Abstract
The chlorophenol degradation pathway in Sphingobium chlorophenolicum is initiated by the pcpB gene product, pentachlorophenol-4-monooxygenase. The distribution of the gene was studied in a phylogenetically diverse group of polychlorophenol-degrading bacteria isolated from contaminated groundwater in Kärkölä, Finland. All the sphingomonads isolated were shown to share pcpB gene homologs with 98.9 to 100% sequence identity. The gene product was expressed when the strains were induced by 2,3,4,6-tetrachlorophenol. A comparative analysis of the 16S rDNA and pcpB gene trees suggested that a recent horizontal transfer of the pcpB gene was involved in the evolution of the catabolic pathway in the Kärkölä sphingomonads. The full-length Kärkölä pcpB gene allele had approximately 70% identity with the three pcpB genes previously sequenced from sphingomonads. It was very closely related to the environmental clones obtained from chlorophenol-enriched soil samples (M. Beaulieu, V. Becaert, L. Deschenes, and R. Villemur, Microbiol. Ecol. 40:345-355, 2000). The gene was not present in polychlorophenol-degrading nonsphingomonads isolated from the Kärkölä source.
Introduction of new xenobiotic chemicals forces microorganisms to develop metabolic pathways in order to exploit new carbon sources and to detoxify toxic compounds. Understanding this evolution is crucial for the successful implementation of bioremediation. Polychlorinated phenols (trichlorophenol [TCP], tetrachlorophenol [TeCP], and pentachlorophenol [PCP]) have been widely used in agriculture and in the timber industry as preservatives against rot and bluestaining of wood since the 1920s. Chlorophenols, particularly those with one or two chlorines, are produced naturally by certain fungi and insects (9), but no evidence of biological sources of PCP has been reported so far. It has been suggested, therefore, that the PCP degradation pathway has been assembled during the past few decades since the anthropogenic introduction of PCP into the environment (5).
The complete degradation pathway and the key enzymes for microbial degradation of PCP are known in detail only in Sphingomonas chlorophenolica (14, 25, 26, 40, 42). The genus Sphingomonas has recently been divided into four genera: Sphingomonas, Sphingobium, Novosphingobium, and Sphingopyxis (34), and Sphingomonas chlorophenolica has been reclassified as Sphingobium. In this study, we refer to these four Sphingomonas-derived genera together and to their close relatives Rhizomonas, Blastomonas, and Erythromonas as sphingomonads. The rate-limiting step for PCP degradation in Sphingobium chlorophenolicum is the parahydroxylation of PCP to tetrachlorohydroquinone (20). This step is catalyzed by the enzyme PCP-4-monooxygenase, encoded by the pcpB gene (26). In addition to PCP, the PCP-4-monooxygenase enzyme can use TCP, TeCP, and many other halogenated phenols as a substrate (41).
Sequence analysis of homologous genes from sphingomonads and other organisms should reveal the evolution of the pcpB gene. Identical pcpB gene sequences have been found in the three of the four known strains of S. chlorophenolicum (ATCC 39723, SR3, and RA2) isolated from chlorophenol-contaminated sites in different regions of the United States (6). A pcpB gene of the fourth S. chlorophenolicum strain (ATCC 33790) differed by 10% from these sequences in a way that did not support the phylogeny drawn from the 16S ribosomal DNA (rDNA) sequences (6). The third variant of the pcpB gene was sequenced from a chlorophenol- and nitrophenol-degrading soil isolate, “Sphingomonas” sp. strain UG30 (3, 15, 16). The sequence similarity between the UG30 and ATCC 39723 pcpB genes was 90% and between the UG30 and ATCC 33790 pcpB genes was 89%. Interestingly, a pcpB gene homolog highly similar (over 98% similarity) to that of S. chlorophenolicum ATCC 39723 has been sequenced from two nondegrading β- and γ-proteobacterial strains isolated from soil samples from a PCP-contaminated wood treatment site (33).
The metabolism of polychlorinated phenols has been thought to be a feature supported by a few bacterial genera (11). However, a previous study on the microbiology of groundwater contaminated over a long period in Kärkölä, Finland, revealed that the ability to degrade polychlorophenols was widely distributed among α-, β- and γ-proteobacteria, the Cytophaga/Flexibacter/Bacteroides group, and gram-positive bacteria (18). The isolates degraded 2,3,4,6-TeCP and 2,4,6-TCP, and some of them also degraded PCP (18). Several chlorophenol-degrading Novosphingobium strains have also been isolated from fluidized-bed bioreactors treating the Kärkölä groundwater (29, 37). Novosphingobium sp. strain MT1 was shown to carry a pcpB gene homolog (37). In the present study, our aim was to assess the role of horizontal gene transfer in the evolution of polychlorophenol-degrading strains in Kärkölä. In addition, a number of other strains were studied to analyze the overall distribution of the pcpB gene. Surprisingly, highly identical pcpB alleles were determined in all the chlorophenol-degrading sphingomonads, suggesting a recent transfer of this gene in situ in the Kärkölä source. However, the pcpB gene transfer was not detected in other bacterial groups.
MATERIALS AND METHODS
Bacterial strains.
The bacterial strains used for the phylogenetic study are presented in Table 1. In addition, the strains listed below were studied in the dot blot hybridization and PCR analyses.
TABLE 1.
Bacterial strains used in this study for phylogenetic analysis
| Bacterial straina | Site of isolation | Reference(s) | EMBL accession no.
|
|
|---|---|---|---|---|
| 16S rDNA | pcpB | |||
| S. chlorophenolicum ATCC 39723 | Minnesota | 22, 28 | X87163 | M98557 |
| S. chlorophenolicum ATCC 33790T | North America | 7, 22 | X87161 | U60175 |
| “Sphingomonas” sp. strain UG30 | Ontario, Canada | 15 | AF170090 | AF059680 |
| Sphingobium sp. strain K74 | Kärkölä, Finland | 18 | AJ009709 | AJ437487c |
| Rhizomonas sp. strain K6 | Kärkölä, Finland | 18 | AJ000918c | AJ437483c |
| Sphingobium sp. strain K40 | Kärkölä, Finland | 18 | AJ009708c | AJ437486c |
| Sphingomonas sp. strain K101 | Kärkölä, Finland | 18 | AJ009706 | AJ437488c |
| N. subarcticum KF1T | Kärkölä,b Finland | 23, 29 | X94102 | AJ437489c |
| Novosphingobium sp. strain MT1 | Kärkölä, Finland | 37 | AJ303009 | AJ319678c |
| Novosphingobium sp. strain K16 | Kärkölä, Finland | 18 | AJ000920c | AJ437484c |
| Novosphingobium sp. strain K39 | Kärkölä, Finland | 18 | AJ009707c | AJ437485c |
Nomenclature of the strains revised according to reference 34.
The strain was isolated from a bioreactor that was fed with Kärkölä groundwater.
Sequences obtained or expanded in this study.
Polychlorophenol-degrading nonsphingomonads isolated from Kärkölä groundwater inside the contaminated plume (18) follow: α-proteobacterial isolates K13 and K31; β-proteobacterial isolates K1, K8, and K33; γ-proteobacterial isolates K27 and K104; isolates belonging to the Cytophaga/Flexibacter/Bacteroides group (K66 and K112); and gram-positive isolates with high G+C content (K44 and K103). Classification of these isolates was based on fatty acid analysis and 16S rDNA sequencing (18).
Other polychlorophenol-degrading strains follow: S. chlorophenolicum RA2 (22, 31), S. chlorophenolicum SR3 (22, 32), Novosphingobium subarcticum NKF1 (23, 29), N. subarcticum KF3 (23, 29), and Mycobacterium chlorophenolicum DSM 43826T (10).
Sphingomonads (nondegraders) isolated from the Kärkölä groundwater outside the plume (19) follow: isolates K203, K210, K213, K222, K228, K230, and K232. These isolates were identified as sphingomonads using fatty acid analysis (19).
Reference sphingomonads not known to degrade polychlorophenols follow: Sphingomonas sp. strain HV1, Sphingomonas sp. strain RW5, Sphingomonas sp. strain RW16, Sphingomonas sp. strain RW100, Sphingomonas sp. strain SS2, Sphingomonas sp. strain SS3, Sphingomonas sp. strain A175, Sphingomonas sp. strain B1, Sphingomonas xenophaga BN6T, Sphingomonas pituitosa EDIVT, Sphingomonas sp. strain HH69-3, Sphingomonas wittichii RW1T, Sphingomonas adhaesiva DSM 7418T, Novosphingobium capsulatum DSM 30196T, Novosphingobium roseae DSM 7285T, Novosphingobium stygium DSM 12445T, Novosphingobium subterraneum DSM 12447T, Sphingopyxis macrogoltabidus IFO 15033T, Sphingomonas mali IFO 15500T, Sphingomonas parapaucimobilis DSM 7463, Sphingomonas paucimobilis ATCC 29837T, Sphingomonas paucimobilis EPA 505, Sphingomonas pruni IFO 15498T, Sphingomonas sanguinis IFO 13937T, Sphingopyxis terrae IFO 15098T, Sphingobium yanoikuyae DSM 7462T, and Sphingomonas asaccharolytica IFO 15499T.
Other reference strains not known to degrade polychlorophenols follow: Alcaligenes faecalis ATCC 11624T, Pseudomonas aureofaciens CCEB 513, Pseudomonas fluorescens ATCC 13525, Escherichia coli DSM 682, Helicobacter pylori 4105, Flavobacterium ferrugineum DSM 30193T, Cytophaga flevensis DSM 1076T, Bacillus pabuli DSM 3036T, Rhodococcus erythropolis DSM 43135T, Sporomusa ovata DSM 2662T, Arthrobacter citreus ATCC 11624T, Arthrobacter viscosus DSM 20159, Fusobacterium nucleatum ATCC 22586, Microcystis sp. strain PCC 7005, Deinococcus proteolyticus DSM 20540T, and Halobacterium salinarum DSM 668.
Bacterial cultivation and DNA extraction.
Bacterial strains were cultivated to the stationary phase in peptone-yeast-glucose medium (0.25 mg of glucose/liter, 0.25 mg of yeast extract/liter, and 0.25 mg of peptone/liter) or in the appropriate growth medium recommended by DSMZ (Deutsche Sammlung von Mikroorganismen und Zellkulturen, Braunschweig, Germany). The cells were crushed by bead milling, and the DNA was purified with phenol-chloroform extractions and isopropanol precipitation, as previously described (18). For dot blot DNA hybridization analysis, the amount of DNA was measured fluorometrically (Fluoroskan; Labsystems, Helsinki, Finland) using Hoechst Dye H33258 (Sigma) as recommended by the manufacturer of the fluorometer. For the other experiments, the amount of DNA was measured spectrophotometrically (GeneQuant; Bio-Rad Laboratories, Hercules, Calif.).
DNA hybridization analyses.
For the dot blot analysis, 250 ng of purified DNA was heated at 95°C for 10 min and transferred with a dot blot apparatus (Schleicher & Schull, Dessel, Germany) onto a Qiabrane Nylon Plus membrane (Qiagen GmbH, Hilden, Germany). For Southern blot analysis, 1 or 2 μg of total EcoRI-digested DNA was electrophoresed on a 1% agarose gel and transferred overnight using an alkaline transfer procedure (4) onto the Qiabrane Nylon Plus membrane. Preparation of the probes, hybridization, and chemiluminescence disodium-3-phenylphosphate detection of the signal were performed with a digoxigenin DNA labeling and detection kit according to the manufacturer's instructions (Boehringer Mannheim, Mannheim, Germany). The double-stranded DNA probe pcpB-5 specific for the PCP-4-monooxygenase gene was generated by PCR using primers that amplified the region corresponding to nucleotides 444 to 616 from the beginning of the reading frame of the pcpB gene (EMBL accession no. M98557). The pcpB template, plasmid pCCL3 (26), was kindly provided by R. Crawford (University of Idaho, Moscow). Overnight hybridization was performed at 65°C for the dot blot hybridizations and 63.5°C for the Southern blot hybridizations.
PCR, sequencing, and phylogenetic analysis.
A fragment of about 700 bp of the pcpB gene was amplified using the degenerate primer pair pcpB-G and pcpB-D2 (Table 2). The product was cloned and sequenced from strains K6, K16, K39, K49, K74, K101, and N. subarcticum KF1T. The partial sequence of the Novosphingobium sp. strain MT1 pcpB gene (37) (EMBL accession no. AJ319678) was extended to full length by cloning the first and last parts of the gene by an inverse PCR method (24). Briefly, genomic DNA was digested with the restriction enzyme PstI and the digestion was subsequently self ligated. The PCR amplification of the self-ligated PstI fragments was facilitated using the MT1 pcpB-specific primer pairs pcpB-D3 and pcpB-E2 and pcpB-D3 and pcpB-E3. Finally, the full-length MT1 pcpB gene was amplified using primers pcpB-F1 and pcpB-F2 designed according to the sequence data yielded from the cloned fragments of the previous steps. Partial 16S rDNA sequences (∼440 bp) previously obtained for the strains K6, K16, K39, K40, and K101 (18) were extended to near-completion by cloning and sequencing PCR-amplified fragments (fragments corresponding to E. coli numbering [2] positions 319 to 1058 and 968 to 1541). DynaZyme F501-L polymerase and the PCR protocol previously described (18) were used in all the PCRs except sequencing. For the sequencing, PCR products were extracted from agarose gel using Ultrafree-DA columns (Millipore, Bedford, Mass.), ligated to the pGEM-T vector (Promega, Madison, Wis.), transformed into E. coli JM109 cells, and analyzed by bidirectional sequencing using the LI-COR DNA4200 sequencer (LI-COR, Lincoln, Nebr.) with vector primers T7 and SP6. Sequences were aligned, and neighbor-joining trees were constructed using ClustalX, version 1.7 (36), and the tree was displayed using the Treeview program (27). Amino acid sequences were predicted and aligned using the DNAman program package (Lynnon Biosoft, Vaudreuil, Quebec, Canada). Nucleic acid and amino acid sequence identity values were analyzed using the programs BLAST and GAP in the GCG program package (Genetic Computer Group, Madison, Wis.).
TABLE 2.
Sequences of PCR primers used for amplification of the pcpB gene
| Primer | Sequence | Direction | Target sitea | Source or reference |
|---|---|---|---|---|
| PcpB-G | 5′-GGSTTCACSTTCAAYTTCGA-3′ | Forward | 262-281 | 1 |
| PcpB-D2 | 5′-TCCTGCATSCCSACRTTCAT-3′ | Reverse | 946-965 | 1 |
| PcpB-D3 | 5′-ACGTTGCATCGAGATGCTG-3 | Reverse | 382-400 | This study |
| PcpB-E2 | 5′-CGACTGGATTCACTATTTCAT-3′ | Forward | 646-666 | This study |
| PcpB-E3 | 5′-CGCTGGCGACCGCGTATC-3′ | Forward | 1259-1276 | This study |
| PcpB-F1 | 5′-CTGCAGTTACACTAACAATG-3′ | Forward | −35-−16 | This study |
| PcpB-F2 | 5′-CGACGTCGATCATATTCG-3′ | Reverse | 1652-1669 | This study |
Corresponds to the nucleotide numbering from the beginning of the open reading frame of the Novosphingobium sp. strain MT1 pcpB gene (AJ319678).
Immunoblot analysis.
Production of PCP-4-monooxygenase was induced in cells grown at room temperature (22 ± 1°C) for 4 days in peptone-yeast-glucose medium in a rotary shaker (125 rpm/min). 2,3,4,6-TeCP was added to the culture mixture to obtain a final concentration of 10 mg/liter, and incubation was continued for 16 h. Boiled bacterial suspensions were assayed by conventional sodium dodecyl sulfate-10% polyacrylamide gel electrophoresis, and the proteins were transferred onto a nitrocellulose filter using a Mini-Protean II apparatus (Bio-Rad). The membranes were then blocked by incubation at room temperature in 5% dry skimmed milk in phosphate-buffered saline containing 0.1% Tween 20 for 1 h. After washing, the blots were incubated with 0.1 μg of affinity-purified polyclonal rabbit anti-PCP-4-monooxygenase/ml (38). The secondary antibody incubation was performed with goat anti-mouse immunoglobulin G horseradish peroxidase conjugate (Bio-Rad) (1:3,000 dilution). Three washes with phosphate-buffered saline containing 0.1% Tween 20 were performed between the incubations. Chemiluminescence detection was performed using SuperSignal substrate (Pierce, Rockford, Ill.).
Nucleotide sequence accession numbers.
The new pcpB gene sequences obtained for the first time in this study were deposited in the EMBL database under accession numbers AJ437483 to AJ43789.
RESULTS
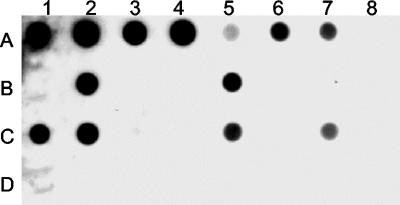
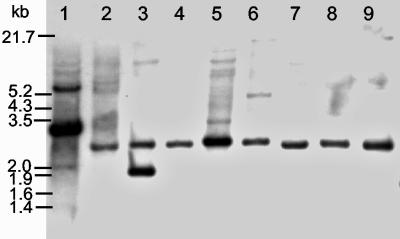
Dot blot hybridization (Fig. 1) and PCR were performed to reveal the existence of the pcpB gene homologs in sphingomonads and other bacteria. All the bacteria mentioned in Materials and Methods and Table 1 were included in the analyses except “Sphingomonas” sp. strain UG30 and Novosphingobium sp. strain MT1, which were not available for us at the time of these analyses. The only pcpB-positive strains in these two tests were the four S. chlorophenolicum strains (ATCC 39723, ATCC 33790T, RA2, and SR3) isolated from chlorophenol-contaminated sites in the United States; the N. subarcticum strains KF1T, KF3, and NKF1, isolated from bioreactors treating the Kärkölä groundwater; and the six polychlorophenol-degrading sphingomonads K6, K16, K39, K40, K74, and K101, isolated from Kärkölä inside the chlorophenol plume. All the strains that were negative in the dot blot assay were also negative in the PCR analysis (data not shown). Selection of the pcpB-positive strains (Fig. 2) and Novosphingobium sp. strain MT1 were assayed using Southern blot hybridization analysis of EcoRI-digested genomic DNA. All the four S. chlorophenolicum strains are known to have the pcpB gene in 3.0-kb EcoRI loci (6), as was again shown here by strain ATCC 39723. No genetic or phenotypic difference between N. subarcticum strains KF1T, KF3, and NKF1 has been shown (23), and therefore the analysis was done only for the type strain of this species. All the pcpB-positive K strains (K6, K16, K39, K40, K74, and K101), N. subarcticum KF1T, and Novosphingobium sp. strain MT1 shared a common hybridizing EcoRI fragment, approximately 2.5 kb in length (Fig. 2). Additionally, strain K6 had a hybridizing EcoRI fragment of about 1.9 kb.
FIG. 1.
Dot blot DNA hybridization analysis to detect strains carrying the pcpB gene. The genomic DNA (250 ng) was spotted onto a nylon membrane, and the blot was probed for the pcpB gene. Lanes: for row A, 1, S. chlorophenolicum ATCC 39723; 2, S. chlorophenolicum ATCC 33790T; 3, S. chlorophenolicum RA2; 4, S. chlorophenolicum SR3; 5, N. subarcticum KF1T; 6, N. subarcticum KF3; and 7, N. subarcticum NKF1; for row B, 1, K1; 2, K6; 3, K8; 4, K13; 5, K16; 6, K27; 7, K31; and 8, K33; for row C, 1, K39; 2, K40, 3, K44; 4, K66; 5, K74; 6, K76; 7, K101; and 8, K103; and for row D, 1; K104; and 2, K112.
FIG. 2.
Southern blot analysis of genomic DNA from chlorophenol-degrading sphingomonads digested with EcoRI. Lane 1, S. chlorophenolicum ATCC 39723; lane 2, N. subarcticum KF1T; lane 3, strain K6; lane 4, strain K16; lane 5, strain K39; lane 6, strain K40; lane 7, strain K74; lane 8, strain K101; and lane 9, Novosphingobium sp. strain MT1. The number of kilobases is given on the left.
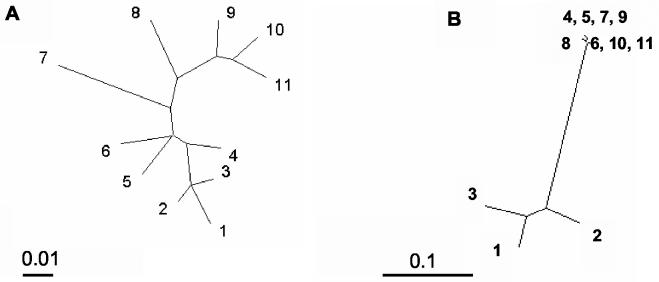
The strains shown in Table 1 were subject for the phylogenetic analysis of 16S rRNA gene and the pcpB gene. Figure 3A and B show phylogenetic trees based on the nucleotide sequences of the 1,369-bp aligned positions of the 16S rRNA gene and 625-bp aligned positions of the pcpB gene. The analysis revealed that the sphingomonads isolated from the Kärkölä groundwater were phylogenetically distinct from each other. In contrast to their positions in the 16S rDNA tree, the strains shared pcpB gene homologs that were highly identical, showing 98.9 to 100% sequence identity within the aligned positions. However, the Kärkölä pcpB allele was different from the three previous alleles of pcpB homologs sequenced from S. chlorophenolicum strains and “Sphingomonas” sp. strain UG30, as shown in Fig. 3B. The Kärkölä pcpB alleles showed great similarity (from 97 to 100% identity) with several environmental pcpB clones from chlorophenol-enriched soil reactors in Canada (1), although it was not known from which organisms these sequences were derived.
FIG. 3.
Phylogenetic trees of chlorophenol-degrading sphingomonads based on differences in the nucleotide sequences of 16S rRNA (A) and the pcpB gene (B). Strain numbering is presented in Table 2. Scale bars indicate 0.01 and 0.1 nucleotide substitutions per position.
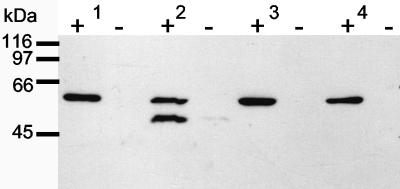
The expression of pcpB gene homologs was confirmed by immunoblot analysis with a polyclonal antibody that was raised using recombinant PCP-4-monooxygenase as an immunogen (38). Following induction by 2,3,4,6-TeCP, a distinct band corresponding to the size of PCP-4-monooxygenase (60 kDa) of S. chlorophenolicum ATCC 39723 was visible in the Kärkölä sphingomonads (strains ATCC 39723, K6, K16, K39, K40, K74, K101, and KF1T were tested). Another, approximately 52-kDa band cross-reacted in N. subarcticum KF1T, as shown in Fig. 4. No band was detected with uninduced bacteria. The results demonstrate that the pcpB genes, which were found in strains isolated from Kärkölä, were functional and that their expression was regulated by chlorophenols.
FIG. 4.
Western blot analysis used to detect expression of PCP-4-monooxygenase with polyclonal antibody. Sample 1, S. chlorophenolicum ATCC 39723; sample 2, N. subarcticum KF1T, sample 3, strain K16, and sample 4, strain K101. Duplicate samples were incubated with (+) and without (−) 2,3,4,6-TeCP. kDa, kilodaltons.
The full-length pcpB gene was sequenced from Novosphingobium sp. strain MT1. The predicted open reading frame of the gene consisted of 1,632 nucleotides, and the deduced amino acid sequence was 543 amino acids long with a predicted molecular mass of 60.2 kDa. The full-length gene had 68.2 to 70.0% sequence identity with the pcpB gene sequences from the S. chlorophenolicum ATCC 39723, ATCC 33790, and “Sphingomonas” sp. strain UG30 strains and 69.1 to 70.3% amino acid sequence identity with the corresponding gene products. p-Hydroxybenzoate hydroxylase has been used as a model by which the structure and function of phenolic monooxygenases have been explained (8). The sequence alignments of the deduced amino acid sequences of p-hydroxybenzoate hydroxylase and PCP-4-monooxygenases revealed two highly conserved domains that have been shown to be involved in binding flavin adenine dinucleotide in p-hydroxybenzoate hydroxylase. The first motif, GlyXGlyXXGly, is located in the NH2-terminal portion of these proteins, and the second domain is located around residue 300 of PCP-4-monooxygenases (sites shown in Fig. 5).
FIG. 5.
Alignment of the predicted amino acid sequences for the S. chlorophenolicum ATCC 39723 (EMBL accession no. M98557) and Novosphingobium sp. strain MT1 (AJ319678) pcpB homologs. The amino acids that are located in the flavin adenine dinucleotide-binding domains and conserved in these two sequences and hydroxybenzoate p-hydroxylase are underlined.
DISCUSSION
Horizontal gene transfer has been considered a principal source of bacterial evolution (13). Potential for the gene transfer can be studied using in vitro experiments, and deductive evidence for gene exchange can be obtained from comparative analyses of nucleotide sequences, codon usage, and enzyme patterns (17). This study suggests deductively that a recent horizontal gene transfer has facilitated chlorophenol degradation among various sphingomonads in the Kärkölä groundwater site. The phylogenetic trees that were based on the 16S rDNA and pcpB gene sequences of the eight Kärkölä strains were incongruous. Similar evidence of natural horizontal transfer of degradative genes has previously been obtained, e.g., in the studies on naphthalene degraders (12) and 2,4-dichlorophenoxyacetic acid degraders (21). However, this is first evidence of the role of horizontal gene transfer in the evolution of polychlorophenol degraders.
Worldwide, four functional pcpB variants are presently known serving in the same role in sphingomonads. The Kärkölä type of pcpB gene is not totally new, since it shows great similarity to the environmental clones sequenced by Beaulieu et al. (1). The sequence data are supported by the results from the hybridization analysis. All the Kärkölä sphingomonads shared a common hybridizing EcoRI band that was 2.5 kb in length, showing conservation of the regions also outside the pcpB gene in these strains.
A large number of other sphingomonads, chlorophenol degraders, and nondegrading control strains were investigated in this study to analyze the distribution of the pcpB gene in various groups of organisms. On the basis of the hybridization and PCR results, the pcpB gene was detected only in chlorophenol-degrading sphingomonads and not in the other bacteria. It seems that a taxonomic barrier has restricted the gene transfer in Kärkölä, since the pcpB gene homolog was not present among the 11 polychlorophenol-degrading nonsphingomonads isolated from the groundwater inside the plume. These strains were selected on a phylogenetic basis to include representative bacteria from α-, β-, and γ-proteobacteria, bacteria from the Cytophaga/Flexibacter/Bacteroides group, and gram-positive bacteria with high G+C content (18). Long-term persistence of horizontally transferred genes is likelier if the genes confer a selective advantage on the recipient organisms (13). It has been suggested that the PCP degradation pathway needs enzymes recruited from two separate catabolic pathways (5). Saboo and Gealt (33) observed pcpB gene sequences in β- and γ-proteobacterial strains, but this property did not lead to the ability to degrade chlorophenols in these organisms. Therefore, in the nonsphingomonads the reason for the “lack” of the observed horizontal transfer may also be a consequence of the lack of the second pathway needed to supply a complete metabolism that could support gene fixation.
The success of chlorophenol attenuation and bioremediation processes in different locations is dependent on the adaptability of the bacteria. In Kärkölä, chlorophenol contamination has been ongoing at least for 25 years since the surrounding sawmill was destructed by fire in 1976. This and other studies (18, 19) on the Kärkölä microbiota have revealed a high diversity in chlorophenol-degrading sphingomonads and other bacteria indicating bacterial adaptation to chlorophenols. The negative hybridization results obtained from the sphingomonads isolated outside the Kärkölä plume support the hypothesis that the ability to degrade polychlorophenols was not initially present in them but arose relatively recently through horizontal gene transfer. As an oligotrophic environment with chlorophenols as the primary carbon source (37), the selective pressure in the Kärkölä groundwater may have been especially favorable for gene selection, transfer, and proliferation. However, the low oxygen concentration of the groundwater seems to be the reason for delayed chlorophenol attenuation (30).
So far it is not known which is the mechanism that has enabled the horizontal transfer of the pcpB gene in Kärkölä sphingomonads. Natural gene transfer among bacteria can occur via plasmids, chromosomal gene mobilization, transduction, transformation, and transposition (39). Many pathways for the degradation of xenobiotic compounds are encoded by plasmids, and involvement of a plasmid in the degradation of pentachlorophenol by a Pseudomonas sp. has recently been suggested (35). However, the pcpB gene was not found to be part of an extensive operon or to be present on the 100-kb endogeneous plasmid of S. chlorophenolicum ATCC 39723 (25), and despite several trials, we did not find catabolic plasmids in the Kärkölä sphingomonads either (unpublished observations). Furthermore, an interesting question raising from the results of this study follows: what is the original host organism of the mobile element carrying the pcpB gene in the uncontaminated environment? This question remains to be studied in the future.
Acknowledgments
This study was supported by the Graduate School for Environmental Ecology, Ecotoxicology and Engineering (University of Jyväskylä), the Academy of Finland, and the Jenny and Antti Wihuri Foundation.
We thank Irene Helkala for technical assistance and Sini Suomalainen for helping in the sequencing.
REFERENCES
- 1.Beaulieu, M., V. Becaert, L. Deschenes, and R. Villemur. 2000. Evolution of bacterial diversity during enrichment of PCP-degrading activated soils. Microbiol. Ecol. 40:345-355. [DOI] [PubMed] [Google Scholar]
- 2.Brosius, J., M. L. Palmer, J. P. Kennedy, and H. F. Noller. 1978. Complete nucleotide sequence of the ribosomal RNA gene from Escherichia coli. Proc. Natl. Acad. Sci. USA 75:4801-4805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cassidy, M. B., H. Lee, J. T. Trevors, and R. B. Zablotowicz. 1999. Chlorophenol and nitrophenol metabolism by Sphingomonas sp. UG30. J. Ind. Microbiol. Biotechnol. 23:232-241. [DOI] [PubMed] [Google Scholar]
- 4.Chomczynski, P. 1992. One-hour downward alkaline capillary transfer for blotting of DNA and RNA. Anal. Biochem. 201:134-139. [DOI] [PubMed] [Google Scholar]
- 5.Copley, S. D. 2000. Evolution of a metabolic pathway for degradation of a toxic xenobiotic: the patchwork approach. Trends Biochem. Sci. 25:261-265. [DOI] [PubMed] [Google Scholar]
- 6.Ederer, M. M., R. L. Crawford, R. P. Herwig, and C. S. Orser. 1997. PCP degradation is mediated by closely related strains of the genus Sphingomonas. Mol. Ecol. 6:39-49. [DOI] [PubMed] [Google Scholar]
- 7.Edgehill, R. U., and R. K. Finn. 1982. Isolation, characterization and growth kinetics of bacteria metabolizing pentachlorophenol. Appl. Microbiol. Biotechnol. 16:179-184. [Google Scholar]
- 8.Entsch, B., and W. J. van Berkel. 1995. Structure and mechanism of para-hydroxybenzoate hydroxylase. FASEB J. 9:476-483. [DOI] [PubMed] [Google Scholar]
- 9.Gribble, G. W. 1996. Naturally occurring organohalogen compounds—a comprehensive study, p. 1-420. In G. W. Herz, R. E. Kirby, R. E. Moore, W. Steglich, and C. H. Tamm (ed.), Progress in the chemistry of organic natural products. Springer-Verlag, Vienna, Austria. [DOI] [PubMed]
- 10.Häggblom, M. M., L. J. Nohynek, and M. S. Salkinoja-Salonen. 1988. Degradation and O-methylation of chlorinated phenolic compounds by Rhodococcus and Mycobacterium strains. Appl. Environ. Microbiol. 54:3043-3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Häggblom, M. M., and R. J. Valo. 1995. Bioremediation of chlorophenol wastes, p. 389-434. In L. Y. Young and C. E. Cerniglia (ed.), Microbial transformation and degradation of toxic organic chemicals. John Wiley & Sons, Inc., New York, N.Y.
- 12.Herrick, J. B., K. G. Stuart-Keil, W. C. Ghiorse, and E. L. Madsen. 1997. Natural horizontal transfer of a naphthalene dioxygenase gene between bacteria native to a coal tar-contaminated field site. Appl. Environ. Microbiol. 63:2330-2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Koonin, E. V., K. S. Makarova, and L. Aravind. 2001. Horizontal gene transfer in prokaryotes: quantification and classification. Annu. Rev. Microbiol. 55:709-742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lange, C. C., B. J. Schneider, and C. S. Orser. 1996. Verification of the role of PCP 4-monooxygenase in chlorine elimination from pentachlorophenol by Flavobacterium sp. strain ATCC 39723. Biochem. Biophys. Res. Commun. 219:146-149. [DOI] [PubMed] [Google Scholar]
- 15.Leung, K. T., M. B. Cassidy, K. W. Shaw, H. Lee, J. T. Trevors, E. M. Lohmeier-Vogel, and H. J. Vogel. 1997. Pentachlorophenol biodegradation by Pseudomonas spp. UG25 and UG30. World J. Microbiol. Biotechnol. 13:305-313. [Google Scholar]
- 16.Leung, K. T., S. Campbell, Y. Gan, D. C. White, H. Lee, and J. T. Trevors. 1999. The role of the Sphingomonas species UG30 pentachlorophenol-4-monooxygenase in p-nitrophenol degradation. FEMS Microbiol. Lett. 173:247-253. [DOI] [PubMed] [Google Scholar]
- 17.Lorenz, M. G., and W. Wackernagel. 1994. Bacterial gene transfer by natural genetic transformation in environment. Microbiol. Rev. 58:563-602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Männistö, M. K., M. A. Tiirola, M. Salkinoja-Salonen, M. S. Kulomaa, and J. A. Puhakka. 1999. Diversity of chlorophenol degrading bacteria isolated from contaminated boreal groundwater. Arch. Microbiol. 171:189-197. [DOI] [PubMed] [Google Scholar]
- 19.Männistö, M. K., M. S. Salkinoja-Salonen, and J. A. Puhakka. 2001. In situ polychlorophenol bioremediation potential of the indigenous bacterial community of boreal groundwater. Water Res. 35:2496-2504. [DOI] [PubMed] [Google Scholar]
- 20.McCarthy, D. L., A. A. Claude, and S. D. Copley. 1997. In vivo levels of chlorinated hydroquinones in a pentachlorophenol-degrading bacterium. Appl. Environ. Microbiol. 63:1883-1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McGowan, C., R. Fulthorpe, A. Wright, and J. M. Tiedje. 1998. Evidence for interspecies gene transfer in the evolution of 2,4-dichlorophenoxyacetic acid degraders. Appl. Environ. Microbiol. 64:4089-4092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nohynek, L. J., E. L. Suhonen, E. L. Nurmiaho-Lassila, J. Hantula, and M. Salkinoja-Salonen. 1995. Description of four pentachlorophenol-degrading bacterial strains as Sphingomonas chlorophenolica sp. nov. Syst. Appl. Microbiol. 18:527-538. [Google Scholar]
- 23.Nohynek, L. J., E. L. Nurmiaho-Lassila, E. L. Suhonen, H. J. Busse, M. Mohammadi, J. Hantula, F. Rainey, and M. S. Salkinoja-Salonen. 1996. Description of chlorophenol-degrading Pseudomonas sp. strains KF1T, KF3, and NKF1 as a new species of the genus Sphingomonas, Sphingomonas subarctica sp. nov. Int. J. Syst. Bacteriol. 46:1042-1055. [DOI] [PubMed] [Google Scholar]
- 24.Ochman, H., A. S. Gerber, and D. L. Hartl. 1988. Genetic applications of an inverse polymerase chain reaction. Genetics 120:621-623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Orser, C. S., and C. C. Lange. 1994. Molecular analysis of pentachlorophenol degradation. Biodegradation 5:277-288. [DOI] [PubMed] [Google Scholar]
- 26.Orser, C. S., C. C. Lange, L. Xun, T. C. Zahrt, and B. J. Schneider. 1993. Cloning, sequence analysis, and expression of the Flavobacterium pentachlorophenol-4-monooxygenase gene in Escherichia coli. J. Bacteriol. 175:411-416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Page, R. D. M. 1996. TREEVIEW: an application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12:357-358. [DOI] [PubMed] [Google Scholar]
- 28.Pignatello, J. J., M. M. Martinson, J. G. Steiert, R. E. Carlson, and R. L. Crawford. 1983. Biodegradation and protolysis of pentachlorophenol in artificial freshwater streams. Appl. Environ. Microbiol. 46:1024-1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Puhakka, J. A., R. P. Herwig, P. M. Koro, G. V. Wolfe, and J. F. Ferguson. 1995. Biodegradation of chlorophenols by mixed and pure cultures from a fluidized-bed reactor. Appl. Microbiol. Biotechnol. 42:951-957. [DOI] [PubMed] [Google Scholar]
- 30.Puhakka, J. A., K. T. Järvinen, J. H. Langwaldt, E. S. Melin, M. K. Männistö, J. M. Salminen, and M. T. Sjölund. 2000. On-site and in situ bioremediation of wood-preservative contaminated groundwater. Water Sci. Technol. 42:371-376. [Google Scholar]
- 31.Radehaus, P., and S. Schmidt. 1992. Characterization of a novel Pseudomonas sp. that mineralizes high concentrations of pentachlorophenol. Appl. Environ. Microbiol. 58:2879-2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Resnick, S., and P. Chapman. 1994. Physiological properties and substrate specificity of a pentachlorophenol-degrading Pseudomonas species. Biodegradation 5:47-54. [DOI] [PubMed] [Google Scholar]
- 33.Saboo, V. M., and M. A. Gealt. 1998. Gene sequences of the pcpB gene of pentachlorophenol-degrading Sphingomonas chlorophenolica found in nondegrading bacteria. Can. J. Microbiol. 44:667-675. [PubMed] [Google Scholar]
- 34.Takeuchi, M., K. Hamana, and A. Hiraishi. 2001. Proposal of the genus Sphingomonas sensu stricto and three new genera, Sphingobium, Novosphingobium and Sphingopyxis, on the basis of phylogenetic and chemotaxonomic analyses. Int. J. Syst. E vol. Microbiol. 51:1405-1417. [DOI] [PubMed] [Google Scholar]
- 35.Thakur, I. S., P. K. Verma, and K. C. Opadhaya. 2001. Involvement of plasmid in degradation of pentachlorophenol by Pseudomonas sp. from a chemostat. Biochem. Biophys. Res. Commun. 286:108-113. [DOI] [PubMed] [Google Scholar]
- 36.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tiirola, M. A., M. K. Männistö, J. A. Puhakka, and M. S. Kulomaa. 2002. Isolation and characterization of Novosphingobium sp. strain MT1, a dominant polychlorophenol-degrading strain in a groundwater bioremediation system. Appl. Environ. Microbiol. 68:173-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang, H., M. A. Tiirola, J. A. Puhakka, and M. S. Kulomaa. 2001. Production and characterization of the recombinant Sphingomonas chlorophenolica pentachlorophenol 4-monooxygenase. Biochem. Biophys. Res. Commun. 289:161-166. [DOI] [PubMed] [Google Scholar]
- 39.Yin, X., and G. Stotzky. 1997. Gene transfer among bacteria in natural environments. Adv. Appl. Microbiol. 45:153-211. [DOI] [PubMed] [Google Scholar]
- 40.Xun, L., and C. S. Orser. 1991. Biodegradation of triiodophenol by cell-free extracts of a pentachlorophenol-degrading Flavobacterium sp. Biochem. Biophys. Res. Commun. 174:43-48. [DOI] [PubMed] [Google Scholar]
- 41.Xun, L., E. Topp, and C. S. Orser. 1992. Diverse substrate range of a Flavobacterium pentachlorophenol hydroxylase and reaction stoichiometries. J. Bacteriol. 174:2898-2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xun, L., J. Bohuslavek, and M. Cai. 1999. Characterization of dichloro-p-hydroquinone 1,2-dioxygenase (pcpA) of Sphingomonas chlorophenolica ATCC 39723. Biochem. Biophys. Res. Commun. 266:322-325. [DOI] [PubMed] [Google Scholar]