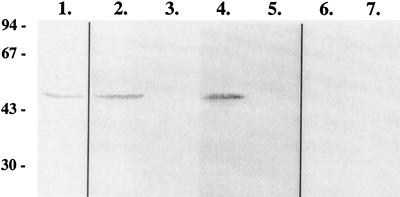
The BGLII protein purified from E. coli was used as an antigen in the immunization of rabbits. An antiserum reacting with the purified BGLII was obtained (Fig. 4, lane 1). The T. reesei strain was grown in minimal medium with glucose, lactose, or cellulose as the carbon source. As shown in Fig. 1, the bgl2 gene is not expressed in a culture grown using glucose but the mRNA is present in mycelia grown on lactose or cellulose. Samples from cell lysates and supernatants of cultures grown on these carbon sources were analyzed by Western blotting with the BGLII antiserum. Consistent with the Northern hybridization results, no signal was obtained from either the cell lysate or the supernatant of the glucose-grown culture (Fig. 4, lanes 6 and 7). In the cell lysates derived from the lactose and cellulose cultures, a protein with about the same mobility as that demonstrated by BGLII in E. coli was detected (Fig. 4, lanes 2 and 4). No signal, however, was obtained from the supernatants of the same cultures (Fig. 4, lanes 3 and 5). These results would indicate that BGLII is not secreted to the culture medium by T. reesei.
FIG. 4.
Localization of BGLII by Western analysis. Samples of BGLII produced in E. coli (lane 1) or T. reesei (lanes 2, 4, and 6) cell lysates or culture supernatants (lanes 3, 5, and 7) are shown. The T. reesei samples were derived from liquid cultures performed with either lactose (lanes 2 and 3), cellulose (lanes 4 and 5), or glucose (lanes 6 and 7) as the carbon source.
Constitutive expression of bgl2 in T. reesei.
The bgl2 cDNA was expressed constitutively from the Aspergillus nidulans gpdA promoter in T. reesei strain QM9414. The transformants were first analyzed by Southern hybridization to detect the transformed construct. Two transformants that had one copy of the construct and the endogenous bgl2 locus intact (data not shown) were chosen for further studies.
The transformants expressing BGLII constitutively were used to show more conclusively the localization of this enzyme. The two transformants and the parental strain were grown in three parallel cultures in minimal medium with either glucose or cellobiose as the carbon source. The glucose cultures were performed to allow study of the β-glucosidase activity derived solely from the transformed construct, because the endogenous bgl2 gene and presumably most of the other β-glucosidase genes are not expressed on this carbon source. Using cell lysate and culture supernatant samples and intact mycelium samples, β-glucosidase activity was examined to reveal secreted, intracellular, and cell-wall-associated activity. The localization of BGLII was also studied by Western blotting using the cell lysates and supernatants.
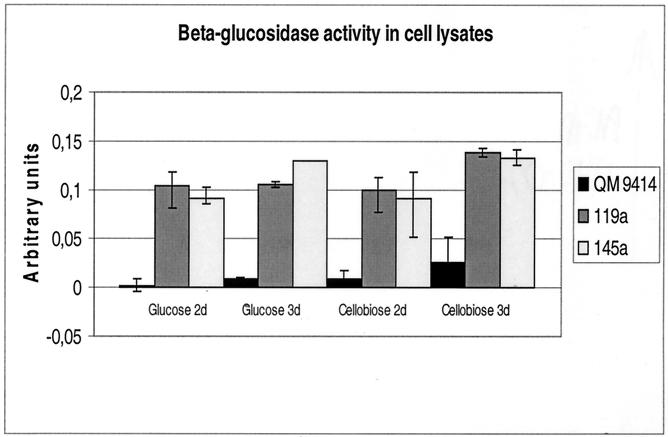
In the cell lysates from the glucose-grown parental strain, very low β-glucosidase activity was found (Fig. 5). The two transformants produced much more activity, presumably originating from the transformed bgl2 construct. The transformants also had much more activity than the parental strain in the lysates of mycelia grown on cellobiose. This suggests that in these conditions, the A. nidulans gpdA promoter reaches a higher expression level than the promoter of the endogenous bgl2 gene. In the supernatant samples of the cultures performed on glucose, practically no β-glucosidase activity was detected from the parental strain or the transformants. A very low level of activity was found in the measurements made with intact mycelia from glucose cultures, but in this assay, the three strains had about equal levels of β-glucosidase (data not shown). This activity could be due to the presence of a constitutive cell-wall-associated β-glucosidase enzyme described earlier (38). Still more proof for the localization of BGLII was obtained from Western blot analysis, carried out using the cell lysate and culture supernatant samples from the glucose culture. The band corresponding to BGLII could be clearly detected in the cell lysate samples from the two transformants but not in those from the parental strain. This band could not be detected in any of the culture medium samples (data not shown).
FIG. 5.
β-Glucosidase activity in cell lysates of T. reesei transformants expressing BGLII constitutively (119a and 145a) and their parental strain (QM 9414). The strains were cultivated with either glucose or cellobiose as the carbon source, and samples are shown for 2- and 3-day-old mycelia.
DISCUSSION
According to the activity measurements carried out in this work, BGLII would appear to be a specific β-glucosidase, cleaving only a β-1-4-bond next to a glucose unit. BGLII had no β-mannosidase, β-xylosidase, β-arabinosidase, or α-arabinofuranosidase activity. Furthermore, no activity was detected towards X-Gal (Table 1) or lactose (Table 2). However, Takashima et al. (35) reported that BGLII has weak activity towards PNP-β-d-galactoside, a substrate analog of lactose. In that study, Humicola grisea BGL4 was found to be clearly more active on this substrate than T. reesei BGLII. Many of the β-glucosidases of glycosyl hydrolase family 1 have been reported to have some side activities, towards lactose, for example (7, 28).
Several of the family 1 β-glucosidases have been shown to possess transglycosylation activity, and research into their use in oligosaccharide synthesis is being carried out (for examples, see the work cited in references 6, 9, 11, and 16). T. reesei BGLII also showed clear transglycosylation activity in our experiments, which were performed with high concentrations of cellobiose and glucose (Fig. 3). The trials carried out in this work did not include any optimization; therefore, the yields of the transglycosylation products were clearly lower than those obtained with, e.g., thermostable bacterial β-glucosidases (7, 9). The reaction products produced by T. reesei BGLII from 40% glucose are of special interest. In these conditions, BGLII produced substantial amounts of sophorose and cellobiose. Thus, it appears that in high glucose concentrations, two glucose units can bind to the BGLII active site and the enzyme can form a glycosidic linkage between them by reverse hydrolysis in a manner previously described (22). The binding of at least one of the glucose units to the enzyme would appear to be flexible, since both β-1-2 and B-1-4 linkages can be formed between the glucose units.
The localization of T. reesei BGLII was studied in this work using Western detection and activity measurements from cell lysates and culture supernatants from an untransformed strain and a strain expressing BGLII constitutively from the A. nidulans gpdA promoter. The protein could not be detected in the supernatants of any of the cultures performed, but it could be clearly detected, both by its activity (Fig. 5) and by Western blot analysis (Fig. 4), in cell lysates. This indicates that BGLII is not secreted by T. reesei. We have also shown that BGLII expressed in S. cerevisiae is not secreted (data not shown). The activity measurements performed with intact T. reesei mycelia did not show any difference between the parental strain and the transformants, in which bgl2 is expressed constitutively, even though the transformants had clearly higher activity in the cell lysate samples than the parental strain (Fig. 5). This would suggest that BGLII is not bound to the cell wall and resides neither in the periplasmic space nor at the plasma membrane, since PNP-β-d-glucoside, which was used as a substrate, most probably can diffuse through the cell wall. Thus, it is very likely that BGLII is a cytoplasmic protein. This is also consistent with the pH profile of BGLII. Unlike the secreted T. reesei cellulases that have acidic pH optima, BGLII is most active at levels of pH close to neutral.
The partial purification of an intracellular β-glucosidase enzyme from T. reesei has been reported (15). The purified enzyme was not characterized thoroughly, but its pH profile matches very well with the one we obtained for BGLII. The molecular mass of the isolated enzyme was determined by gel filtration as 98 kDa. The molecular mass of BGLII, as determined in this work by SDS-gel electrophoresis, was 50 kDa (Fig. 2 and 4). We have no proof of dimerization for BGLII, but if it is dimeric, it could be the same enzyme as the one isolated previously (15).
In previous work on BGLII, Takashima et al. (35) first isolated the gene encoding a secreted Humicola grisea β-glucosidase BGL4 and then used this gene as a hybridization probe and isolated the T. reesei bgl2 gene, assuming that this gene product would also be secreted. Both of the isolated genes were expressed in Aspergillus oryzae. T. reesei BGLII was not secreted from A. oryzae when its protein-coding region was solely in the expression construct. Only when T. reesei BGLII was linked to the H. grisea BGL4 signal sequence was it secreted. This is consistent with our finding that BGLII is intracellular in T. reesei. The N-terminal amino acid sequences of T. reesei BGLII and H. grisea BGL4 are highly similar (Fig. 6), but neither of them is recognized as a signal sequence by a signal sequence prediction program (39). This could be due to the presence of an aspartate residue found close to the N terminus. Acidic residues are not commonly found in the amino termini of signal sequences; instead, they usually have one or two positively charged amino acids. Both T. reesei BGLII and H. grisea BGL4 have a lysine close to the aspartate. In T. reesei BGLII, the lysine is immediately before the aspartate, and in H. grisea, it is positioned two residues after the aspartate (Fig. 6). The difference in position of the lysine with respect to that of the aspartate could explain the fact that this sequence from H. grisea works as a signal sequence and the one in T. reesei BGLII does not.
FIG. 6.
Alignment of the N-terminal amino acid sequences of T. reesei BGLII, Humicola grisea BGL4, and Bacillus polymyxa BglA. Identical amino acids are indicated by asterisks, and similar ones are indicated by dots. The lysine and aspartate residues mentioned in the Discussion section are in bold.
It is interesting that the putative signal sequence region of H. grisea BGL4 shows clear amino acid similarity to those of other β-glucosidases of family 1 from plants and bacteria (an example is shown in Fig. 6). This was an unexpected finding, because signal sequence regions are not conserved between species although they have some common features. Furthermore, this conserved region in Bacillus polymyxa BglA and other enzymes with known crystal structures is a part of the active enzyme and forms a β-strand and an α-helix into the structure (Fig. 6) (33)(http://afmb.cnrs-mrs.fr/∼cazy/CAZY/index.html). Thereforeit would seem likely that H. grisea BGL4 was also originally an intracellular enzyme and that a mutation at its N-terminal region would have turned this region into a signal sequence.
Acknowledgments
We thank Riitta Nurmi for skillful technical assistance, Maija Tenkanen for useful discussions, and Marjukka Perttula for HPLC analysis.
A part of the work was financed by Genencor International Inc. and the National Technology Agency of Finland (Tekes).
REFERENCES
- 1.Aalto, M. K., H. Ronne, and S. Keränen. 1993. Yeast syntaxins Sso1p and Sso2p belong to a family of related membrane proteins that function in vesicular transport. EMBO J. 12:4095-4103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aro, N., A. Saloheimo, M. Ilmén, and M. Penttilä. 2001. ACEII, a novel transcriptional activator involved in regulation of cellulase and xylanase genes of Trichoderma reesei. J. Biol. Chem. 276:24309-24314. [DOI] [PubMed] [Google Scholar]
- 3.Barnett, C. C., R. M. Berka, and T. Fowler. 1991. Cloning and amplification of the gene encoding an extracellular β-glucosidase from Trichoderma reesei: evidence for improved rates of saccharification of cellulosic substrates. Biotechnology 9:562-567. [DOI] [PubMed] [Google Scholar]
- 4.Chirgwin, J., A. Przybyla, R. MacDonald, and W. Rutter. 1979. Isolation of biologically active ribonucleic acid from sources enriched in ribonuclease. Biochemistry 18:5294-5299. [DOI] [PubMed] [Google Scholar]
- 5.Chirico, W. J., and R. D. Brown, Jr. 1987. Purification and characterization of a β-glucosidase from Trichoderma reesei. Eur. J. Biochem. 165:333-341. [DOI] [PubMed] [Google Scholar]
- 6.Christakopoulos, P., P. W. Goodenough, D. Kekos, B. J. Macris, M. Claeyssens, and M. K. Bhat. 1994. Purification and characterisation of an extracellular β-glucosidase with transglycosylation and exo-glucosidase activities from Fusarium oxysporium. Eur. J. Biochem. 224:379-385. [DOI] [PubMed] [Google Scholar]
- 7.Dion, M., L. Fourage, J. N. Hallet, and B. Colas. 1999. Cloning and expression of a beta-glucosidase from Thermus thermophilus. Sequence and biochemical characterisation of the encoded enzyme. Glycoconj. J. 16:27-37. [DOI] [PubMed] [Google Scholar]
- 8.Fowler, T., and R. D. Brown, Jr. 1992. The bgl1 gene encoding extracellular β-glucosidase from Trichoderma reesei is required for rapid induction of the cellulase complex. Mol. Microbiol. 6:3225-3235. [DOI] [PubMed] [Google Scholar]
- 9.Hansson, T., T. Kaper, J. van der Oost, W. de Vos, and P. Adlercreutz. 2001. Improved oligo-saccharide synthesis by protein engineering of β-glucosidase CelB from hyperthermophilic Pyrococcus furiosus. Biotechnol. Bioeng. 73:203-210. [DOI] [PubMed] [Google Scholar]
- 10.Harman, G., and C. Kubicek. 1998. Trichoderma and Gliocladium. Taylor & Francis, London, United Kingdom.
- 11.Hays, W. S., D. J. van der Jagt, B. Bose, A. S. Serianni, and R. H. Glew. 1998. Catalytic mechanism and specificity for hydrolysis and transglycosylation reactions of cytosolic β-glucosidase from guinea pig liver. J. Biol. Chem. 273:34941-34948. [DOI] [PubMed] [Google Scholar]
- 12.Hofer, F., E. Weissinger, H. Mischak, R. Messner, B. Meixner-Monori, D. Blaas, J. Visser, and C. P. Kubicek. 1989. A monoclonal antibody against the alkaline extracellular β-glucosidase from Trichoderma reesei: reactivity with other Trichoderma β-glucosidases. Biochim. Biophys. Acta 992:298-306. [Google Scholar]
- 13.Ilmen, M., C. Thrane, and M. Penttilä. 1996. The glucose repressor gene cre1 of Trichoderma: isolation and expression of a full-length and a truncated mutant form. Mol. Gen. Genet. 251:451-460. [DOI] [PubMed] [Google Scholar]
- 14.Ilmen, M., A. Saloheimo, M.-L. Onnela, and M. Penttilä. 1997. Regulation of cellulase gene expression in filamentous fungus Trichoderma reesei. Appl. Environ. Microbiol. 63:1298-1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Inglin, M., B. A. Feinberg, and J. R. Loewenberg. 1980. Partial purification and characterization of a new intracellular β-glucosidase of Trichoderma reesei. Biochem. J. 185:515-519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kono, H., S. Kawano, K. Tajima, T. Erata, and M. Takai. 1999. Structural analysis of new tri- and tetrasaccharides produced from disaccharides by transglycosylation of purified Trichoderma viride β-glucosidase. Glycoconj. J. 16:415-423. [DOI] [PubMed] [Google Scholar]
- 17.Kubicek, C. P., R. Messner, F. Gruber, M. Mandels, and E. M. Kubicek-Pranz. 1993. Triggering of cellulase biosynthesis by cellulose in Trichoderma reesei. J. Biol. Chem. 268:19364-19368. [PubMed] [Google Scholar]
- 18.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 19.Loewenberg, J. R. 1984. Sophorose induction of an intracellular β-glucosidase in Trichoderma. Arch. Microbiol. 137:53-57. [DOI] [PubMed] [Google Scholar]
- 20.Lowry, O. H., N. J. Rosebrough, A. L. Farr, and R. J. Randall. 1951. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 193:265-275. [PubMed] [Google Scholar]
- 21.Mach, R. L., B. Seiboth, A. Myasnikov, R. Gonzalez, J. Strauss, A. M. Harkki, and C. P. Kubicek. 1995. The bgl1 gene of Trichoderma reesei QM9414 encodes an extracellular, cellulose-inducible β-glucosidase involved in cellulase induction by sophorose. Mol. Microbiol. 16:687-697. [DOI] [PubMed] [Google Scholar]
- 22.Mackenzie, L. F., Q. Wang, R. A. J. Warren, and S. G. Withers. 1998. Glycosynthases: mutant glycosidases for oligosaccharide synthesis. J. Am. Chem. Soc. 120:5583-5584. [Google Scholar]
- 23.Mandels, M., J. Weber, and R. Parizek. 1971. Enhanced cellulase production by a mutant of Trichoderma viride. Appl. Microbiol. 21:152-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Margolles-Clark, E., M. Tenkanen, T. Nakari-Setälä, and M. Penttilä. 1996. Cloning of genes encoding α-l-arabinofuranosidase and β-xylosidase from Trichoderma reesei by expression in Saccharomyces cerevisiae. Appl. Environ. Microbiol. 21:3840-3846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Margolles-Clark, E., M. Ilmen, and M. Penttilä. 1997. Expression patterns of ten hemicellulase genes of the filamentous fungus Trichoderma reesei on various carbon sources. J. Biotechnol. 57:167-179. [Google Scholar]
- 26.Messner, R., and C. P. Kubicek. 1991. Evidence for a single, specific β-glucosidase in the cell walls from Trichoderma reesei QM9414. Enzyme Microb. Technol. 12:685-690. [Google Scholar]
- 27.Montenecourt, B. S., and D. E. Eveleigh. 1979. Selective screening methods for the isolation of high yielding cellulase mutants of Trichoderma reesei. Adv. Chem. Ser. 181:289-301. [Google Scholar]
- 28.Paavilainen, S., J. Hellman, and T. Korpela. 1993. Purification, characterization, gene cloning, and sequencing of a new β-glucosidase from Bacillus circulans subsp. alkalophilus. Appl. Environ. Microbiol. 59:927-993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Penttilä, M., H. Nevalainen M. Rättö, E. Salminen, and J. K. C. Knowles. 1987. A versatile transformation system for the cellulolytic filamentous fungus Trichoderma reesei. Gene 61:155-164. [DOI] [PubMed] [Google Scholar]
- 30.Punt, P. J., M. A. Dingemanse, A. Kuyvenhoven, R. D. Soede, P. H. Pouwels, and C. A. M. J. J. van den Hondel. 1990. Functional elements in the promoter region of the Aspergillus nidulans gpdA gene encoding glyceraldehyde-3-phosphate dehydrogenase. Gene 93:101-109. [DOI] [PubMed] [Google Scholar]
- 31.Saloheimo, A., B. Henrissat, A.-M. Hoffren, O. Teleman, and M. Penttilä. 1994. A novel, small endoglucanase gene, egl5, from Trichoderma reesei isolated by expression in yeast. Mol. Microbiol. 13:219-228. [DOI] [PubMed] [Google Scholar]
- 32.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 33.Sanz-Aparicio, J., J. A. Hermoso, M. Martinez-Ripoll, J. L. Lequerica, and J. Polaina. 1998. Crystal structure of beta-glucosidase from Bacillus polymyxa: insights into the catalytic activity in family 1 glycosyl hydrolases. J. Mol. Biol. 275:491-502. [DOI] [PubMed] [Google Scholar]
- 34.Sheir-Ness, G., and B. S. Montenecourt. 1984. Characterization of the secreted cellulases of Trichoderma reesei wild type and mutants during controlled fermentations. Appl. Microbiol. Biotechnol. 20:46-53. [Google Scholar]
- 35.Takashima, S., A. Nakamura, M. Hidaka, H. Msaki, and T. Uozumi. 1999. Molecular cloning and expression of the novel fungal β-glucosidase genes from Humicola grisea and Trichoderma reesei. J. Biochem. 125:728-736. [DOI] [PubMed] [Google Scholar]
- 36.Takkinen, K., M.-L. Laukkanen, D. Sizmann, K. Alftan, T. Immonen, L. Vanne, M. Kaartinen, J. K. C. Knowles, and T. T. Teeri. 1991. An active single-chain antibody containing a cellulase linker domain is secreted from Escherichia coli. Protein Eng. 4:837-841. [DOI] [PubMed] [Google Scholar]
- 37.Tenkanen, M., M. Makkonen, M. Perttula, L. Viikari, and A. Teleman. 1997. Action of Trichoderma reesei mannanase on galactoglucomannan in pine kraft pulp. J. Biotechnol. 55:191-204. [DOI] [PubMed] [Google Scholar]
- 38.Umile, C., and C. P. Kubicek. 1986. A constitutive, plasma membrane-bound β-glucosidase in Trichoderma reesei. FEMS Microbiol. Lett. 34:291-295. [Google Scholar]
- 39.von Heijne, G. 1986. A new method for predicting signal sequence cleavage sites. Nucleic Acids Res. 14:4683-4690. [DOI] [PMC free article] [PubMed] [Google Scholar]